Rapid Spread and Genetic Characterisation of a Recently Emerged Recombinant Lumpy Skin Disease Virus in Thailand
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
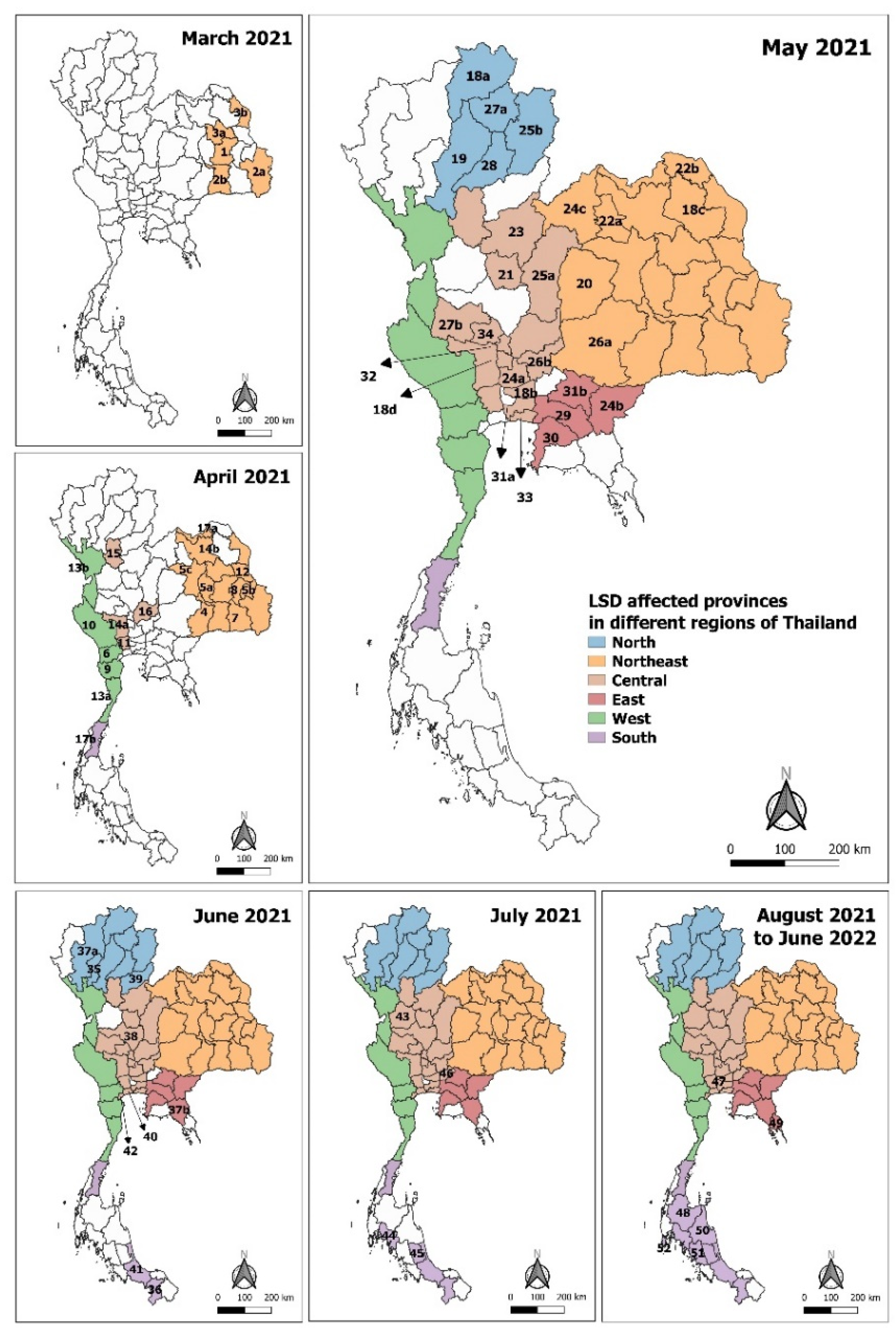
2.2. Geographical Mapping of the LSD Outbreaks
2.3. Real-Time PCR
2.4. Virus Isolation and Detection
2.5. DNA Sequencing and Phylogenetic Analysis
2.6. Recombination Analysis
2.7. Statistical Analysis
3. Results
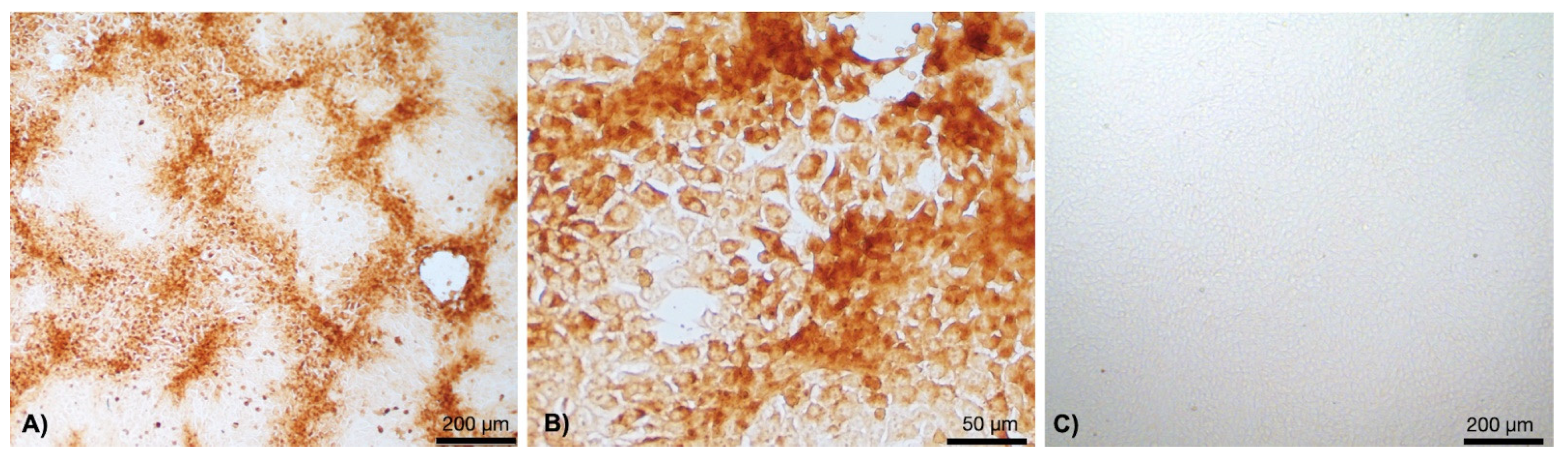
3.1. Clinical Cases and Histopathological Findings
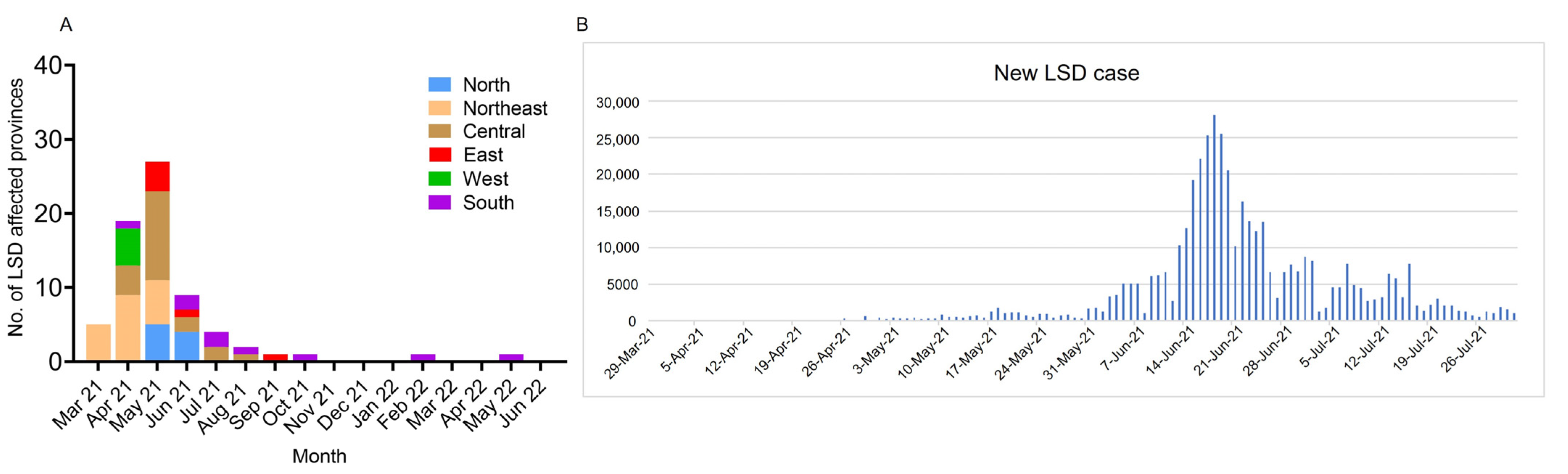
3.2. Distribution of LSD
3.3. LSDV Diagnosis
3.4. DNA Sequencing and Phylogenetic Analysis
3.5. Recombination Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coetzer, J.A.W.; Tustin, R.C. Infectious Diseases of Livestock, 2nd ed.; Oxford University Press: Cape Town, South Africa, 2004; Volume 3, pp. 1268–1276. [Google Scholar]
- Ahmed, E.M.; Eltarabilli, M.M.; Shahein, M.A.; Fawzy, M. Lumpy skin disease outbreaks investigation in Egyptian cattle and buffaloes: Serological evidence and molecular characterization of genome termini. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101639. [Google Scholar] [CrossRef]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Sur, J.-H.; Sandybaev, N.T.; Kerembekova, U.Z.; Zaitsev, V.L.; Kutish, G.F.; Rock, D.L. The genomes of sheeppox and goatpox viruses. J. Virol. 2002, 76, 6054–6061. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. Genome of lumpy skin disease virus. J. Virol. 2001, 75, 7122–7130. [Google Scholar] [CrossRef]
- Tuppurainen, E.S.; Oura, C.A. Review: Lumpy skin disease: An emerging threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 2012, 59, 40–48. [Google Scholar] [CrossRef]
- MacDonald, R.A.S. Pseudo-urticaria of cattle. In Annual Report for 1930; Department of Animal Health, Northern Rhodesia, His Majesty’s Stationery Office: London, UK, 1931; pp. 20–21. [Google Scholar]
- Rweyemamu, M.; Paskin, R.; Benkirane, A.; Martin, V.; Roeder, P.; Wojciechowski, K. Emerging diseases of Africa and the Middle East. Ann. N. Y. Acad. Sci. 2000, 916, 61–70. [Google Scholar] [CrossRef]
- Davies, F.G. Lumpy skin disease, an African capripox virus disease of cattle. Br. Vet. J. 1991, 147, 489–503. [Google Scholar] [CrossRef]
- Alkhamis, M.A.; VanderWaal, K. Spatial and Temporal Epidemiology of Lumpy Skin Disease in the Middle East, 2012–2015. Front. Vet. Sci. 2016, 3, 19. [Google Scholar] [CrossRef]
- Tasioudi, K.E.; Antoniou, S.E.; Iliadou, P.; Sachpatzidis, A.; Plevraki, E.; Agianniotaki, E.I.; Fouki, C.; Mangana-Vougiouka, O.; Chondrokouki, E.; Dile, C. Emergence of lumpy skin disease in Greece, 2015. Transbound. Emerg. Dis. 2016, 63, 260–265. [Google Scholar] [CrossRef]
- Sprygin, A.; Artyuchova, E.; Babin, Y.; Prutnikov, P.; Kostrova, E.; Byadovskaya, O.; Kononov, A. Epidemiological characterization of lumpy skin disease outbreaks in Russia in 2016. Transbound. Emerg. Dis. 2018, 65, 1514–1521. [Google Scholar] [CrossRef]
- Gupta, T.; Patial, V.; Bali, D.; Angaria, S.; Sharma, M.; Chahota, R. A review: Lumpy skin disease and its emergence in India. Vet. Res. Commun. 2020, 44, 111–118. [Google Scholar] [CrossRef]
- Lu, G.; Xie, J.; Luo, J.; Shao, R.; Jia, K.; Li, S. Lumpy skin disease outbreaks in China, since 3 August 2019. Transbound. Emerg. Dis. 2021, 68, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Nguyen, C.T.; Chu, N.T.; Van Hoang, T.; Nguyen, H.T.; Nguyen, V.T.; Dang, H.V. Lumpy skin disease outbreaks in Vietnam, 2020. Transbound. Emerg. Dis. 2021, 68, 977–980. [Google Scholar] [CrossRef] [PubMed]
- Badhy, S.C.; Chowdhury, M.G.A.; Settypalli, T.B.K.; Cattoli, G.; Lamien, C.E.; Fakir, M.A.U.; Akter, S.; Osmani, M.G.; Talukdar, F.; Begum, N.; et al. Molecular characterization of lumpy skin disease virus (LSDV) emerged in Bangladesh reveals unique genetic features compared to contemporary field strains. BMC Vet. Res. 2021, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Maw, M.T.; Khin, M.M.; Hadrill, D.; Meki, I.K.; Settypalli, T.B.K.; Kyin, M.M.; Myint, W.W.; Thein, W.Z.; Aye, O.; Palamara, E.; et al. First Report of Lumpy Skin Disease in Myanmar and Molecular Analysis of the Field Virus Isolates. Microorganisms 2022, 10, 897. [Google Scholar] [CrossRef]
- Weiss, K. Lumpy Skin Disease Virus. Cytomegaloviruses. Rinderpest Virus. Lumpy Skin Disease Virus; Springer: Berlin/Heidelberg, Germany, 1968; pp. 111–131. [Google Scholar]
- Yeruham, I.; Perl, S.; Nyska, A.; Abraham, A.; Davidson, M.; Haymovitch, M.; Zamir, O.; Grinstein, H. Adverse reactions in cattle to a capripox vaccine. Vet. Rec. 1994, 135, 330–332. [Google Scholar] [CrossRef]
- Chihota, C.M.; Rennie, L.F.; Kitching, R.P.; Mellor, P.S. Mechanical transmission of lumpy skin disease virus by Aedes aegypti (Diptera: Culicidae). Epidemiol. Infect. 2001, 126, 317–321, (print). [Google Scholar] [CrossRef]
- Aleksandr, K.; Olga, B.; David, W.B.; Pavel, P.; Yana, P.; Svetlana, K.; Alexander, N.; Vladimir, R.; Dmitriy, L.; Alexander, S. Non-vector-borne transmission of lumpy skin disease virus. Sci. Rep. 2020, 10, 7436. [Google Scholar] [CrossRef]
- Arjkumpa, O.; Suwannaboon, M.; Boonrawd, M.; Punyawan, I.; Laobannu, P.; Yantaphan, S.; Bungwai, A.; Ponyium, V.; Suwankitwat, N.; Boonpornprasert, P.; et al. First emergence of lumpy skin disease in cattle in Thailand, 2021. Transbound. Emerg. Dis. 2021, 68, 3002–3004. [Google Scholar] [CrossRef]
- Sariya, L.; Paungpin, W.; Chaiwattanarungruengpaisan, S.; Thongdee, M.; Nakthong, C.; Jitwongwai, A.; Taksinoros, S.; Sutummaporn, K.; Boonmasawai, S.; Kornmatitsuk, B. Molecular detection and characterization of lumpy skin disease viruses from outbreaks in Thailand in 2021. Transbound. Emerg. Dis. 2022, 69, e2145–e2152. [Google Scholar] [CrossRef]
- Singhla, T.; Boonsri, K.; Kreausukon, K.; Modethed, W.; Pringproa, K.; Sthitmatee, N.; Punyapornwithaya, V.; Vinitchaikul, P. Molecular Characterization and Phylogenetic Analysis of Lumpy Skin Disease Virus Collected from Outbreaks in Northern Thailand in 2021. Vet. Sci. 2022, 9, 194. [Google Scholar] [CrossRef]
- WOAH. Chapter 3.4.12 Lumpy Skin Disease: World Organisation for Animal Health (WOAH). 2021. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.12_LSD.pdf (accessed on 18 August 2021).
- Gharban, H.A.J.; Al-Shaeli, S.J.J.; Al-Fattli, H.H.H.; Altaee, M.N.K. Molecular and histopathological confirmation of clinically diagnosed lumpy skin disease in cattle, Baghdad Province of Iraq. Vet. World 2019, 12, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Amin, D.M.; Shehab, G.; Emran, R.; Hassanien, R.T.; Alagmy, G.N.; Hagag, N.M.; Abd-El-Moniem, M.I.I.; Habashi, A.R.; Ibraheem, E.M.; Shahein, M.A. Diagnosis of naturally occurring lumpy skin disease virus infection in cattle using virological, molecular, and immunohistopathological assays. Vet. World 2021, 14, 2230–2237. [Google Scholar] [CrossRef] [PubMed]
- Lubinga, J.C.; Clift, S.J.; Tuppurainen, E.S.; Stoltsz, W.H.; Babiuk, S.; Coetzer, J.A.; Venter, E.H. Demonstration of lumpy skin disease virus infection in Amblyomma hebraeum and Rhipicephalus appendiculatus ticks using immunohistochemistry. Ticks Tick Borne Dis. 2014, 5, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Bowden, T.R.; Babiuk, S.L.; Parkyn, G.R.; Copps, J.S.; Boyle, D.B. Capripoxvirus tissue tropism and shedding: A quantitative study in experimentally infected sheep and goats. Virology 2008, 371, 380–393. [Google Scholar] [CrossRef]
- Kumar, N.; Chander, Y.; Kumar, R.; Khandelwal, N.; Riyesh, T.; Chaudhary, K.; Shanmugasundaram, K.; Kumar, S.; Kumar, A.; Gupta, M.K.; et al. Isolation and characterization of lumpy skin disease virus from cattle in India. PLoS ONE 2021, 16, e0241022. [Google Scholar] [CrossRef]
- Salnikov, N.; Usadov, T.; Kolcov, A.; Zhivoderov, S.; Morgunov, Y.; Gerasimov, V.; Gogin, A.; Titov, I.; Yurkov, S.; Malogolovkin, A.; et al. Identification and characterization of lumpy skin disease virus isolated from cattle in the Republic of North Ossetia-Alania in 2015. Transbound. Emerg. Dis. 2018, 65, 916–920. [Google Scholar] [CrossRef]
- Lekcharoensuk, P.; Wiriyarat, W.; Petcharat, N.; Lekcharoensuk, C.; Auewarakul, P.; Richt, J.A. Cloned cDNA of A/swine/Iowa/15/1930 internal genes as a candidate backbone for reverse genetics vaccine against influenza A viruses. Vaccine 2012, 30, 1453–1459. [Google Scholar] [CrossRef][Green Version]
- Haegeman, A.; De Leeuw, I.; Mostin, L.; Van Campe, W.; Aerts, L.; Vastag, M.; De Clercq, K. An Immunoperoxidase Monolayer Assay (IPMA) for the detection of lumpy skin disease antibodies. J. Virol. Methods 2020, 277, 113800. [Google Scholar] [CrossRef] [PubMed]
- Gelaye, E.; Belay, A.; Ayelet, G.; Jenberie, S.; Yami, M.; Loitsch, A.; Tuppurainen, E.; Grabherr, R.; Diallo, A.; Lamien, C.E. Capripox disease in Ethiopia: Genetic differences between field isolates and vaccine strain, and implications for vaccination failure. Antivir. Res. 2015, 119, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; WF Freeman & Co.: San Francisco, CA, USA, 1973. [Google Scholar]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using The Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Lemey, P.; Lott, M.; Moulton, V.; Posada, D.; Lefeuvre, P. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics 2010, 26, 2462–2463. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Posada, D.; Crandall, K.; Williamson, C. A Modified Bootscan Algorithm for Automated Identification of Recombinant Sequences and Recombination Breakpoints. AIDS Res. Hum. Retrovir. 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Amin, A.; El-Nahas, E.; El-Mashed, A.-E. Pathological and Virological Studies on an Outbreak of Lumpy Skin Disease among Cattle in Kalubia Governorate-Egypt. J. Adv. Vet. Res. 2015, 5, 165–175. [Google Scholar]
- Roche, X.; Rozstalnyy, A.; TagoPacheco, D.; Pittiglio, C.; Kamata, A.; Beltran Alcrudo, D.; Bisht, K.; Karki, S.; Kayamori, J.; Larfaoui, F.; et al. Introduction and Spread of Lumpy Skin Disease in South, East and Southeast Asia: Qualitative Risk Assessment and Management; FAO Animal Production and Health: Rome, Italy, 2020; pp. 1–183. [Google Scholar]
- Buamithup, N.; Pamaranon, N.; Charoenlarp, W.; Panyasomboonying, P.; Kuatako, N.; Luangtrakool, D.; Wannakee, H.; Suwannaboon, M.; Arjkumpa, O.; Suwankitwat, N. Outbreak of Lumpy Skin Disease of the Cattle in Thailand: An UPDATE. In Proceedings of the 22nd KKU Veterinary Annual International Conference 2021 “International Conference on Transboundary Animal Diseases, Khon Kaen, Thailand, 22–23 July 2021; 23 July 2021. [Google Scholar]
- Tuppurainen, E.; Dietze, K.; Wolff, J.; Bergmann, H.; Beltran-Alcrudo, D.; Fahrion, A.; Lamien, C.E.; Busch, F.; Sauter-Louis, C.; Conraths, F.J.; et al. Review: Vaccines and Vaccination against Lumpy Skin Disease. Vaccines 2021, 9, 1136. [Google Scholar] [CrossRef] [PubMed]
- Calistri, P.; Declercq, K.; De Vleeschauwer, A.; Gubbins, S.; Klement, E.; Stegeman, A.; Abrahantes, J.C.; Antoniou, S.-E.; Broglia, A.; Gogin, A. Lumpy skin disease: Scientific and technical assistance on control and surveillance activities. EFSA J. 2018, 16, e05452. [Google Scholar] [PubMed]
- Tuppurainen, E.S.; Venter, E.H.; Coetzer, J.A. The detection of lumpy skin disease virus in samples of experimentally infected cattle using different diagnostic techniques. Onderstepoort J. Vet. Res. 2005, 72, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.S.; Haga, I.R.; Wijesiriwardana, N.; Hawes, P.C.; Simpson, J.; Morrison, L.R.; MacIntyre, N.; Brocchi, E.; Atkinson, J.; Haegeman, A.; et al. Lumpy Skin Disease Is Characterized by Severe Multifocal Dermatitis With Necrotizing Fibrinoid Vasculitis Following Experimental Infection. Vet. Pathol. 2020, 57, 388–396. [Google Scholar] [CrossRef]
- Ireland, D.C.; Binepal, Y.S. Improved detection of Capripoxvirus in biopsy samples by PCR. J. Virol. Methods 1998, 74, 1–7. [Google Scholar] [CrossRef]
- Sprygin, A.; Babin, Y.; Pestova, Y.; Kononova, S.; Wallace, D.B.; VAN Schalkwyk, A.; Byadovskaya, O.; Diev, V.; Lozovoy, D.; Kononov, A. Analysis and insights into recombination signals in lumpy skin disease virus recovered in the field. PLoS ONE 2018, 13, e0207480. [Google Scholar] [CrossRef]
- Sprygin, A.; Van Schalkwyk, A.; Shumilova, I.; Nesterov, A.; Kononova, S.; Prutnikov, P.; Byadovskaya, O.; Kononov, A. Full-length genome characterization of a novel recombinant vaccine-like lumpy skin disease virus strain detected during the climatic winter in Russia, 2019. Arch. Virol. 2020, 165, 2675–2677. [Google Scholar] [CrossRef]
- Balinsky, C.A.; Delhon, G.; Afonso, C.L.; Risatti, G.R.; Borca, M.V.; French, R.A.; Tulman, E.R.; Geary, S.J.; Rock, D.L. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J. Virol. 2007, 81, 11392–11401. [Google Scholar] [CrossRef]
- Ma, J.; Yuan, Y.; Shao, J.; Sun, M.; He, W.; Chen, J.; Liu, Q. Genomic characterization of lumpy skin disease virus in southern China. Transbound. Emerg. Dis. 2021, 1–12. [Google Scholar] [CrossRef]
- Babiuk, S.; Bowden, T.R.; Parkyn, G.; Dalman, B.; Manning, L.; Neufeld, J.; Embury-Hyatt, C.; Copps, J.; Boyle, D.B. Quantification of lumpy skin disease virus following experimental infection in cattle. Transbound. Emerg. Dis 2008, 55, 299–307. [Google Scholar] [CrossRef]
- Flannery, J.; Shih, B.; Haga, I.R.; Ashby, M.; Corla, A.; King, S.; Freimanis, G.; Polo, N.; Tse, A.C.; Brackman, C.J.; et al. A novel strain of lumpy skin disease virus causes clinical disease in cattle in Hong Kong. Transbound. Emerg. Dis. 2021, 27, e336–e343. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-W.; Ting, L.-J.; Liu, Y.-P.; Lin, Y.-J.; Lee, F.; Chiou, C.-J. Complete Coding Sequence of Lumpy Skin Disease Virus Isolated from Kinmen Island, Taiwan, in 2020. Microbiol. Resour. Announc. 2022, 11, e01204–e01221. [Google Scholar] [CrossRef] [PubMed]
- Azeem, S.; Sharma, B.; Shabir, S.; Akbar, H.; Venter, E. Lumpy skin disease is expanding its geographic range: A challenge for Asian livestock management and food security. Vet. J. 2022, 279, 105785. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Mathijs, E.; Philips, W.; Saduakassova, M.; De Leeuw, I.; Sultanov, A.; Haegeman, A.; De Clercq, K. Recombinant LSDV Strains in Asia: Vaccine Spillover or Natural Emergence? Viruses 2022, 14, 1429. [Google Scholar] [CrossRef] [PubMed]
- Haegeman, A.; De Leeuw, I.; Saduakassova, M.; Van Campe, W.; Aerts, L.; Philips, W.; Sultanov, A.; Mostin, L.; De Clercq, K. The importance of quality control of LSDV live attenuated vaccines for its safe application in the field. Vaccines 2021, 9, 1019. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; Bourgeois Luethi, N.; Huachun, L.; Naing Oo, K.; Phonvisay, A.; Premashthira, S.; Abila, R.; Widders, P.; Kukreja, K.; Miller, C. Movement Pathways and Market Chains of Large Ruminants in the Greater Mekong Sub-Region; School of Agricultural, Forest and Food Sciences HAFL: Zollikofen, Switzerland; OIE World Organisation for Animal Health: Paris, France, 2015; p. 28. [Google Scholar]
- Chaisirirat, T.; Sangthong, P.; Arunvipas, P.; Petcharat, N.; Thangthamniyom, N.; Chumsing, W.; Lekcharoensuk, P. Molecular characterization of bovine ephemeral fever virus in Thailand between 2013 and 2017. Vet. Microbiol. 2018, 227, 1–7. [Google Scholar] [CrossRef]
- Cocks, P.; Abila, R.; Black, P.; Edwards, J.; Robertson, I. Livestock trade and marketing networks in Malaysia, Thailand and Myanmar. Report for AusAID—DAFF SPS Capacity Building Project. 2009. [Google Scholar]
- Tuppurainen, E.; Alexandrov, T.; Beltrán-Alcrudo, D. Lumpy Skin Disease Field Manual—A Manual for Veterinarians; FAO Animal Production and Health Manual No.20; Food and Agriculture Oraganization of the United Nations (FAO): Rome, Italy, 2017. [Google Scholar]




| Skin | Blood | Total | |
|---|---|---|---|
| Positive | 103 | 62 | 165 |
| Negative | 8 | 49 | 57 |
| Total | 111 | 111 | 222 |
| Recombination Event Number | Breakpoint Positions in Alignment | Size (bp) | ORF | Detection Methods | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Begin | End | RDP | GENECONV | Bootscan | Maxchi | Chimaera | SiScan | 3Seq | |||
| 1 | 67,096 | 84,072 | 16,977 | 075–089 | NS | 4.95 × 10−35 | 4.27 × 10−20 | 2.30 × 10−16 | 4.98 × 10−17 | NS | 2.22 × 10−16 |
| 2 | 22 * | 4792 | 4771 | 001–007 | NS | 2.01 × 10−18 | 1.31 × 10−16 | 2.80 × 10−15 | 2.80 × 10−15 | NS | NS |
| 3 | 95,124 * | 99,574 | 4451 | 101–105 | NS | 1.19 × 10−16 | 1.13 × 10−16 | 4.94 × 10−10 | 4.94 × 10−10 | NS | 5.24 × 10−14 |
| 4 | 120,668 * | 122,753 | 2086 | 133 | NS | 1.95 × 10−17 | 4.66 × 10−20 | 8.64 × 10−13 | 8.64 × 10−13 | NS | 1.67 × 10−15 |
| 5 | 140,127 | 144,246 | 4120 | 146–149 | 3.41 × 10−7 | 7.09 × 10−13 | 1.97 × 10−9 | 1.24 × 10−14 | 1.24 × 10−14 | NS | 5.55 × 10−15 |
| 6 | 19,579 | 23,622 | 4044 | 027–030 | NS | 2.54 × 10−14 | 1.34 × 10−15 | 1.29 × 10−8 | 1.29 × 10−8 | NS | 2.74 × 10−11 |
| 7 | 108,572 | 111,696 * | 3125 | 116–119 | NS | 2.01 × 10−11 | 1.55 × 10−11 | 6.20 × 10−5 | 6.20 × 10−5 | NS | NS |
| 8 | 85,944 * | 89,483 | 3540 | 091–094 | NS | 2.95 × 10−12 | 9.72 × 10−11 | 6.37 × 10−9 | 6.37 × 10−9 | NS | NS |
| 9 | 45,480 | 53,842 | 8363 | 050–060 | NS | 1.57 × 10−8 | 1.35 × 10−5 | 3.59 × 10−9 | 3.59 × 10−9 | NS | 6.29 × 10−12 |
| 10 | 102,225 | 103,207 * | 983 | 110 | NS | 2.99 × 10−4 | 3.01 × 10−5 | NS | NS | NS | 2.96 × 10−3 |
| 11 | 149,094 | 151,035 * | 1942 | 154–156 | NS | 3.17 × 10−4 | 1.88 × 10−4 | NS | NS | NS | NS |
| Event | ORF | Protein | Amino Acid Differences from the LSDV Strain SIS-Lumpyvax Vaccine (Format: YST/Protein Position/SIS) |
|---|---|---|---|
| 1 | 075 | RNA polymerase-associated protein | V324A |
| 076 | late transcription factor VLTF-4 | V64A, D96DNDN, N101D, D102N, D151G | |
| 079 | mRNA capping enzyme, large subunit | I206T, D295N, T374P | |
| 080 | Virion protein | I26M, T93I | |
| 081 | Virion protein | H17N, S227N | |
| 082 | Uracil DNA glycosylase | R54Q | |
| 083 | NTPase; DNA replication | S3G, S49T, G106D, I135M, I253L, N663K, I708T | |
| 084 | Early transcription factor VETF transmembrane | L353V, D581N | |
| 085 | RNA polymerase subunit RPO18 | M136T | |
| 086a | mut T motif | E121D, L191F, 207 extension 3 amino acid residues due to frameshift mutation | |
| 087a | mut T motif; gene expression regulator | D12G, V46I,196 extension 53 amino acid residues due to frameshift mutation, S199F, N200L | |
| 088 | NPH-I; transcription termination factor | V24I | |
| 089 | mRNA capping enzyme, small subunit; VITF | V171I | |
| 2 | 1 | A52R-like family protein, SP | D129N, I144M |
| 3 | ER-localised apoptosis regulator, SP, TM | S93T, A100S | |
| 5 | IL-10, SP, TM | I14IF, A24V, I25V | |
| 6 | IL-1 receptor, SP | F13L, S61L, AS94A, I111S, S216N | |
| 7 | IFN-g | Q58K, S200T, N295D | |
| 3 | 101 | Virion core protein P4a | E223D |
| 102 | TM | A61N, L115S, T162A | |
| 103 | Virion core protein | T50N, P72T, S89G | |
| 4 | 133 | DNA ligase-like protein | V165I, D200E, S275N, L312S, I344T, D347N, S514A |
| 5 | 146 | Phospholipase D-like protein | T285S |
| 147 | Ankyrin repeat protein | I487M | |
| 148 | Ankyrin repeat protein | G40S, G51D, I102L, N167D, E169D, V351M, K361Q, C397Y, K413E, A418T, N439S. | |
| 149 | Serpin-like protein CDS | K139R | |
| 6 | 027 | EEV maturation protein | I440F, D328N, K197R, 153D, A61V |
| 028 | Palmitylated EEV membrane lycoprotein | G263S, A135T | |
| 030 | Hypothetical protein | H34N | |
| 7 | 116 | RNA polymerase subunit (RPO132) | N1154S |
| 118 | Hypothetical protein, TM | I10V | |
| 119 | RNA polymerase subunit, RPO35 | A291S | |
| 8 | 093 | Hypothetical protein | D60N |
| 094 | Virion core protein P4b, TM | D93N, F607L | |
| 9 | 050 | Metalloprotease, virion morphogenesis | NT376T |
| 054 | Hypothetical protein | S54G, M184I | |
| 056 | Hypothetical protein | K171R, N174D | |
| 057 | Virion core protein, TM | V372I | |
| 059 | Myristylated protein | Q125K | |
| 10 | 110 | DNA helicase; transcriptional elongation factor | A64T, R100L, L344F |
| 11 | 154 | ER-localised apoptosis regulator | S93T, A100S |
| 156 | A52R-like family protein, SP | D129N, I144M |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suwankitwat, N.; Songkasupa, T.; Boonpornprasert, P.; Sripipattanakul, P.; Theerawatanasirikul, S.; Deemagarn, T.; Suwannaboon, M.; Arjkumpa, O.; Buamithup, N.; Hongsawat, A.; et al. Rapid Spread and Genetic Characterisation of a Recently Emerged Recombinant Lumpy Skin Disease Virus in Thailand. Vet. Sci. 2022, 9, 542. https://doi.org/10.3390/vetsci9100542
Suwankitwat N, Songkasupa T, Boonpornprasert P, Sripipattanakul P, Theerawatanasirikul S, Deemagarn T, Suwannaboon M, Arjkumpa O, Buamithup N, Hongsawat A, et al. Rapid Spread and Genetic Characterisation of a Recently Emerged Recombinant Lumpy Skin Disease Virus in Thailand. Veterinary Sciences. 2022; 9(10):542. https://doi.org/10.3390/vetsci9100542
Chicago/Turabian StyleSuwankitwat, Nutthakarn, Tapanut Songkasupa, Prakit Boonpornprasert, Phurida Sripipattanakul, Sirin Theerawatanasirikul, Taweewat Deemagarn, Minta Suwannaboon, Orapun Arjkumpa, Noppawan Buamithup, Akkarapol Hongsawat, and et al. 2022. "Rapid Spread and Genetic Characterisation of a Recently Emerged Recombinant Lumpy Skin Disease Virus in Thailand" Veterinary Sciences 9, no. 10: 542. https://doi.org/10.3390/vetsci9100542
APA StyleSuwankitwat, N., Songkasupa, T., Boonpornprasert, P., Sripipattanakul, P., Theerawatanasirikul, S., Deemagarn, T., Suwannaboon, M., Arjkumpa, O., Buamithup, N., Hongsawat, A., Jindajang, S., Nipaeng, N., Aunpomma, D., Molee, L., Puangjinda, K., Lohlamoh, W., Nuansrichay, B., Narawongsanont, R., Arunvipas, P., & Lekcharoensuk, P. (2022). Rapid Spread and Genetic Characterisation of a Recently Emerged Recombinant Lumpy Skin Disease Virus in Thailand. Veterinary Sciences, 9(10), 542. https://doi.org/10.3390/vetsci9100542







