Effect of Api-Bioxal® and ApiHerb® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods
Abstract
1. Introduction
2. Materials and Methods
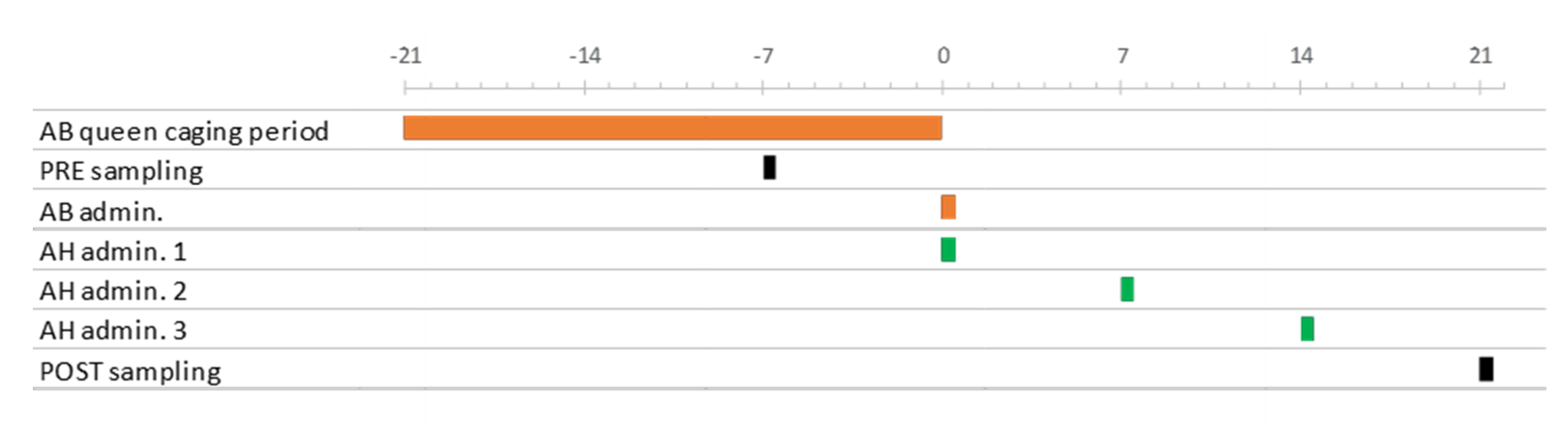
2.1. Experimental Design
2.2. DNA Extraction and qPCR Analysis
2.3. Statistical Analysis
3. Results
3.1. N. ceranae Infection in Test Colonies
3.2. Treatment Effect
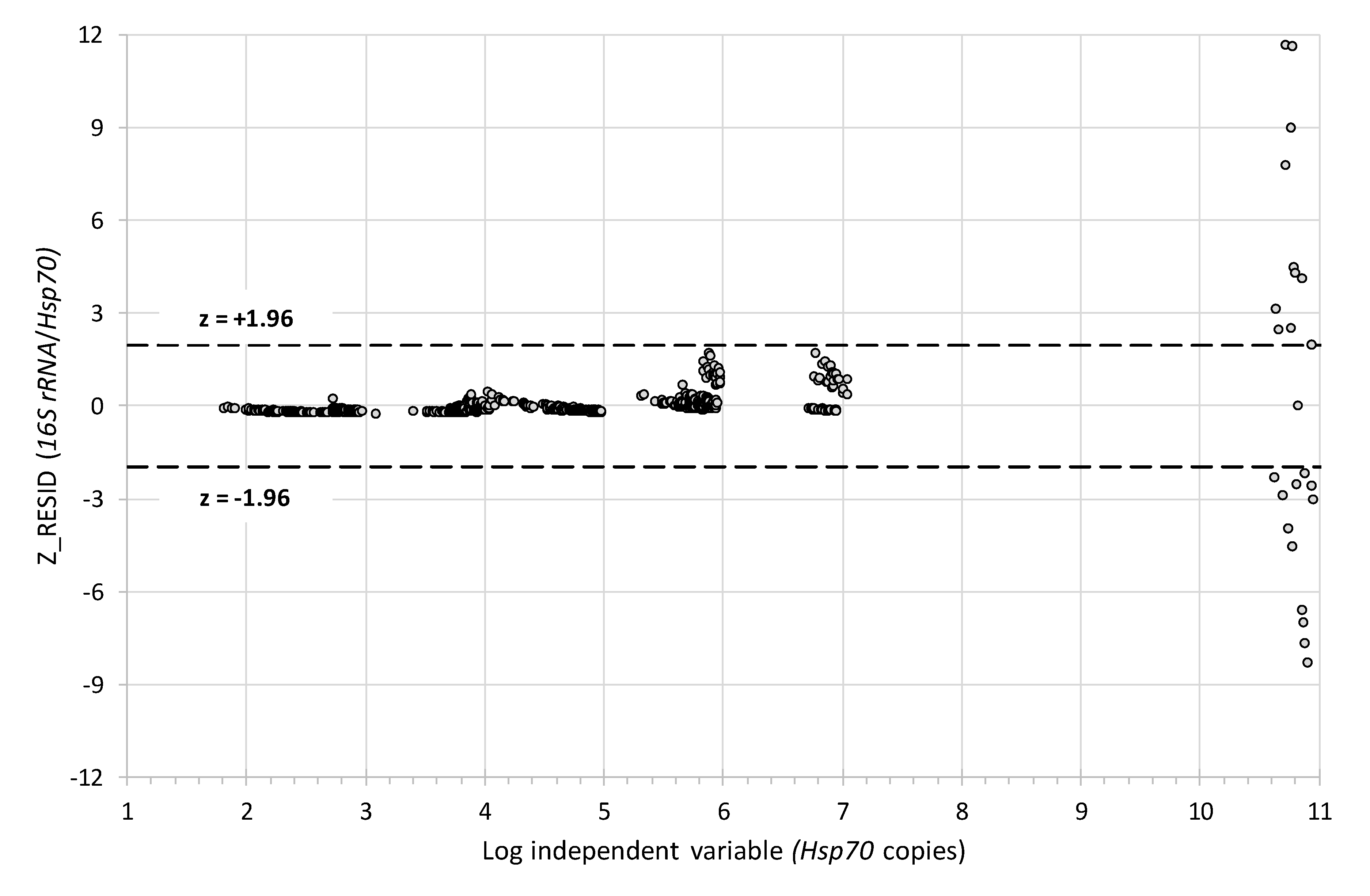
3.3. Comparability of the 16S rRNA and Hsp70 Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fries, I.; Chauzat, M.-P.; Chen, Y.-P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Fries, I.; Feng, F.; da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Botías, C.; Anderson, D.L.; Meana, A.; Garrido-Bailón, E.; Martín-Hernández, R.; Higes, M. Further evidence of an oriental origin for Nosema ceranae (Microsporidia: Nosematidae). J. Invertebr. Pathol. 2012, 110, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.-L.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Martín-Hernández, R.; Meana, A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 2007, 94, 211–217. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Urbieta, A.; Nanetti, A.; Martín-Hernández, R. Nosema apis and Nosema ceranae tissue tropism in worker honey bees (Apis mellifera). Vet. Pathol. 2019. [Google Scholar] [CrossRef]
- Eiri, D.M.; Suwannapong, G.; Endler, M.; Nieh, J.C. Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS ONE 2015, 10, e0126330. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; González-Porto, A.V.; García-Palencia, P.; Meana, A.A.; del Nozal, M.J.; Mayo, R.; Bernal, J.L. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Rep. 2009, 1, 110–113. [Google Scholar] [CrossRef]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef] [PubMed]
- Ravoet, J.; De Smet, L.; Meeus, I.; Smagghe, G.; Wenseleers, T.; De Graaf, D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014, 122, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Plischuk, S.; Lange, C.E. Bombus brasiliensis Lepeletier (Hymenoptera, Apidae) infected with Nosema ceranae (Microsporidia). Rev. Bras. Entomol. 2016, 60, 347–351. [Google Scholar] [CrossRef]
- Plischuk, S.; Martín-Hernández, R.; Prieto, L.; Lucía, M.; Botías, C.; Meana, A.; Abrahamovich, A.H.; Lange, C.; Higes, M. South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera). Environ. Microbiol. Rep. 2009, 1, 131–135. [Google Scholar] [CrossRef]
- Porrini, M.P.; Porrini, L.P.; Garrido, P.M.; de Melo e Silva Neto, C.; Porrini, D.P.; Muller, F.; Nuñez, L.A.; Alvarez, L.; Iriarte, P.F.; Eguaras, M.J. Nosema ceranae in South American native stingless bees and social wasp. Microb. Ecol. 2017, 74, 761–764. [Google Scholar] [CrossRef]
- Cilia, G.; Cardaio, I.; dos Santos, P.E.J.; Ellis, J.D.; Nanetti, A. The first detection of Nosema ceranae (Microsporidia) in the small hive beetle, Aethina tumida Murray (Coleoptera: Nitidulidae). Apidologie 2018, 49, 619–624. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; Botías, C.; García-Palencia, P.; Meana, A. Regurgitated pellets of Merops apiaster as fomites of infective Nosema ceranae (Microsporidia) spores. Environ. Microbiol. 2008, 10, 1374–1379. [Google Scholar] [CrossRef]
- Graystock, P.; Goulson, D.; Hughes, W.O.H. Parasites in bloom: Flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. Biol. Sci. 2015, 282. [Google Scholar] [CrossRef]
- Purkiss, T.; Lach, L. Pathogen spillover from Apis mellifera to a stingless bee. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191071. [Google Scholar] [CrossRef]
- Williams, G.R.; Sampson, M.A.; Shutler, D.; Rogers, R.E.L. Does fumagillin control the recently detected invasive parasite Nosema ceranae in western honey bees (Apis mellifera)? J. Invertebr. Pathol. 2008, 99, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; del Nozal, M.J.; Alvaro, A.; Barrios, L.; Meana, A.; Martín-Hernández, R.; Bernal, J.L.; Bernal, J. The stability and effectiveness of fumagillin in controlling Nosema ceranae (Microsporidia) infection in honey bees (Apis mellifera) under laboratory and field conditions. Apidologie 2011, 42, 364–377. [Google Scholar] [CrossRef]
- Huang, W.-F.; Solter, L.F.; Yau, P.M.; Imai, B.S. Nosema ceranae escapes fumagillin control in honey bees. PLoS Pathog. 2013, 9, e1003185. [Google Scholar] [CrossRef] [PubMed]
- Botías, C.; Martín-Hernández, R.; Meana, A.; Higes, M. Screening alternative therapies to control Nosemosis type C in honey bee (Apis mellifera iberiensis) colonies. Res. Vet. Sci. 2013, 95, 1041–1045. [Google Scholar] [CrossRef]
- Maistrello, L.; Lodesani, M.; Costa, C.; Leonardi, F.; Marani, G.; Caldon, M.; Mutinelli, F.; Granato, A. Screening of natural compounds for the control of Nosema disease in honeybees (Apis mellifera). Apidologie 2008, 39, 436–445. [Google Scholar] [CrossRef]
- Bravo, J.; Carbonell, V.; Sepúlveda, B.; Delporte, C.; Valdovinos, C.E.; Martín-Hernández, R.; Higes, M. Antifungal activity of the essential oil obtained from Cryptocarya alba against infection in honey bees by Nosema ceranae. J. Invertebr. Pathol. 2017, 149, 141–147. [Google Scholar] [CrossRef]
- Michalczyk, M.; Sokół, R.; Koziatek, S. Evaluation of the effectiveness of selected treatments of Nosema spp. infection by the hemocytometric method and duplex PCR. Acta Vet. 2016, 66, 115–124. [Google Scholar] [CrossRef][Green Version]
- Michalczyk, M.; Sokół, R. Estimation of the influence of selected products on co-infection with N. apis/N. ceranae in Apis mellifera using real-time PCR. Invertebr. Reprod. Dev. 2018, 62, 92–97. [Google Scholar] [CrossRef]
- Charriére, J.-D.; Imdorf, A. Oxalic acid treatment by trickling against Varroa destructor: Recommendations for use in central Europe and under temperate climate conditions. Bee World 2002, 83, 51–60. [Google Scholar] [CrossRef]
- Milani, N. Activity of oxalic and citric acids on the mite Varroa destructor in laboratory assays. Apidologie 2001, 32, 127–138. [Google Scholar] [CrossRef]
- Nanetti, A.; Büchler, R.; Charriere, J.D.; Friesd, I.; Helland, S.; Imdorf, A.; Korpela, S.; Kristiansen, P. Oxalic acid treatments for varroa control (review). Apiacta 2003, 38, 81–87. [Google Scholar]
- Nanetti, A.; Rodriguez-García, C.; Meana, A.; Martín-Hernández, R.; Higes, M. Effect of oxalic acid on Nosema ceranae infection. Res. Vet. Sci. 2015, 102, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Porrini, M.P.; Garrido, P.M.; Silva, J.; Cuniolo, A.; Román, S.; Iaconis, D.; Eguaras, M. Ácido oxálico: Potencial antiparasitario frente a Nosema ceranae por administración oral y exposición total. In Proceedings of the I Workshop Latinoamericano en Sanidad Apicola, Mar de Plata, Argentina, 25–26 October 2018; p. 64. [Google Scholar]
- Büchler, R.; Uzunov, A.; Kovačić, M.; Prešern, J.; Pietropaoli, M.; Hatjina, F.; Pavlov, B.; Charistos, L.; Formato, G.; Galarza, E.; et al. Summer brood interruption as integrated management strategy for effective Varroa control in Europe. J. Apic. Res. 2020, 1–10. [Google Scholar] [CrossRef]
- Botías, C.; Martín-Hernández, R.; Días, J.; García-Palencia, P.; Matabuena, M.; Juarranz, Á.; Barrios, L.; Meana, A.; Nanetti, A.; Higes, M. The effect of induced queen replacement on Nosema spp. infection in honey bee (Apis mellifera iberiensis) colonies. Environ. Microbiol. 2012, 14, 845–859. [Google Scholar] [CrossRef]
- Bourgeois, A.L.; Rinderer, T.E.; Beaman, L.D.; Danka, R.G. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invertebr. Pathol. 2010, 103, 53–58. [Google Scholar] [CrossRef]
- Cilia, G.; Cabbri, R.; Maiorana, G.; Cardaio, I.; Dall’Olio, R.; Nanetti, A. A novel TaqMan® assay for Nosema ceranae quantification in honey bee, based on the protein coding gene Hsp70. Eur. J. Protistol. 2018, 63, 44–50. [Google Scholar] [CrossRef]
- Cilia, G.; Fratini, F.; Tafi, E.; Turchi, B.; Mancini, S.; Sagona, S.; Nanetti, A.; Cerri, D.; Felicioli, A. Microbial profile of the Ventriculum of honey bee (Apis mellifera ligustica Spinola, 1806) fed with veterinary drugs, dietary supplements and non-protein amino acids. Vet. Sci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Pai, S.T.; Platt, M.W. Antifungal effects of Allium sativum (garlic) extract against the Aspergillus species involved in otomycosis. Lett. Appl. Microbiol. 1995, 20, 14–18. [Google Scholar] [CrossRef]
- Li, W.-R.; Shi, Q.-S.; Liang, Q.; Huang, X.-M.; Chen, Y.-B. Antifungal effect and mechanism of garlic oil on Penicillium funiculosum. Appl. Microbiol. Biotechnol. 2014, 98, 8337–8346. [Google Scholar] [CrossRef]
- Asehraou, A.; Mohieddine, S.; Faid, M.; Serhrouchni, M. Use of antifungal principles from garlic for the inhibition of yeasts and moulds in fermenting green olives. Grasas Aceites 1997, 48, 68–73. [Google Scholar] [CrossRef]
- Singh, U.P.; Pandey, V.N.; Wagner, K.G.; Singh, K.P. Antifungal activity of ajoene, a constituent of garlic (Allium sativum). Can. J. Bot. 1990, 68, 1354–1356. [Google Scholar] [CrossRef]
- Devi, A.G.; Sharma, S.D.; Balavenkatasubbaiah, M.; Chandrasekharan, K.; Nayaka, A.R.N.; Kumar, J.J. Identification of botanicals for the suppression of pebrine disease of the silkworm, Bombyx mori L. Uttar Pradesh J. Zool. 2010, 173–179. [Google Scholar]
- Porrini, M.P.; Fernández, N.J.; Garrido, P.M.; Gende, L.B.; Medici, S.K.; Eguaras, M.J. In vivo evaluation of antiparasitic activity of plant extracts on Nosema ceranae (Microsporidia). Apidologie 2011, 42, 700–707. [Google Scholar] [CrossRef]
- Borges, D.; Guzman-Novoa, E.; Goodwin, P.H. Control of the microsporidian parasite Nosema ceranae in honey bees (Apis mellifera) using nutraceutical and immuno-stimulatory compounds. PLoS ONE 2020, 15, e0227484. [Google Scholar] [CrossRef] [PubMed]
- Bessi, E.; Nanetti, A. Evaluation of tree different strategies to Nosema control. In Proceedings of the 39th Apimondia International Apicultural Congress, Dublin, Ireland, 21–26 August 2005. [Google Scholar]
- Sóstenes Rodríguez Dehaibes, R.; Luna-Olivares, G.; Chávez-Hernández, E. Resultados preliminares de ApiHerb para el control de Nosema spp. en México. Vida Apic. 2018, 207, 38–39. [Google Scholar]
- Nanetti, A.; Martín-Hernández, R.; Gómez-Moracho, T.; Cabbri, R.; Higes, M. Api-Herb en el control orgánico de la Nosemosis tipo C (Nosema ceranae, Microsporidia). In Proceedings of the III Congreso Ibérico de Apicultura, Mirandela, Portugal, 13–15 April 2015; Vilas-Boas, M., Dias, L.G., Moreira, L.M., Eds.; Instituto Politécnico de Bragança: Bragança, Portugal, 2014; p. 36. [Google Scholar]
- Cilia, G.; Sagona, S.; Giusti, M.; Jarmela dos Santos, P.E.; Nanetti, A.; Felicioli, A. Nosema ceranae infection in honeybee samples from Tuscanian Archipelago (Central Italy) investigated by two qPCR methods. Saudi J. Biol. Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef]
- Sagastume, S.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Genotype diversity in the honey bee parasite Nosema ceranae: Multi-strain isolates, cryptic sex or both? BMC Evol. Biol. 2016, 16, 1–11. [Google Scholar] [CrossRef]
- Sagastume, S.; Del Águila, C.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Polymorphism and recombination for rDNA in the putatively asexual microsporidian Nosema ceranae, a pathogen of honeybees. Environ. Microbiol. 2011, 13, 84–95. [Google Scholar] [CrossRef]
- O’Mahony, E.M.; Tay, W.T.; Paxton, R.J. Multiple rRNA variants in a single spore of the microsporidian Nosema bombi. J. Eukaryot. Microbiol. 2007, 54, 103–109. [Google Scholar] [CrossRef]
- Gomez-Moracho, T.; Maside, X.; Martin-Hernandez, R.; Higes, M.; Bartolomé, C. High levels of genetic diversity in Nosema ceranae within Apis mellifera colonies. Parasitology 2014, 141, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ru, Y.; Liu, W.; Wang, D.; Zhou, J.; Jiang, Y.; Shi, S.; Qin, L. Morphological characterization and HSP70-, IGS-based phylogenetic analysis of two microsporidian parasites isolated from Antheraea pernyi. Parasitol. Res. 2017, 116, 971–977. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Guideline on Veterinary Medicinal Products Controlling Varroa Destructor Parasitosis in Bees; European Medicines Agency: London, UK, 2011; p. 9. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-veterinary-medicinal-products-controlling-varroa-destructor-parasitosis-bees_en.pdf (accessed on 27 July 2020).
| Gene | Primers | Sequence (5′–3′) | Reference |
|---|---|---|---|
| 16S rRNA | Forward | AAGAGTGAGACCTATCAGCTAGTTG | [36] |
| Reverse | CCGTCTCTCAGGCTCCTTCTC | ||
| TaqMan Probe | ACCGTTACCCGTCACAGCCTTGTT | ||
| Hsp70 | Forward | GGGATTACAAGTGCTTAGAGTGATT | [37] |
| Reverse | TGTCAAGCCCATAAGCAAGTG | ||
| TaqMan Probe | TGAGCCTACTGCGGC |
| 16S rRNA | Hsp70 | |||||
|---|---|---|---|---|---|---|
| Colony | Group | N | Mean +/− s.e. | SD | Mean +/− s.e. | SD |
| 3 | AB | 25 | 13.45 +/− 12.55 | 13.25 | 10.80 +/− 9.45 | 10.15 |
| 4 | AB | 25 | 6.73 +/− 5.00 | 5.70 | 5.72 +/− 4.24 | 4.94 |
| 5 | AB | 25 | 4.73 +/− 3.15 | 3.85 | 3.86 +/− 2.23 | 2.93 |
| 8 | AB | 25 | 4.75 +/− 3.09 | 3.79 | 3.86 +/− 2.22 | 2.92 |
| 10 | AB | 25 | 7.75 +/− 6.26 | 6.96 | 5.92 +/− 4.22 | 4.92 |
| 13 | AB | 25 | 7.09 +/− 5.85 | 6.55 | 5.85 +/− 4.30 | 4.99 |
| 14 | AB | 25 | 4.54 +/− 3.06 | 3.76 | 3.84 +/− 2.39 | 3.09 |
| F1 | AH | 25 | 7.13 +/− 5.67 | 6.36 | 5.84 +/− 4.39 | 5.09 |
| F3 | AH | 25 | 7.17 +/− 5.88 | 6.58 | 5.79 +/− 4.29 | 4.99 |
| 16 | AH | 25 | 5.51 +/− 3.93 | 4.63 | 4.30 +/− 3.23 | 3.93 |
| 22 | AH | 25 | 6.77 +/− 5.15 | 5.85 | 5.58 +/− 4.24 | 4.94 |
| 24 | AH | 25 | 5.20 +/− 3.84 | 4.53 | 3.93 +/− 2.40 | 3.10 |
| 25 | AH | 25 | 4.14 +/− 3.31 | 4.01 | 3.70 +/− 2.40 | 3.10 |
| F4 | C | 25 | 5.68 +/− 4.07 | 4.77 | 4.68 +/− 3.35 | 4.05 |
| F7 | C | 25 | 5.43 +/− 3.90 | 4.60 | 4.66 +/− 3.29 | 3.99 |
| F9 | C | 25 | 4.90 +/− 3.00 | 3.70 | 3.91 +/− 2.43 | 3.12 |
| 17 | C | 25 | 5.57 +/− 3.92 | 4.62 | 4.78 +/− 3.07 | 3.77 |
| 19 | C | 25 | 6.94 +/− 5.02 | 5.72 | 5.75 +/− 4.26 | 4.96 |
| 26 | C | 25 | 7.64 +/− 6.12 | 6.82 | 6.86 +/− 5.40 | 6.10 |
| 16S rRNA | Hsp70 | |||||
| Colony | Group | N | Mean +/− s.e. | SD | Mean +/− s.e. | SD |
| 3 | AB | 25 | 4.45 +/− 3.36 | 4.06 | 3.76 +/− 2.08 | 2.78 |
| 4 | AB | 25 | 3.54 +/− 2.68 | 3.38 | 2.77 +/− 1.01 | 1.71 |
| 5 | AB | 25 | 2.87 +/− 1.42 | 2.12 | 2.31 +/− 1.08 | 1.78 |
| 8 | AB | 25 | 2.86 +/− 1.45 | 2.15 | 2.24 +/− 1.11 | 1.81 |
| 10 | AB | 25 | 3.55 +/− 2.45 | 3.15 | 2.86 +/− 1.23 | 1.93 |
| 13 | AB | 25 | 3.53 +/− 2.50 | 3.20 | 2.86 +/− 1.35 | 2.04 |
| 14 | AB | 25 | 2.73 +/− 1.36 | 2.06 | 2.35 +/− 0.92 | 1.62 |
| Group AB | 175 | 3.76 +/− 3.57 | 4.00 | 3.08 +/− 2.89 | 3.31 | |
| F1 | AH | 25 | 2.85 +/− 1.11 | 1.81 | 2.51 +/− 1.64 | 2.34 |
| F3 | AH | 25 | 2.86 +/− 1.07 | 1.77 | 2.47 +/− 1.39 | 2.09 |
| 16 | AH | 25 | 0.34 +/− 0.11 | 0.81 | −* | −* |
| 22 | AH | 25 | 2.87 +/− 1.14 | 1.84 | 2.25 +/− 1.16 | 1.86 |
| 24 | AH | 25 | 0.56 +/− 0.30 | 1.00 | −* | −* |
| 25 | AH | 25 | −* | −* | −* | −* |
| Group AH | 150 | 2.56 +/− 2.21 | 2.60 | 2.12 +/− 1.80 | 2.19 | |
| F4 | C | 25 | 5.56 +/− 4.37 | 5.07 | 4.93 +/− 3.06 | 3.76 |
| F7 | C | 25 | 5.58 +/− 4.25 | 4.95 | 4.93 +/− 3.07 | 3.77 |
| F9 | C | 25 | 4.90 +/− 3.45 | 4.15 | 4.00 +/− 2.18 | 2.88 |
| 17 | C | 25 | 5.46 +/− 4.07 | 4.77 | 4.89 +/− 3.03 | 3.73 |
| 19 | C | 25 | 6.84 +/− 5.28 | 5.98 | 5.81 +/− 4.06 | 4.76 |
| 26 | C | 25 | 8.69 +/− 7.33 | 8.03 | 6.92 +/− 5.49 | 6.19 |
| Group C | 150 | 7.92 +/− 7.91 | 8.30 | 6.18 +/− 6.13 | 6.52 | |
| AB | AH | C | |
|---|---|---|---|
| AB | - | 0.051 | 1.000 |
| AH | 0.049 | - | 0.064 |
| C | 1.000 | 0.062 | - |
| 16S rRNA | Hsp70 | |||||
|---|---|---|---|---|---|---|
| Group | Mean +/− s.e. | SD | 95% CI | Mean +/− s.e. | SD | 95% CI |
| AB | –99.38 +/− 0.28 | 0.75 | −100.07, −98.69 | –98.75 +/− 0.56 | 0.56 | −100.12, −97.37 |
| AH | –100.00 +/− 0.00 | 0.00 | −100.00, −99.99 | –99.98 +/− 0.01 | 0.03 | −100.00, −99.94 |
| C | +164.80 +/− 170.50 | 417.64 | −273.49, +603.09 | +40.52 +/− 12.98 | 31.80 | −7.14, +73.89 |
| Model | F(1873) = 1627.70, p = 0.000, Adj. R2 = 0.650 |
| Intercept | 12.82 +/− 1.80 s.e. (95% CI = 9.28, 16.35), t(873) = 7.119, p = 0.000 |
| Slope | 6.68 +/− 0.16 s.e. (95% CI = 6.36, 7.00) × 10−9, t(873) = 40.345, p = 0.000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilia, G.; Garrido, C.; Bonetto, M.; Tesoriero, D.; Nanetti, A. Effect of Api-Bioxal® and ApiHerb® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods. Vet. Sci. 2020, 7, 125. https://doi.org/10.3390/vetsci7030125
Cilia G, Garrido C, Bonetto M, Tesoriero D, Nanetti A. Effect of Api-Bioxal® and ApiHerb® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods. Veterinary Sciences. 2020; 7(3):125. https://doi.org/10.3390/vetsci7030125
Chicago/Turabian StyleCilia, Giovanni, Claudia Garrido, Martina Bonetto, Donato Tesoriero, and Antonio Nanetti. 2020. "Effect of Api-Bioxal® and ApiHerb® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods" Veterinary Sciences 7, no. 3: 125. https://doi.org/10.3390/vetsci7030125
APA StyleCilia, G., Garrido, C., Bonetto, M., Tesoriero, D., & Nanetti, A. (2020). Effect of Api-Bioxal® and ApiHerb® Treatments against Nosema ceranae Infection in Apis mellifera Investigated by Two qPCR Methods. Veterinary Sciences, 7(3), 125. https://doi.org/10.3390/vetsci7030125






