Development of a Novel, Non-Invasive Saliva Sampling Method for the Detection of Bovine Respiratory Viruses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection
2.3. RNA/DNA Extraction and PCR Tests
2.4. Development of RT-PCRs for BRAV and BRBV
2.5. Sequencing and Sequence Analysis
2.6. Data Analysis
3. Results
3.1. Virus Detection in Deep Nasal Swabs
3.2. Virus Detection in Saliva Samples
3.3. Sequencing Results
3.4. Comparison of Deep Nasal Swabs and Saliva Samples
- Positive result in deep nasal swab and saliva sample (+/+);
- Negative result in deep nasal swab and saliva sample (−/−);
- Positive result in deep nasal swab and negative result in saliva sample (+/−);
- Negative result in deep nasal swab and positive result in saliva sample (−/+).
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BRD | Bovine Respiratory Disease |
| PCR | Polymerase Chain Reaction |
| Abbrev. | Abbreviation |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
| BRSV | Bovine Respiratory Syncytial Virus |
| BPIV3 | Bovine Parainfluenzavirus Type 3 |
| BoAHV1 | Bovine Alphaherpesvirus 1 |
| BoHV | Bovine Herpesvirus |
| BoCV | Bovine Coronavirus |
| BAdV | Bovine Adenovirus |
| BAdV-4 | Bovine Adenovirus 4 |
| BAdV-5 | Bovine Adenovirus 5 |
| BAdV-8 | Bovine Adenovirus 8 |
| BAdV-7 | Bovine Adenovirus 7 |
| BAdV-6 | Bovine Adenovirus 6 |
| BAdV-1 | Bovine Adenovirus 1 |
| BAdV-3 | Bovine Adenovirus 3 |
| BAdV-10 | Bovine Adenovirus 10 |
| IDV | Influenza D Virus |
| IAV | Influenza A Virus |
| BRAV | Bovine Rhinitis A Virus |
| BRAV-1 | Bovine Rhinitis A Virus 1 |
| BRAV-2 | Bovine Rhinitis A Virus 2 |
| BRBV | Bovine Rhinitis B Virus |
| BRBV-1 | Bovine Rhinitis B Virus 1 |
| BRBV-2 | Bovine Rhinitis B Virus 2 |
| BRBV-3 | Bovine Rhinitis B Virus 3 |
| BRBV-4 | Bovine Rhinitis B Virus 4 |
| BRBV-5 | Bovine Rhinitis B Virus 5 |
| BVDV | Bovine Viral Diarrhea Virus |
| BoGHV6 | Bovine Gammaherpesvirus 6 |
| ddH2O | Double-Distilled H2O |
References
- Bell, R.L.; Turkington, H.L.; Cosby, S.L. The Bacterial and Viral Agents of BRDC: Immune Evasion and Vaccine Developments. Vaccines 2021, 9, 337. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, S.A.; van Eenennaam, A.L.; Aly, S.S.; Karle, B.M.; Rossitto, P.V.; Overton, M.W.; Lehenbauer, T.W.; Fadel, J.G. Preweaning cost of bovine respiratory disease (BRD) and cost-benefit of implementation of preventative measures in calves on California dairies: The BRD 10K study. J. Dairy Sci. 2020, 103, 1583–1597. [Google Scholar] [CrossRef]
- Chai, J.; Capik, S.F.; Kegley, B.; Richeson, J.T.; Powell, J.G.; Zhao, J. Bovine respiratory microbiota of feedlot cattle and its association with disease. Vet. Res. 2022, 53, 4. [Google Scholar] [CrossRef]
- Gaudino, M.; Nagamine, B.; Ducatez, M.F.; Meyer, G. Understanding the mechanisms of viral and bacterial coinfections in bovine respiratory disease: A comprehensive literature review of experimental evidence. Vet. Res. 2022, 53, 70. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can. Vet. J. 2010, 51, 1095–1102. [Google Scholar] [PubMed]
- Padalino, B.; Cirone, F.; Zappaterra, M.; Tullio, D.; Ficco, G.; Giustino, A.; Ndiana, L.A.; Pratelli, A. Factors Affecting the Development of Bovine Respiratory Disease: A Cross-Sectional Study in Beef Steers Shipped from France to Italy. Front. Vet. Sci. 2021, 8, 627894. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, W.; Yang, M.; Lin, J.; Xue, F.; Zhu, Y.; Yin, X. Development of a One-Step Multiplex Real-Time PCR Assay for the Detection of Viral Pathogens Associated with the Bovine Respiratory Disease Complex. Front. Vet. Sci. 2022, 9, 825257. [Google Scholar] [CrossRef]
- Chiapponi, C.; Faccini, S.; Fusaro, A.; Moreno, A.; Prosperi, A.; Merenda, M.; Baioni, L.; Gabbi, V.; Rosignoli, C.; Alborali, G.L.; et al. Detection of a New Genetic Cluster of Influenza D Virus in Italian Cattle. Viruses 2019, 11, 1110. [Google Scholar] [CrossRef]
- Fulton, R.W. Viruses in Bovine Respiratory Disease in North America: Knowledge Advances Using Genomic Testing. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 321–332. [Google Scholar] [CrossRef]
- Pardon, B.; Buczinski, S. Bovine Respiratory Disease Diagnosis: What Progress Has Been Made in Infectious Diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 425–444. [Google Scholar] [CrossRef]
- Centeno-Martinez, R.E.; Glidden, N.; Mohan, S.; Davidson, J.L.; Fernández-Juricic, E.; Boerman, J.P.; Schoonmaker, J.; Pillai, D.; Koziol, J.; Ault, A.; et al. Identification of bovine respiratory disease through the nasal microbiome. Anim. Microbiome 2022, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, S.; Fecteau, G.; Dubuc, J.; Francoz, D.; Rousseau, M.; Roy, J.-P.; Buczinski, S. Scoping review on clinical definition of bovine respiratory disease complex and related clinical signs in dairy cows. J. Dairy Sci. 2021, 104, 7095–7108. [Google Scholar] [CrossRef] [PubMed]
- Buczinski, S.; Pardon, B. Bovine Respiratory Disease Diagnosis: What Progress Has Been Made in Clinical Diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 399–423. [Google Scholar] [CrossRef]
- Doyle, D.; Credille, B.; Lehenbauer, T.W.; Berghaus, R.; Aly, S.S.; Champagne, J.; Blanchard, P.; Crossley, B.; Berghaus, L.; Cochran, S.; et al. Agreement Among 4 Sampling Methods to Identify Respiratory Pathogens in Dairy Calves with Acute Bovine Respiratory Disease. J. Vet. Intern. Med. 2017, 31, 954–959. [Google Scholar] [CrossRef]
- Kliučinskas, R.; Lukauskas, K.; Milius, J.; Vyšniauskis, G.; Kliučinskas, D.; Šalomskas, A. Detection of bovine viral diarrhoea virus in saliva samples. Bull. Vet. Inst. Pulawy 2008, 52, 31–37. [Google Scholar]
- Wellehan, J.F.X.; Johnson, A.J.; Harrach, B.; Benkö, M.; Pessier, A.P.; Johnson, C.M.; Garner, M.M.; Childress, A.; Jacobson, E.R. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 2004, 78, 13366–13369. [Google Scholar] [CrossRef]
- VanDevanter, D.R.; Warrener, P.; Bennett, L.; Schultz, E.R.; Coulter, S.; Garber, R.L.; Rose, T.M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 1996, 34, 1666–1671. [Google Scholar] [CrossRef] [PubMed]
- Vilcek, S.; Elvander, M.; Ballagi-Pordány, A.; Belák, S. Development of nested PCR assays for detection of bovine respiratory syncytial virus in clinical samples. J. Clin. Microbiol. 1994, 32, 2225–2231. [Google Scholar] [CrossRef]
- Maidana, S.S.; Lomonaco, P.M.; Combessies, G.; Craig, M.I.; Diodati, J.; Rodriguez, D.; Parreño, V.; Zabal, O.; Konrad, J.L.; Crudelli, G.; et al. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes (Bubalus bubalis) in Argentina. BMC Vet. Res. 2012, 8, 83. [Google Scholar] [CrossRef]
- Parvin, R.; Shehata, A.A.; Heenemann, K.; Gac, M.; Rueckner, A.; Halami, M.Y.; Vahlenkamp, T.W. Differential replication properties among H9N2 avian influenza viruses of Eurasian origin. Vet. Res. 2015, 46, 75. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Amer, H.M.; Almajhdi, F.N. Development of a SYBR Green I based real-time RT-PCR assay for detection and quantification of bovine coronavirus. Mol. Cell. Probes 2011, 25, 101–107. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Kondov, N.O.; Deng, X.; van Eenennaam, A.; Neibergs, H.L.; Delwart, E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015, 89, 5340–5349. [Google Scholar] [CrossRef]
- Lehmkuhl, H.D.; Smith, M.H.; Dierks, R.E. A bovine adenovirus type 3: Isolation, characterization, and experimental infection in calves. Arch. Virol. 1975, 48, 39–46. [Google Scholar] [CrossRef]
- Thompson, K.G.; Thomson, G.W.; Henry, J.N. Alimentary Tract Manifestations of Bovine Adenovirus Infections. Can. Vet. J. 1981, 22, 68–71. [Google Scholar]
- Rosato, G.; Ruiz Subira, A.; Al-Saadi, M.; Michalopoulou, E.; Verin, R.; Dettwiler, M.; Nordgren, H.; Chiers, K.; Groβmann, E.; Köhler, K.; et al. Gammaherpesvirus Infections in Cattle in Europe. Viruses 2021, 13, 233. [Google Scholar] [CrossRef]
- Headley, S.A.; Dall Agnol, A.M.; Oliveira, T.E.S.; Bon, V.R.; Scuisato, G.S.; Xavier, A.A.C.; Yasumitsu, C.Y.; Alfieri, A.F.; Alfieri, A.A. Possible Association of Bovine Gammaherpesvirus 6 with Pulmonary Disease in a Cow. Animals 2023, 13, 417. [Google Scholar] [CrossRef]
- Fabian, R.; Rosato, G.; Stewart, J.P.; Kipar, A. Bovine Gammaherpesvirus 6 Tropism in the Natural Host. Viruses 2024, 16, 1730. [Google Scholar] [CrossRef]
- Makoschey, B.; Berge, A.C. Review on bovine respiratory syncytial virus and bovine parainfluenza—Usual suspects in bovine respiratory disease—A narrative review. BMC Vet. Res. 2021, 17, 261. [Google Scholar] [CrossRef]
- Berge, A.C.; Vertenten, G. Bovine Coronavirus Prevalence and Risk Factors in Calves on Dairy Farms in Europe. Animals 2024, 14, 2744. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Saif, L.J. Bovine Coronavirus and the Associated Diseases. Front. Vet. Sci. 2021, 8, 643220. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, S.; Lin, C.-M.; Temeeyasen, G.; Palinski, R.; Li, F.; Kaushik, R.S.; Hause, B.M. Bovine rhinitis B virus is highly prevalent in acute bovine respiratory disease and causes upper respiratory tract infection in calves. J. Gen. Virol. 2022, 103, 1714. [Google Scholar] [CrossRef]
- Zhang, M.; Hill, J.E.; Fernando, C.; Alexander, T.W.; Timsit, E.; van der Meer, F.; Huang, Y. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound. Emerg. Dis. 2019, 66, 1379–1386. [Google Scholar] [CrossRef]
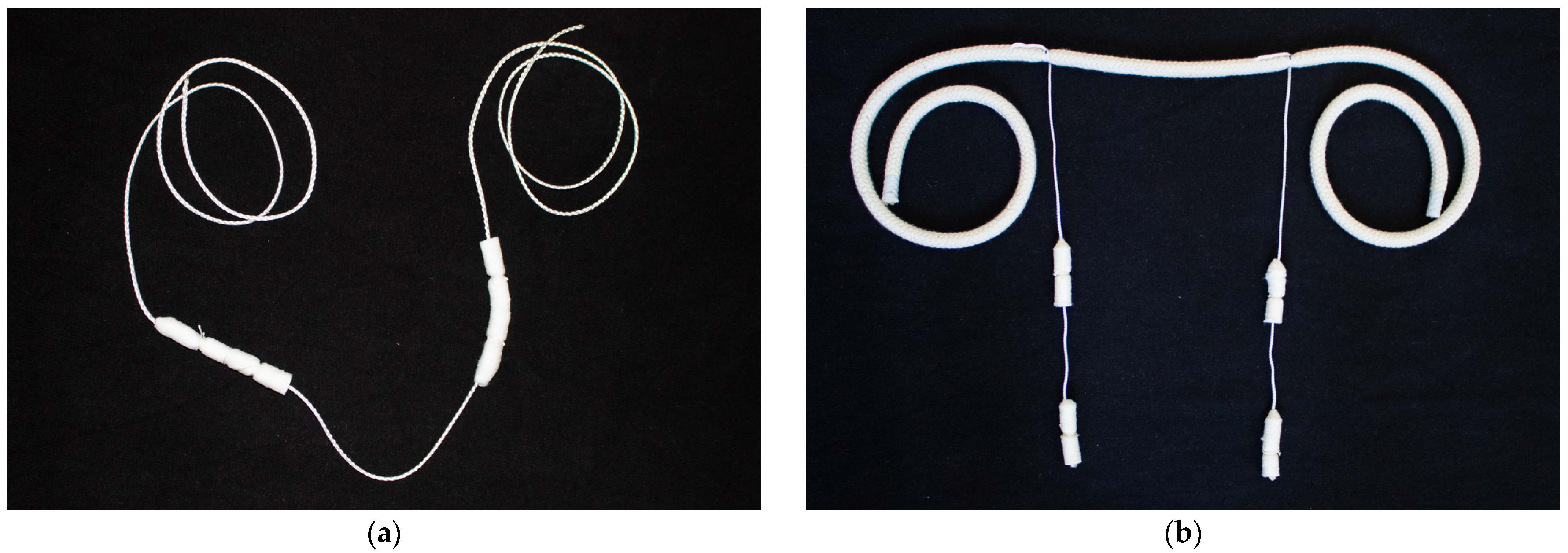
| Family | Genus | Species | Virus Name | Abbrev. |
|---|---|---|---|---|
| Pneumoviridae | Orthopneumovirus | Orthopneumovirus bovis | Bovine Respiratory Syncytial Virus | BRSV |
| Paramyxoviridae | Respirovirus | Respirovirus bovis | Bovine Parainfluenza Virus 3 | BPIV3 |
| Orthoherpesviridae | Varicellovirus | Varicellovirus bovinealpha1 | Bovine Aalphaherpesvirus 1 | BoAHV1 |
| Coronaviridae | Betacoronavirus | Betacoronavirus gravedinis | Bovine Coronavirus | BoCV |
| Adenoviridae | Barthadenovirus | Barthadenovirus bosquartum | Bovine Adenovirus 4, 5 and 8 | BAdV-4/5/8 |
| Barthadenovirus bosseptimum | Bovine Adenovirus 7 | BAdV-7 | ||
| Barthadenovirus bossextum | Bovine Adenovirus 6 | BAdV-6 | ||
| Mastadenovirus | Mastadenovirus bosprimum | Bovine Adenovirus 1 | BAdV-1 | |
| Mastadenovirus bostertium | Bovine Adenovirus 3 | BAdV-3 | ||
| Mastadenovirus bosdecimum | Bovine Adenovirus 10 | BAdV-10 | ||
| Orthomyxoviridae | Deltainfluenzavirus | Deltainfluenzavirus influenzae | Influenza D Virus | IDV |
| Orthomyxoviridae | Alphainfluenzavirus | Alphainfluenzavirus influenzae | Influenza A Virus | IAV |
| Picornaviridae | Aphthovirus | Aphthovirus bogeli | Bovine Rhinitis A Virus 1 | BRAV-1 |
| Bovine Rhinitis A Virus 2 | BRAV-2 | |||
| Aphthovirus reedi | Bovine Rhinitis B Virus 1, 2, 3, 4 and 5 | BRBV-1/2/3/4/5 |
| Group of Animals | Sample Material | |
|---|---|---|
| Deep Nasal Swab | Saliva | |
| 5× Individual cows | 5× Deep nasal swabs (EYDAM) | 5× Saliva sampling system for cows |
| One group of cows | 5× random Deep nasal swabs (EYDAM) =pooled to one sample | 2× Saliva sampling system for cows =pooled to one sample |
| 5× Individual calves | 5× Deep nasal swabs with UTM™ | 5× Saliva sampling system for calves |
| One group of calves | 5× random Deep nasal swabs with UTM™ =pooled to one sample | 2× Saliva sampling system for calves =pooled to one sample |
| Target Virus | Target Gene | PCR Method | PCR Kit | Primer Name | Primer Sequence (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|---|---|---|---|
| Adenovirus | DNA polymerase gene | consensus nested PCR | DreamTaq DNA Polymerase | pol Fouter | TNMGNGGNGGNMGNTGYTAYCC | 320 | [16] |
| pol Router | GTDGCRAANSHNCCRTABARNGMRTT | ||||||
| pol Finner | GTNTWYGAYATHTGYGGHATGTAYGC | ||||||
| pol Rinner | CCANCCBCDRTTRTGNARNGTRA | ||||||
| Herpesvirus | DNA polymerase gene | consensus nested PCR | DreamTaq DNA Polymerase | KG1 | GTCTTGCTCACCAGNTCNACNCCYTT | 210 | [17] |
| DFA | GAYTTYGCNAGYYTNTAYCC | ||||||
| ILK | TCCTGGACAAGCAGCARNYSGCNMTNAA | ||||||
| TGV | TGTACCTCGGTGTAYGGNTTYACNGGNGT | ||||||
| IYG | CACAGAGTCCGTRTCNCCRTACAT | ||||||
| BRSV | G gene | one-step nested RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase and Platinum™ Taq DNA-Polymerase High Fidelity | B5A | CCACCCTAGCAATGATAACCTTGAC | 371 | [18] |
| B6A | AAGAGAGGATGCYTTGCTGTGG | ||||||
| B7A | CATCAATCCAAAGCACCACACTGTC | ||||||
| B8 | GCTAGTTCTGTGGTGGATTGTTGTC | ||||||
| BPIV3 | M gene | one-step RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase | M1 | AGTGATCTAGATGATGATCCA | 328 | [19] |
| M2 | GTTATTGATCCAATTGCTGT | ||||||
| IAV | M gene | realtime RT-PCR | RevertAid H Minus Reverse Transcriptase and QuantiTect® SYBR® Green PCR Kit (QIAGEN, Hilden, Germany) | Uni 12 | AGCAAAAGCAGG | 149 | [20,21] |
| M1F | GATGTYTTTGCAGGRAAGAAC | ||||||
| M2R | AABCGTCTACGCTGCAGTCC | ||||||
| IDV | PB1 gene | one-step RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase | F | TGGATGGAGAGTGCTGCTTC | 110 | [8] |
| R | GCCAATGCTTCCTCCCTGTA | ||||||
| BoCV | N gene | one-step RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase | F | TTGGATCAAGATTAGAGTTGGC | 237 | [22] |
| R | CCTTGTCCATTCTTCTGACC | ||||||
| BRAV | 3D polymerase gene | one-step nested RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase and Platinum™ Taq DNA-Polymerase High Fidelity | F | GCCGTGTTCTCGGAYGAG | 232 | this study |
| R1 | TARGCAACCATMACATARTCRTC | ||||||
| R2 | TTRAARTCAAAATCRTGGTCYGA | ||||||
| BRBV | polyprotein gene | one-step RT-PCR | SuperScript™ III One-Step RT-PCR/Platinum™ Taq High Fidelity DNA-Polymerase | F | TACTYGGWCAYACYATAACACC | 294 | this study |
| R | ACCTGTATGAYGGWATCTCRAAACT |
| Target Virus | Target Gene | Primer Name | Primer Sequence (5′-3′) | Position in Nucleotide | Reference Sequence (GenBank Acc.-No.) |
| BRAV | 3D polymerase gene | F | GCCGTGTTCTCGGAYGAG | 6615-6632 | NC_038303.1 |
| R 1 | TARGCAACCATMACATARTCRTC | 6825-6847 | |||
| R 2 | TTRAARTCAAAATCRTGGTCYGA | 6870-6892 | |||
| BRBV | polyprotein gene | F | TACTYGGWCAYACYATAACACC | 7185-7206 | NC_010354.1 |
| R | ACCTGTATGAYGGWATCTCRAAACT | 7454-7478 |
| Cows | Calves | |||||||
|---|---|---|---|---|---|---|---|---|
| Individuals | Groups | Individuals | Groups | |||||
| Swab 1 | Saliva 2 | Swab 1 | Saliva 2 | Swab 3 | Saliva 4 | Swab 3 | Saliva 4 | |
| Farm 1 | 5 | 2 | 5 (1) | 2 (1) | 5 | 1 | 5 (1) | 2 (1) |
| Farm 2 | 4 | 5 | 5 (1) | 2 (1) | 5 | 3 | 5 (1) | 0 |
| Farm 3 | 5 | 5 | 5 (1) | 1 (1) | 5 | 4 | 5 (1) | 0 |
| Farm 4 | 5 | 4 | 5 (1) | 2 (1) | 5 | 5 | 5 (1) | 2 (1) |
| Farm 5 | 5 | 5 | 5 (1) | 2 (1) | 5 | 5 | 6 (2) | 2 (2) |
| Total | 24 | 21 | 25 (5) | 5 (5) | 25 | 18 | 26 (6) | 6 (4) |
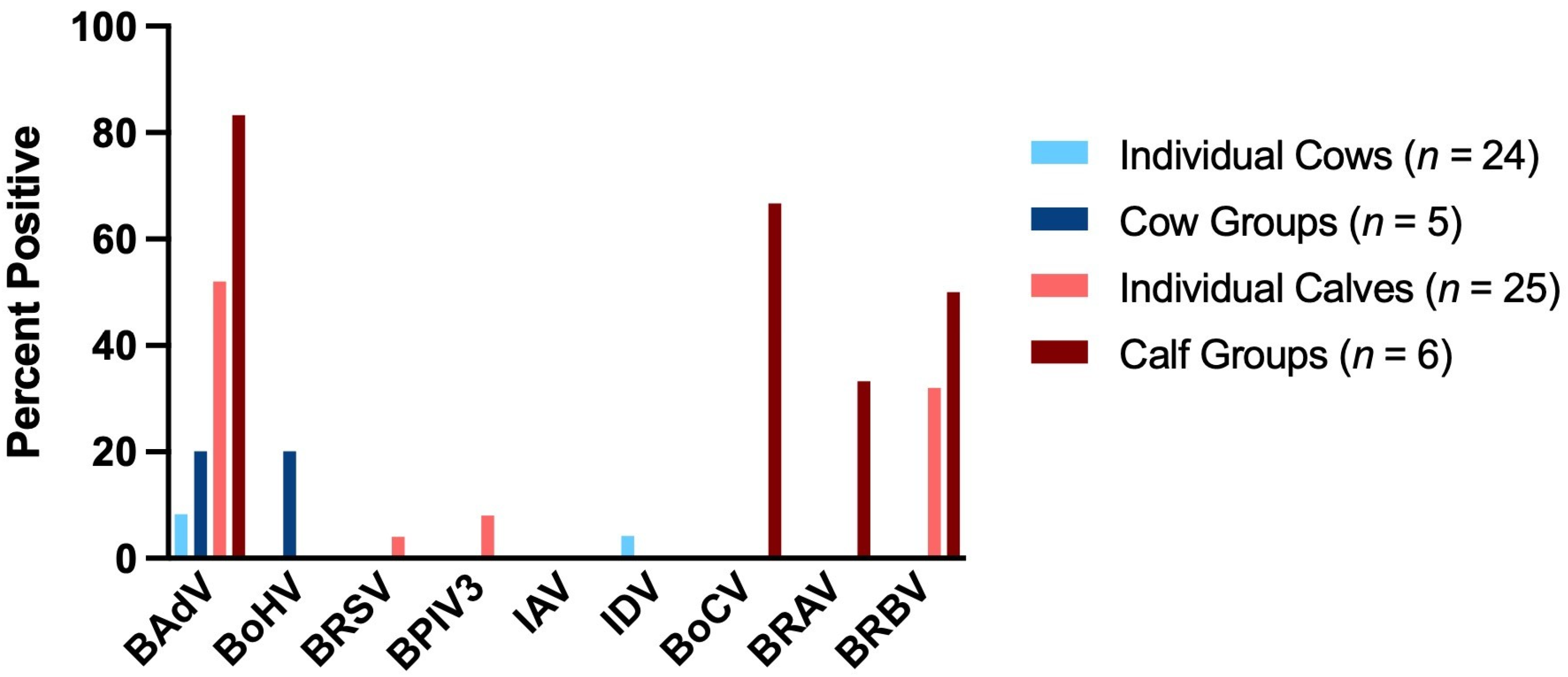
| Group of Animals | BAdV | BoHV | BRSV | BPIV3 | IAV | IDV | BoCV | BRAV | BRBV |
|---|---|---|---|---|---|---|---|---|---|
| Individual Cows (n = 24) | 8.33% [2/24] | 0.0% [0/24] | 0.0% [0/24] | 0.0% [0/24] | 0.0% [0/24] | 4.17% [1/24] | 0.0% [0/24] | 0.0% [0/24] | 0.0% [0/24] |
| Cow Groups (n = 5) | 20.0% [1/5] | 20.0% [1/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] |
| Individual Calves (n = 25) | 52.0% [13/25] | 0.0% [0/25] | 4.0% [1/25] | 8.0% [2/25] | 0.0% [0/25] | 0.0% [0/25] | 0.0% [0/25] | 0.0% [0/25] | 32.0% [8/25] |
| Calf Groups (n = 6) | 83.33% [5/6] | 0.0% [0/6] | 0.0% [0/6] | 0.0% [0/6] | 0.0% [0/6] | 0.0% [0/6] | 66.67% [4/6] | 33.33% [2/6] | 50.0% [3/6] |
| Total (n = 60) | 35.0% [21/60] | 1.67% [1/60] | 1.67% [1/60] | 3.33% [2/60] | 0.0% [0/60] | 1.67% [1/60] | 6.67% [4/60] | 3.33% [2/60] | 18.33% [11/60] |
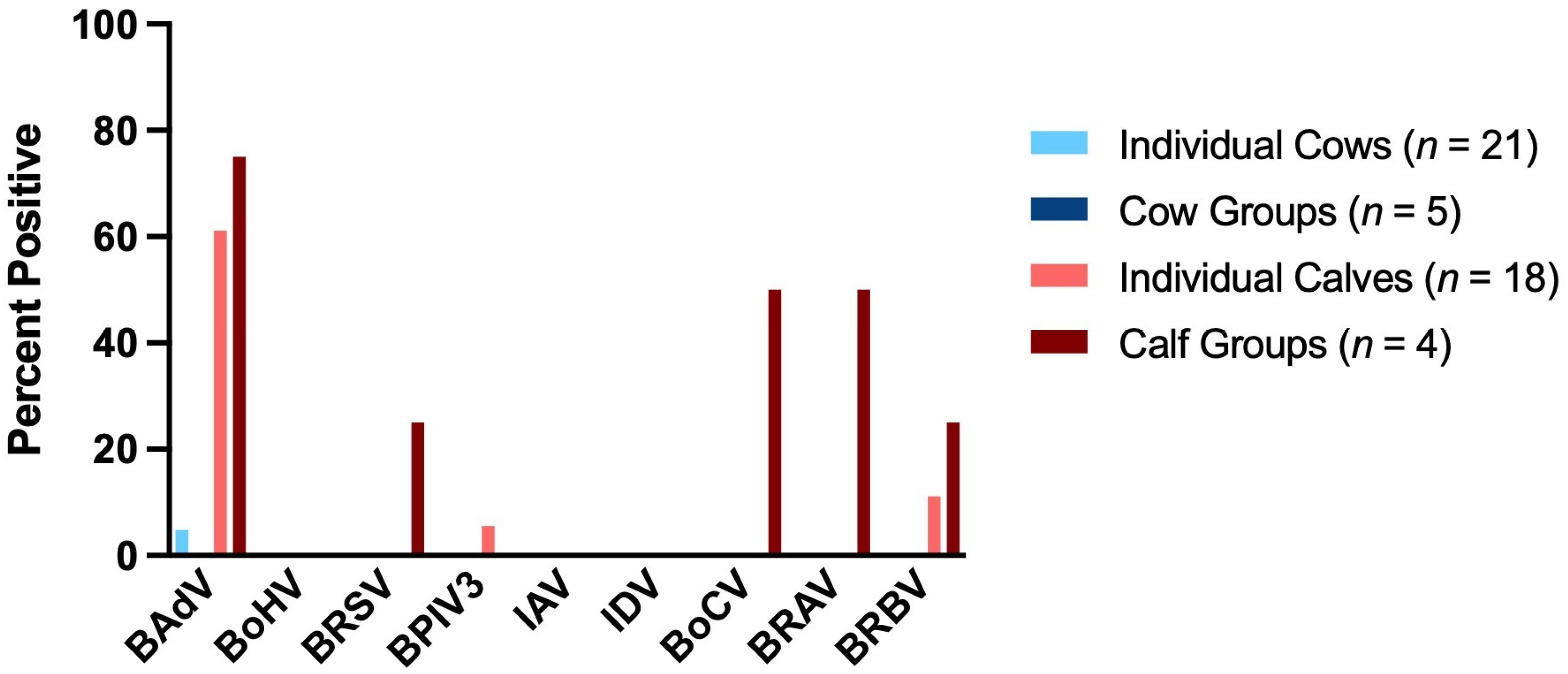
| Group of Animals | BAdV | BoHV | BRSV | BPIV3 | IAV | IDV | BoCV | BRAV | BRBV |
| Individual Cows (n = 21) | 4.76% [1/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] | 0.0% [0/21] |
| Cow Groups (n = 5) | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] | 0.0% [0/5] |
| Individual Calves (n = 18) | 61.11% [11/18] | 0.0% [0/18] | 0.0% [0/18] | 5.56% [1/18] | 0.0% [0/18] | 0.0% [0/18] | 0.0% [0/18] | 0.0% [0/18] | 11.11% [2/18] |
| Calf Groups (n = 4) | 75.0% [3/4] | 0.0% [0/4] | 25.0% [1/4] | 0.0% [0/4] | 0.0% [0/4] | 0.0% [0/4] | 50.0% [2/4] | 50.0% [2/4] | 25.0% [1/4] |
| Total (n = 48) | 31.25% [15/48] | 0.0% [0/48] | 2.08% [1/48] | 2.08% [1/48] | 0.0% [0/48] | 0.0% [0/48] | 4.17% [2/48] | 4.17% [2/48] | 8.33% [4/48] |
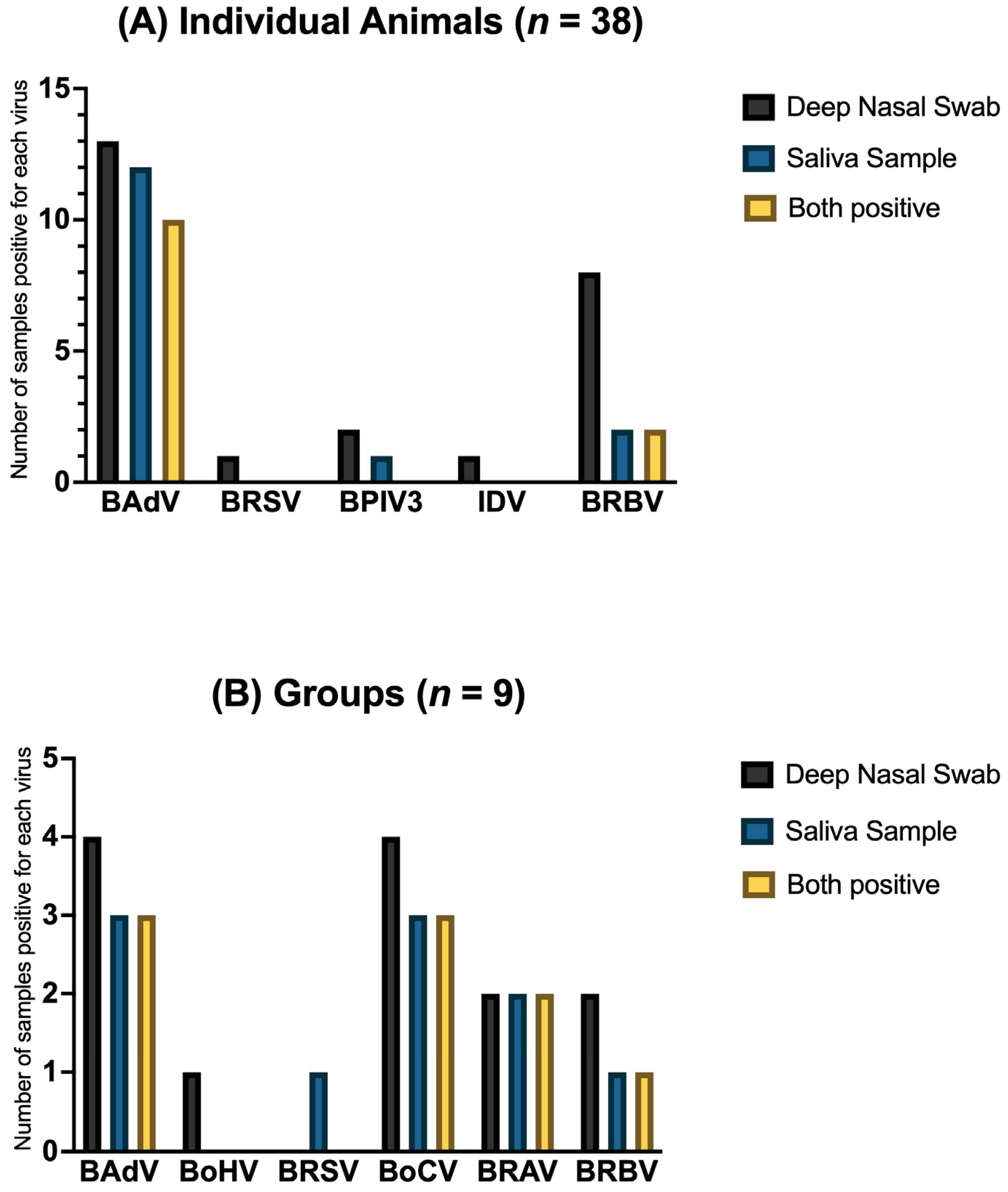
| Virus | Individual Animals (n = 38) | Groups (n = 9) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| +/+ | −/− | Sum | +/− | −/+ | +/+ | −/− | Sum | +/− | −/+ | |
| BAdV | 26.32% | 60.53% | 86.86% | 7.89% | 5.26% | 33.33% | 55.56% | 88.89% | 11.11% | 0.0% |
| BoHV | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% | 0.0% | 88.89% | 88.89% | 11.11% | 0.0% |
| BRSV | 0.0% | 97.37% | 97.37% | 2.63% | 0.0% | 0.0% | 88.89% | 88.89% | 0.0% | 11.11% |
| BPIV3 | 0.0% | 92.11% | 92.11% | 5.26% | 2.63% | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% |
| IAV | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% |
| IDV | 0.0% | 97.37% | 97.37% | 2.63% | 0.0% | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% |
| BoCV | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% | 33.33% | 55.56% | 88.89% | 11.11% | 0.0% |
| BRAV | 0.0% | 100.0% | 100.0% | 0.0% | 0.0% | 22.22% | 77.78% | 100.0% | 0.0% | 0.0% |
| BRBV | 5.26% | 78.95% | 85.21% | 15.79% | 0.0% | 11.11% | 77.78% | 88.89% | 11.11% | 0.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumann, S.; Euring, B.; Harzer, M.; Eibisch, M.; Lindner, A.; Vahlenkamp, T.W.; Heenemann, K. Development of a Novel, Non-Invasive Saliva Sampling Method for the Detection of Bovine Respiratory Viruses. Vet. Sci. 2025, 12, 637. https://doi.org/10.3390/vetsci12070637
Baumann S, Euring B, Harzer M, Eibisch M, Lindner A, Vahlenkamp TW, Heenemann K. Development of a Novel, Non-Invasive Saliva Sampling Method for the Detection of Bovine Respiratory Viruses. Veterinary Sciences. 2025; 12(7):637. https://doi.org/10.3390/vetsci12070637
Chicago/Turabian StyleBaumann, Simona, Belinda Euring, Maxi Harzer, Mandy Eibisch, Andrea Lindner, Thomas W. Vahlenkamp, and Kristin Heenemann. 2025. "Development of a Novel, Non-Invasive Saliva Sampling Method for the Detection of Bovine Respiratory Viruses" Veterinary Sciences 12, no. 7: 637. https://doi.org/10.3390/vetsci12070637
APA StyleBaumann, S., Euring, B., Harzer, M., Eibisch, M., Lindner, A., Vahlenkamp, T. W., & Heenemann, K. (2025). Development of a Novel, Non-Invasive Saliva Sampling Method for the Detection of Bovine Respiratory Viruses. Veterinary Sciences, 12(7), 637. https://doi.org/10.3390/vetsci12070637






