Simple Summary
Pseudomonas aeruginosa (P. aeruginosa) is an aerobic, non-fermentative, oxidase-positive, small Gram-negative bacterium typically found in single pairs. It is a prevalent opportunistic pathogen in clinical practice that can cause healthcare-associated infections in both humans and animals, including goat, dog, cat, cow, forest musk deer, mink, blue fox, and so on. P. aeruginosa plays a significant role in diseases such as intestinal infections. We confirmed that P. aeruginosa was identified as the pathogen in the diarrhea feces of a cynomolgus monkey through morphological analysis, biochemical tests, 16S rRNA gene sequencing, and animal pathogenicity experiments. Additionally, we performed drug sensitivity testing on P. aeruginosa. So far, this is the first case of P. aeruginosa isolated from the diarrhea feces of a cynomolgus monkey. This study provides an important scientific foundation for isolating and identifying P. aeruginosa as well as for preventing and treating bacterial diseases in experimental monkeys.
Abstract
In this study, we isolated and identified bacteria from the feces of a diarrheal cynomolgus monkey. The results showed that the isolated strain was P. aeruginosa, named PA/CM-101101. Morphological observations indicated that when cultured on Luria–Bertani (LB) nutrient agar at 37 °C for 24 h, the strain formed smooth, slightly elevated colonies with neat and wavy edges. On acetamide agar at the same temperature and duration, the colonies appeared flat with irregular edges and a faint pink periphery, while the medium changed to rose-red; in LB broth at 37 °C for 24 h, the medium became turbid and yellowish-green. Gram staining revealed that it was negative and rod-shaped, without sporulation characteristics. The 16S rRNA gene sequence analysis showed that the sequence identity of the strain shared more than 98.4% similarity with 11 strains of P. aeruginosa from various sources in GenBank. The animal toxicity test showed that it had a strong pathogenic effect on mice. The results of drug sensitivity tests showed that strain PA/CM-101101 was sensitive to amikacin, azithromycin, cefoperazone, ceftazidime, ceftriaxone, ciprofloxacin, gentamicin, imipenem, levofloxacin, meropenem, norfloxacin, ofloxacin, and polymyxin B; however, it displayed resistance to ampicillin, cefadroxil, cefazolin, erythromycin, and vancomycin. The research findings provide valuable insights for diagnosis and treatment strategies for cynomolgus monkeys. It also provides a reference for molecular epidemiological studies. To our knowledge, this is the first time P. aeruginosa isolated from the diarrhea feces of cynomolgus monkey has been reported.
1. Introduction
Macaques, as the animal model closest to human beings, play an important role in basic research and biomedical research [1]. However, macaques living in open environments are susceptible to infections caused by various pathogenic microorganisms, leading to disease development [2]. Intestinal infections can stimulate the gastrointestinal mucosa and impair both secretion and absorption functions of intestinal fluids. The primary clinical manifestation is diarrhea, a prevalent health problem in experimental monkeys [3,4]. Once diarrhea occurs, it will cause malnutrition in macaques, reduce the reproductive rate, and even lead to death, which seriously threatens the feeding and management of primates and is not conducive to the output of high-quality laboratory animals. Bacteria represent one of the main groups of pathogenic microorganisms responsible for intestinal infections in macaques, due to their close evolutionary relationship with humans and due to the similar gut microbiota observed in developing countries, so there is a significant risk of zoonotic transfer from these pathogens [5,6].
P. aeruginosa belongs to the genus Pseudomonas and is an aerobic Gram-negative bacillus with strong environmental adaptability. It is widely present in the external environment such as drinking water and soil, as well as in animals and plants [7], and is an important zoonotic pathogen. It can cause infections in humans and animals, and is a major healthcare-associated pathogen worldwide [8,9]. It can result in skin and soft tissue infection [10,11], keratitis [12], pneumonia [13,14], pulmonary cystic fibrosis [15], urinary tract infection [16], bronchiectasis [17,18], bacteremia [19], and other diseases. In the aquaculture industry, contaminated water, cages, and feed tanks serve as reservoirs of P. aeruginosa and are the origin of P. aeruginosa outbreaks [20,21]. The epidemic outbreak of P. aeruginosa not only leads to significant losses in the livestock and poultry industry but also has a considerable impact on the health of experimental and wild animals and their reproductive derivatives. P. aeruginosa can infect humans [22], goats (Capra aegagrus hircus) [23], dogs (Canis lupus familiaris) and cats (Felis silvestris catus) [24,25], cows (Bos taurus) [26], forest musk deer (Moschus berezovskii) [27], mink (Mustelidae) [28], and blue fox (Alopex lagopus) [29]. Non-human primates (NHPs) such as rhesus monkeys, squirrel monkeys, and African green monkeys are also susceptible to P. aeruginosa infection, which may cause bronchitis and myocarditis [30,31,32]. However, to our knowledge, systematic research on the morphology and molecular identification of P. aeruginosa isolated from the feces of diarrheal cynomolgus monkeys has not been reported.
2. Materials and Methods
To identify potential causes of severe diarrhea in a cynomolgus monkey (Macaca fascicularis), the bacteria was identified by morphological examination, physiological and biochemical tests, molecular biological analysis, a drug sensitivity test, and animal pathogenicity experiments.
2.1. Samples
The cynomolgus monkey employed in this study originated from Kunming University of Science and Technology [SYXK (Yunnan) K2022-0001]. The clinical manifestations of the monkey were as follows: watery stools, listlessness, dehydration, and decreased appetite. After sterilizing the skin around the anus with iodophor, feces were collected in a sterile 2 mL centrifuge tube via a sterile disposable sampling swab.
2.2. Bacterial Isolation and Morphological Observation
Anal swab samples were thoroughly mixed in sterile saline solution. The resultant suspension was inoculated onto LB nutrient agar (Hopebio, Qingdao, China). The plates were incubated at 37 °C for a period of 24 h. Single colonies were selected for streaking and subculturing on acetamide agar medium (Hopebio, Qingdao, China). Additionally, three colonies were isolated and inoculated into 5 mL of LB broth medium (Hopebio, Qingdao, China). The cultures on the plates were maintained at 37 °C for a duration of 18–24 h to observe bacterial growth characteristics. Purified and cultured isolates were collected for smear staining (Gram stain kit, Solarbio, Beijing, China) and observed under a microscope.
2.3. Physiological and Biochemical Identification
The colonies to be detected were selected in normal saline, and the bacterial suspension of 0.5 McFarland (approximately 108 CFU/mL) was prepared. Under sterile conditions, 100 µL of bacterial suspension was picked out with an inoculation rod and inoculated into the biochemically encoded identification tube CYZ-15E for the Enterobacteriaceae kit (Binhe Microorganism Reagent, Hangzhou, China) and sealed. Based on the instructions of the kit, the inoculated identification tubes were incubated at 37 °C for 18–24 h and the results were observed. The physiological and biochemical characteristics of the obtained strains were compared with the results of the standard strains in the Manual of Identification of Common Bacterial Systems to preliminarily determine the strain species.
2.4. Bacterial 16S rRNA Amplification and Sequencing
Bacterial 16S rRNA gene universal primers were used [33] (27F, 5′-AGAGTTTGATCCTGGCTCAG-3′, and 1492R, 5′-CGGCTACCTTGTTACGACTT-3′) for PCR identification. The PCR reaction system was as follows: 12.5 µL of 2× Taq PCR Master Mix (Tiangen, Beijing, China), the upper and lower primers (10 µmol/L) comprised 1 µL each and were supplemented with distilled water up to a total volume of 25 µL, and a small quantity of bacteria was used as the PCR template. The PCR amplification protocol included an initial denaturation step at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min. A final extension step occurred at 72 °C for 5 min. The PCR products were identified by agarose gel electrophoresis and recovered, and sequencing was carried out by Beijing Tsingke Biotechnology Co., Ltd., in Beijing, China.
2.5. Sequence Alignment Analysis and Construction of Phylogenetic Tree
For sequence alignment analysis and phylogenetic tree construction purposes, the acquired gene sequences underwent homology comparison via BLAST search against related sequences available in NCBI GenBank (MegAlign 7.0.26). Subsequently, MEGA11 software facilitated the construction of a phylogenetic tree aimed at analyzing and comparing the genetic relationships among P. aeruginosa strains derived from various sources.
2.6. Antibiotic Susceptibility Testing
A drug susceptibility test was performed according to the Kirby–Bauer disk diffusion method. A total of 100 µL of the purified P. aeruginosa bacteria suspension was spread on the ordinary agar medium, and the drug-sensitive tablets of 24 antibiotics were pasted on the medium. After 18 h of incubation at 37 °C, the inhibition zone diameter of each antibiotic was measured.
2.7. Bacterial Artificial Regression Infection Test
The isolated and purified strain of P. aeruginosa (PA/CM-101101) was inoculated onto a slant of nutrient agar and incubated at 28 °C for a duration of 18–24 h. Subsequently, the bacterial colony was washed with sterile saline. A total of ten healthy mice aged 9 to 10 weeks were randomly and evenly allocated into an experimental group and a blank control group. Five mice in the experimental group received an intraperitoneal injection of 0.1 mL of bacteria suspension that had been diluted using a 10-fold dilution method from an initial concentration ranging from 1 × 109 to 1 × 105 CFU/mL in sterile phosphate-buffered saline (PBS), while the control group consisted of five mice that were administered the same volume of sterile PBS buffer via intraperitoneal injection. After injecting the bacteria, the disease symptoms and the number of dead mice were observed and recorded every morning and evening. The mice underwent normal feeding management for 7 days.
3. Results
3.1. Morphological Characteristics of Bacteria
The anal swab was cultured on LB agar medium, revealing colony growth in only one morphological form. The isolated bacteria exhibited smooth, slightly elevated colonies after being cultured on LB agar medium for 24 h, and these colonies had neat and wavy edges (Figure 1A). When transferred to acetamide agar at 37 °C for an additional 24 h, the colonies appeared flat with irregular edges surrounded by slightly pink coloration; concurrently, the color change in the medium resulted in rose-red pigmentation (Figure 1B). Culturing in LB broth for another day led to turbidity accompanied by yellowish-green coloration (Figure 1C). Gram staining results indicated negative reactions characterized by blunt ends without spore formation (Figure 1D).
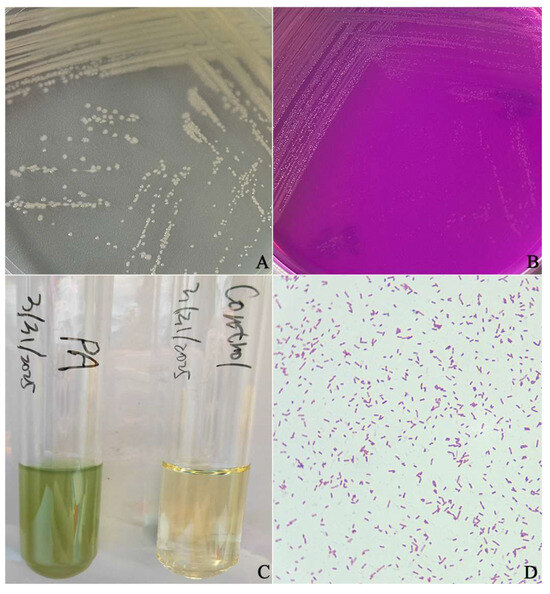
Figure 1.
The morphology of P. aeruginosa isolated from a cynomolgus monkey. (A) Morphological observations of the P. aeruginosa isolate on an LB agar plate. (B) Morphological characteristics of the isolate on acetamide agar. (C) Morphological observations of the P. aeruginosa isolate in LB broth. (D) Morphological observations of the P. aeruginosa and Gram staining microscopy results.
3.2. Results of Bacterial Biochemical Identification
Physiological and biochemical identification revealed that the isolated strain could catabolize glucose and urea, but could not metabolize mannitol or raffinose. The methyl red test, Voges–Proskauer test, and indole test yielded negative results; no hydrogen sulfide production was observed. However, both the motility and the citrate tests revealed positive results, aligning with the known biochemical characteristics of P. aeruginosa (Table 1).

Table 1.
Physiological and biochemical characteristics of strain PA/CM-101101.
3.3. 16S rRNA Gene Sequencing, Homology Comparison, and Phylogenetic Analysis
The amplified PCR products were subjected to analysis via 1.5% agarose gel electrophoresis to isolate the target gene band of the anticipated size (1000–2000 bp) (Figure 2). The sequence of the 16S rRNA gene obtained from the P. aeruginosa strain isolated from the cynomolgus monkey was determined to be 1396 bp in length (Supplementary Materials).
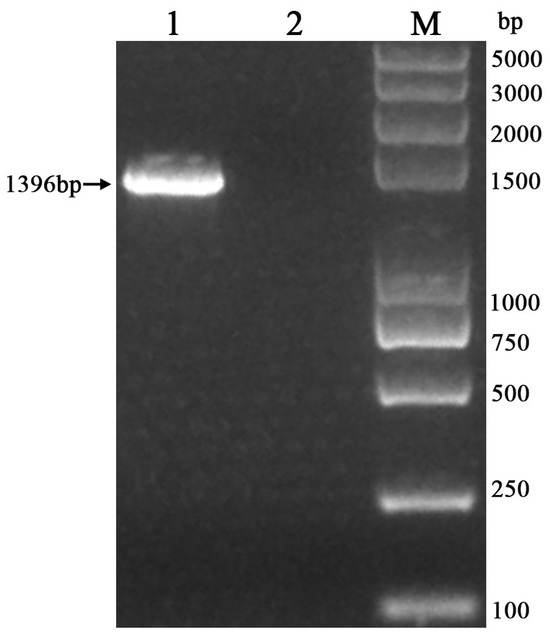
Figure 2.
Identification of PCR-amplified P. aeruginosa with 16S rRNA gene in a cynomolgus monkey (M: DL-2K marker; 1: sample; 2: negative control).
The sequencing of the obtained 16S rRNA was conducted using the Sanger dideoxy method, and the results have been reported in the NCBI (GenBank Accession No.: PV608028). Sequence alignment analysis was performed using the Clustalw method, and the results indicated a coincidence rate of over 95% with P. aeruginosa, thereby confirming that the isolate was indeed P. aeruginosa. The 16S rRNA gene sequence analysis showed that the sequence identity of strain PA/CM-101101 shared more than 98.4% similarity with 11 strains of P. aeruginosa from various sources in GenBank, showing maximum homology with elephant (Elephantidae) (GenBank Accession No.: FN433043), parrot fish (Amphilophus) (GenBank Accession No.: HM224410), mink (Mustelidae) (GenBank Accession No.: MG547342), human (Homo sapiens) (GenBank Accession No.: MH378333), and chicken (Gallus domesticus) (GenBank Accession No.: OM021866) at a similarity rate of 99.9% (Figure 3).
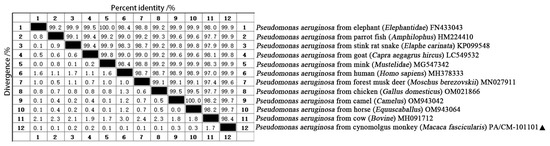
Figure 3.
Comparison of similarity between P. aeruginosa of a cynomolgus monkey (Macaca fascicularis) (PA/CM-101101) and other strains. “▲” is the isolated strain in this study.
The Clustalw method in MegAlign (version 11.1.0.59) software was used to align the sequences and construct the phylogenetic evolutionary tree. The results indicate that bacterial strain PA/CM-101101 isolated from the anal secretion of the cynomolgus monkey formed a distinct branch alongside human (Homo sapiens) (MH378333) within the phylogeny of P. aeruginosa (Figure 4). Furthermore, it clustered together with strains from forest musk deer (Moschus berezovskii) (MN027911), parrot fish (Amphilophus) (HM224410), chicken (Gallus domesticus) (OM021866), camel (Camelus) (OM943042), horse (Equuscaballus) (OM943064), elephant (Elephantidae) (FN433043), stink rat snake (Elaphe carinata) (KP099548), mink (Mustelidae) (MG547342), and goat (Capra aegagrus hircus) (LC549532) strains within P. aeruginosa phylogeny, while forming a separate branch from that of cow (Bovine) (MH091712). Therefore, we conclude that the pathogenic bacterium isolated from the cynomolgus monkey (Macaca fascicularis) was indeed identified as P. aeruginosa based on 16S rRNA gene sequence analyses. It was most closely related to the P. aeruginosa from human (Homo sapiens) (MH378333), while it had a distant relationship with P. aeruginosa of the cow (Bovine) (MH091712).
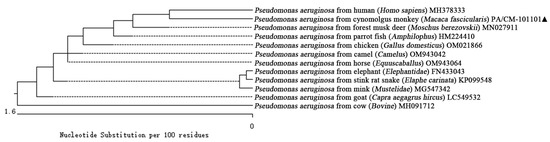
Figure 4.
Phylogenetic tree of 16S rRNA sequences among different reported strains of P. aeruginosa, including cynomolgus monkey (Macaca fascicularis) (PA/CM-101101). “▲” is the isolated strain in this study.
3.4. Antimicrobial Susceptibility Test Results of the Isolated Strain
To evaluate antibiotic sensitivity profiles for strain PA/CM-101101 against 24 different chemical agents, we employed the Kirby–Bauer disc diffusion methodology. The results indicate that this isolated strain exhibited sensitivity towards amikacin, azithromycin, cefoperazone, ceftazidime, ceftriaxone, ciprofloxacin, gentamicin, imipenem, levofloxacin, meropenem, norfloxacin, ofloxacin, and polymyxin B; moderate sensitivity was observed towards amoxicillin, bacitracin, cotrimoxazole, furazolidone, lincomycin, rifampicin; and resistant sensitivity was observed towards ampicillin, cefadroxil, cefazolin, erythromycin, and vancomycin (Table 2).

Table 2.
Drug sensitivity analysis results of isolated strain.
3.5. Results of Animal Pathogenicity Tests
Four hours after the injection of the bacterial liquid, the mice in the experimental group began to show symptoms such as listlessness, slow activity, loss of appetite, and diarrhea, and all died within 48 h. Culture characteristics and morphological features consistent with strain PA/CM-101101 were isolated from the dead mice. All the mice in the control group were healthy for 7 days. No gross lesions were found in the internal organs during post-mortem examination.
4. Discussion
P. aeruginosa is an opportunistic pathogen in clinical practice that can cause healthcare-associated infections in both humans and animals [34], and can even lead to mortality [35]. It seriously threatens the health of humans and animals. However, recent reports on P. aeruginosa have primarily focused on nosocomial infections as well as infections in livestock and poultry. So far, P. aeruginosa has not been reported to cause intestinal infection in cynomolgus monkey.
Bacterial diseases spread rapidly and have a high mortality rate, so it is necessary to isolate and identify the pathogenic bacteria in order to study their effects on animal health [36]. Utilizing 16S rRNA molecular markers for bacterial taxonomic identification proves highly effective [37]. In this study, bacterial strain PA/CM-101101 was isolated from the feces of a diarrheal cynomolgus monkey. The morphological characteristics and biochemical identification were consistent with those typical of P. aeruginosa. The isolated strain showed high homology with P. aeruginosa from different species, and was the closest to the P. aeruginosa strain from Homo sapiens (MH378333). Further studies are needed to determine whether the genetic characteristics of P. aeruginosa are related to different species and sources. Pathogenicity tests conducted on mice showed symptoms such as listlessness, slow activity, loss of appetite, and diarrhea, culminating in mortality within 48 h post-infection, which confirmed that P. aeruginosa was the pathogen causing diarrhea in the cynomolgus monkey.
Currently, antibiotic therapy remains the primary treatment option due to its widespread application and rapid efficacy; however, many bacteria have developed resistance against it [38]. Long-term overuse of antibiotics not only results in residues but also fosters antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) [39]. It has been reported that P. aeruginosa is highly resistant to antibiotics [20,40,41], particularly concerning carbapenem resistance, which poses life-threatening risks that severely impact public health safety [42]. Consequently, in order to develop a more rational use of drugs for P. aeruginosa, there is an ongoing effort towards standardizing rational use protocols for antibiotics aimed at minimizing indiscriminate usage while reducing environmental pollution and animal casualties. In this study, we conducted a drug susceptibility test on P. aeruginosa, and the results indicate that the isolates exhibited good sensitivity to most antibiotics. We hypothesize that this finding may be related to the frequency of daily antibiotic use at this location. It has certain guiding significance for the breeding and management of cynomolgus monkeys in the future. In the aquaculture industry, contaminated water, cages, feed tanks, etc. are reservoirs of P. aeruginosa and are a source of outbreaks of P. aeruginosa [20,21]. Therefore, maintaining clean environmental hygiene during the feeding and management of experimental monkeys plays an important role in the prevention of P. aeruginosa.
5. Conclusions
In conclusion, P. aeruginosa was identified as a pathogen in the feces of a diarrheal cynomolgus monkey through morphological analysis, biochemical tests, 16S rRNA gene sequencing, and animal pathogenicity experiments. The isolated strain showed high homology with P. aeruginosa from different species, and was the closest to the P. aeruginosa strain from Homo sapiens (MH378333). The isolated strains demonstrated sensitivity to 13 antibiotics, including imipenem and meropenem. This study provides an important scientific foundation for isolating and identifying P. aeruginosa as well as for preventing and treating bacterial diseases in experimental monkeys.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci12070636/s1.
Author Contributions
H.L. and H.W. designed the experiments and wrote the paper. H.L. and Z.Q. performed the experiments and prepared the figures. H.W. and Y.Y. reviewed drafts of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Natural Science Foundation of Yunnan Province (202102AA100053, 202001BC070001).
Institutional Review Board Statement
This study was reviewed and approved by the ethics committee of the Institutional Animal Care and Use Committee of Kunming University of Science and Technology, approval code KUST202404003, date 23 March 2024.
Informed Consent Statement
The cynomolgus monkey’s owner provided written informed consent to participate in this study.
Data Availability Statement
All data in this study are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Belmonte, J.C.I.; Callaway, E.M.; Churchland, P.; Caddick, S.J.; Feng, G.; Homanics, G.E.; Lee, K.F.; Leopold, D.A.; Miller, C.T.; Mitchell, J.F.; et al. Brains, Genes, and Primates. Neuron 2015, 86, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Chen, Z.G.; Ning, Q.; Zong, F.L.; Wang, H. Isolation and Identification of Morganella morganii from Rhesus Monkey (Macaca mulatta) in China. Vet. Sci. 2024, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Maaskant, A.; Voermans, B.; Levin, E.; de Goffau, M.C.; Plomp, N.; Schuren, F.; Remarque, E.J.; Smits, A.; Langermans, J.A.M.; Bakker, J.; et al. Microbiome signature suggestive of lactose-intolerance in rhesus macaques (Macaca mulatta) with intermittent chronic diarrhea. Anim. Microbiome 2024, 6, 53. [Google Scholar] [CrossRef]
- Maaskant, A.; Blees, N.R.; Smits, A.; Corbee, R.J.; Bakker, J.; Langermans, J.A.M.; Remarque, E.J. Evaluation of commercial diets on fecal consistency and defecation frequency in rhesus macaques (Macaca mulatta) with chronic intermittent idiopathic diarrhea. Lab. Anim. Res. 2025, 41, 15. [Google Scholar] [CrossRef]
- Keita, M.B.; Hamad, I.; Bittar, F. Looking in apes as a source of human pathogens. Microb Pathog. 2014, 77, 149–154. [Google Scholar] [CrossRef]
- Rhoades, N.; Barr, T.; Hendrickson, S.; Prongay, K.; Haertel, A.; Gill, L.; Garzel, L.; Whiteson, K.; Slifka, M. Maturation of the infant rhesus macaque gut microbiome and its role in the development of diarrheal disease. Genome Biol. 2019, 20, 173. [Google Scholar] [CrossRef]
- Palleroni, N.J. The Pseudomonas Story. Environ. Microbiol. 2010, 12, 1377–1383. [Google Scholar] [CrossRef]
- Fred, C.; Nicolau, D.P.; Gill, C.M. Carbapenemaseproducing Pseudomonas aeruginosa—An emerging challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar]
- Chung, J.; Eisha, S.; Park, S.; Morris, A.J.; Martin, I. How three self-secreted biofilm exopolysaccharides of Pseudomonas aeruginosa, psl, pel, and alginate, can each be exploited for antibiotic adjuvant effects in cystic fibrosis lung infection. Int. J. Mol. Sci. 2023, 24, 8709. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, L.; Yang, Y.T.; Yin, Z.H.; Guo, B.L. Biodegradable gelatin/silver nanoparticle composite cryogel with excellent antibacterial and antibiofilm activity and hemostasis for Pseudomonas aeruginosa-infected burn wound healing. J. Colloid Interface Sci. 2022, 608, 2278–2289. [Google Scholar] [CrossRef]
- Thuenauer, R.; Landi, A.; Trefzer, A.; Altmann, S.; Wehrum, S.; Eierhoff, T.; Diedrich, B.; Dengjel, J.; Nyström, A.; Imberty, A.; et al. The Pseudomonas aeruginosa lectin LecB causes integrin internalization and inhibits epithelial wound healing. mBio 2020, 11, e03260-19. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Stapleton, F.; Summers, S.; Rice, S.A.; Willcox, M.D.P. Antibiotic resistance characteristics of Pseudomonas aeruginosa isolated from keratitis in Australia and India. Antibiotics 2020, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.; Baishya, J.; Wakeman, C.A. Pseudomonas aeruginosa polymicrobial interactions during lung infection. Curr. Opin. Microbiol. 2020, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alemán, S.; Bustamante, A.E.; Jimenez-Valdes, R.J.; González, G.M.; Sánchez-González, A. Pseudomonas aeruginosa isolates from cystic fibrosis patients induce neutrophil extracellular traps with different morphologies that could correlate with their disease severity. Int. J. Med. Microbiol. 2020, 310, 151451. [Google Scholar] [CrossRef]
- Plebani, R.; Potla, R.; Soong, M.; Bai, H.Q.; Izadifar, Z.; Jiang, A.; Travis, R.N.; Belgur, C.; Dinis, A.; Cartwright, M.J.; et al. Modeling pulmonary cystic fibrosis in a human lung airway-on-a-chip. J. Cyst. Fibros. 2022, 21, 606–615. [Google Scholar] [CrossRef]
- Gajdács, M. Carbapenem-Resistant but Cephalosporin-Susceptible Pseudomonas aeruginosa in Urinary Tract Infections: Opportunity for Colistin Sparing. Antibiotics 2020, 9, 153. [Google Scholar] [CrossRef]
- McVey, M.J.; Maishan, M.; Foley, A.; Turki, R.; Roach, E.J.; Deschler, R.; Weidenfeld, S.; Goldenberg, N.M.; Khursigara, C.M.; Kuebler, W.M. Pseudomonas aeruginosa membrane vesicles cause endothelial barrier failure and lung injury. Eur. Respir. J. 2022, 59, 2101500. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Polverino, E.; Crichton, M.L.; Ringshausen, F.C.; De Soyza, A.; Vendrell, M.; Burgel, P.R.; Haworth, C.S.; Loebinger, M.R.; Dimakou, K.; et al. Bronchiectasis in Europe data on disease characteristics from the European Bronchiectasis registry (EMBARC). Lancet Respir. Med. 2023, 11, 637–649. [Google Scholar] [CrossRef]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-Negative Bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef]
- Rodrigues, Y.C.; Furlaneto, I.P.; Maciel, A.H.P.; Quaresma, A.J.P.G.; De Matos, E.C.O.; Conceiçao, M.L.; Vieira, M.C.D.; Brabo, G.L.D.; Sarges, E.D.N.F.; Lima, L.N.G.C.; et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS ONE 2020, 15, e0238741. [Google Scholar] [CrossRef]
- Bonnet, R.; Sampaio, J.L.M.; Labia, R.; De Champs, C.; Sirot, D.; Chanal, C.; Sirot, J. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 2000, 44, 1936–1942. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Bour, M.; Châtre, P.; Madec, J.-Y.; Plésiat, P.; Jeannot, K. Resistance of animal strains of Pseudomonas aeruginosa to carbapenems. Front. Microbiol. 2017, 8, 1847. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.J.; Wilson, D.J. Pseudomonas aeruginosa mastitis in two goats associated with an essential oil-based teat dip. J. Vet. Diagn. Investig. 2016, 28, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Darwich, L.; Seminati, C.; Burballa, A.; Nieto, A.; Durán, I.; Tarradas, N.; Molina-López, R.A. Antimicrobial susceptibility of bacterial isolates from urinary tract infections in companion animals in Spain. Vet. Rec. 2021, 188, e60. [Google Scholar] [CrossRef]
- Rubin, J.; Walker, R.D.; Blickenstaff, K.; Bodeis-Jones, S.; Zhao, S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomon asaeruginosa isolated from canine infections. Vet. Microbiol. 2008, 131, 164–172. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. Fems. Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Zhao, K.L.; Ma, J.N.; Wang, X.R.; Guo, Y.D.; Yue, B.S.; Chu, Y.W. Population divergence of Pseudomonas aeruginosa can lead to the coexistencewith Escherichia coli in animal suppurative lesions. Vet. Microbiol. 2019, 231, 169–176. [Google Scholar] [CrossRef]
- Zhu, Q.; Peng, H.; Li, H.; Yang, L.Z.; Zhang, B.S.; Zhu, J.; Guo, W.L.; Wang, N.; Jiang, S.J.; Xie, Z.J. Serotypes and virulence genes of Pseudomonas aeruginosa isolated from mink and its pathogenicity in mink. Microb. Pathog. 2020, 139, 103904. [Google Scholar]
- Gierløff, B. Pseudomonas aeruginosa. IV. IV. Pyocine typing of strains isolated from the blue fox (Alopex lagopus), mink (Mustela vison), and dog (Canis familiaris) and from their environment. Nord. Vet. Med. 1980, 32, 147–160. [Google Scholar]
- Cheung, A.T.; Moss, R.B.; Leong, A.B.; Novick, W.J. Chronic Pseudomonas aeruginosa endobronchitis in rhesus monkeys: I. Effects of pentoxifylline on neutrophil influx. J. Med. Primatol. 1992, 21, 357–362. [Google Scholar] [CrossRef]
- Lausen, N.C.; Richter, A.G.; Lage, A.L. Pseudomonas aeruginosa infection in squirrel monkeys. JAVMA-J. Am. Vet. Med. A 1986, 189, 1216–1218. [Google Scholar] [CrossRef]
- Stills, H.F.; Bond, M.G.; Bullock, B.C. Bacterial myocarditis in African green monkeys (Cercopithecus aethiops). Vet. Pathol. 1979, 16, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, M.; Thangavelu, R.; Pushpakanth, P.; Muthubharathi, K.; Ramesh, R.; Selvarajan, R.; Uma, S. First report of rhizome rot of banana caused by Klebsiella variicola in India. Plant Dis. 2021, 105, 2011. [Google Scholar] [CrossRef]
- Veetilvalappil, V.V.; Manuel, A.; Aranjani, J.M.; Tawale, R.; Koteshwara, A. Pathogenic arsenal of Pseudomonas aeruginosa: An update on virulence factors. Future Microbiol. 2022, 17, 465–481. [Google Scholar] [CrossRef]
- Wang, Z.W.; Tian, W.T.; Sun, S.Y.; Chen, X.; Wang, H.F. Genomic and proteomic analysis of Pseudomonas aeruginosa isolated from industrial wastewater to assess its resistance to antibiotics. Separations 2023, 10, 549. [Google Scholar] [CrossRef]
- Wamala, P.S.; Mugimba, K.K.; Mutoloki, S.; Evensen, Ø.; Mdegela, R.; Byarugaba, D.K. Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fish. Aquat. Sci. 2018, 21, 6. [Google Scholar] [CrossRef]
- Quang, T.; Diem-Trang, P.; Vinhthuy, P. Using 16S rRNA gene asmarker to detect unknown bacteria in microbial communities. BMC Bioinform. 2017, 18, 499. [Google Scholar]
- Sabnis, A.; Hagart, K.L.H.; Klöckner, A.; Becce, M.; Evans, L.E.; Furniss, R.C.D.; Mavridou, D.A.I.; Murphy, R.; Stevens, M.M.; Davies, J.C.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife 2021, 10, e65836. [Google Scholar] [CrossRef]
- Yuan, X.; Lv, Z.Q.; Zhang, Z.Y.; Han, Y.; Liu, Z.Q.; Zhang, H.J. A review of antibiotics, antibiotic resistant bacteria, and resistance genes in aquaculture: Occurrence, contamination, and transmission. Toxics 2023, 11, 420. [Google Scholar] [CrossRef]
- Roulová, N.; Mot’ková, P.; Brozková, I.; Pejchalová, M. Antibiotic resistance of Pseudomonas aeruginosa isolated from hospital wastewater in the Czech Republic. J. Water Health 2022, 20, 692–701. [Google Scholar] [CrossRef]
- Shariati, A.; Azimi, T.; Ardebili, A.; Chirani, A.S.; Bahramian, A.; Pormohammad, A.; Sadredinamin, M.; Erfanimanesh, S.; Bostanghadiri, N.; Shams, S.; et al. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microb. New Infect. 2017, 20, 75–80. [Google Scholar] [CrossRef]
- Papanikolopoulou, A.; Gargalianos-Kakolyris, P.; Stoupis, A.; Moussas, N.; Pangalis, A.; Theodoridou, K.; Chronopoulou, G.; Pantazis, N.; Kantzanou, M.; Maltezou, H.C.; et al. Carbapenem-resistant Pseudomonas aeruginosa bacteremia, through a six-year infection control program in a hospital. Microorganisms 2023, 11, 1315. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).