Companion Animals as Reservoirs of Multidrug Resistance—A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Presentation
2.2. Nucleic Acid Extraction and Whole Genome Sequencing
2.3. Sequencing Analysis
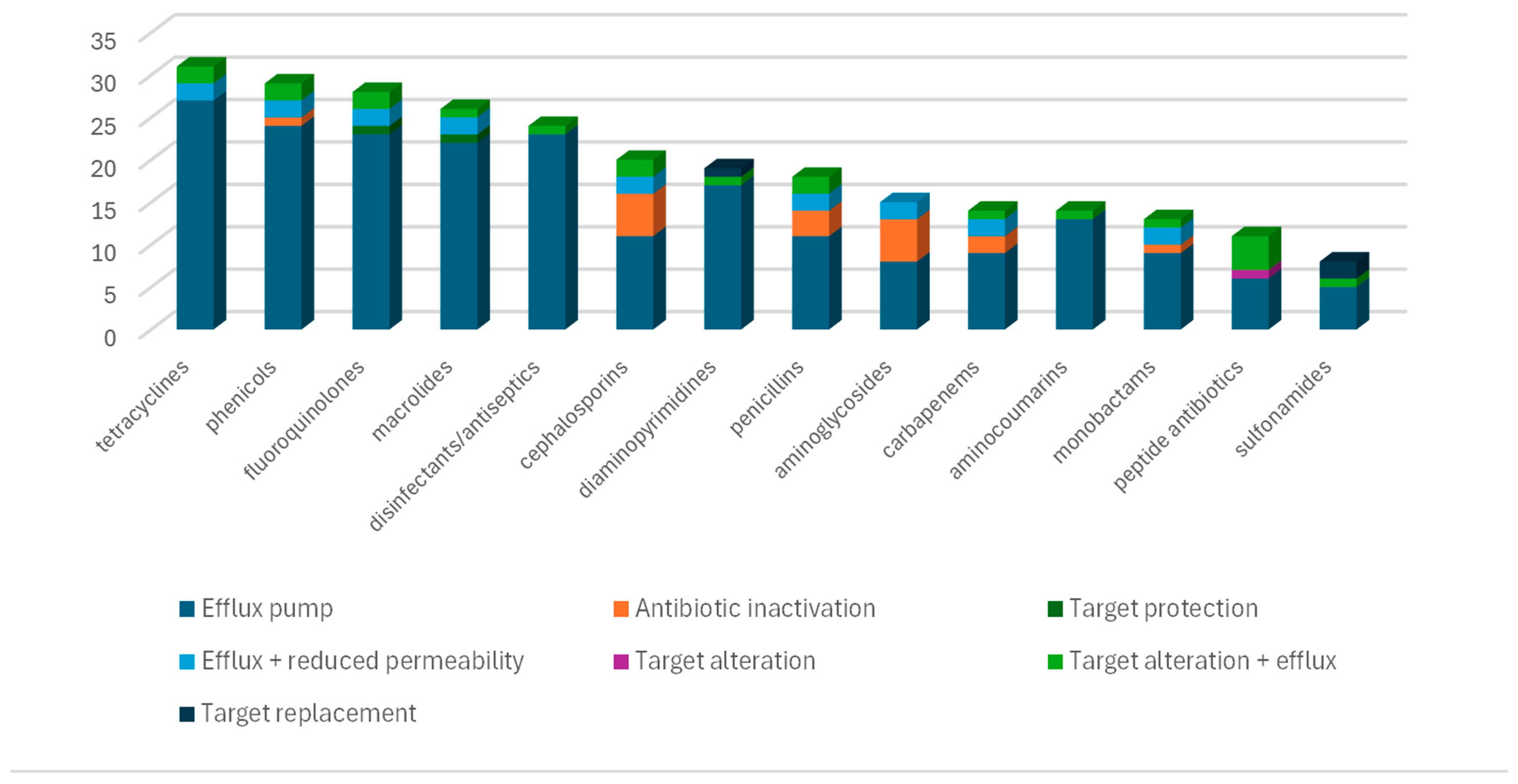
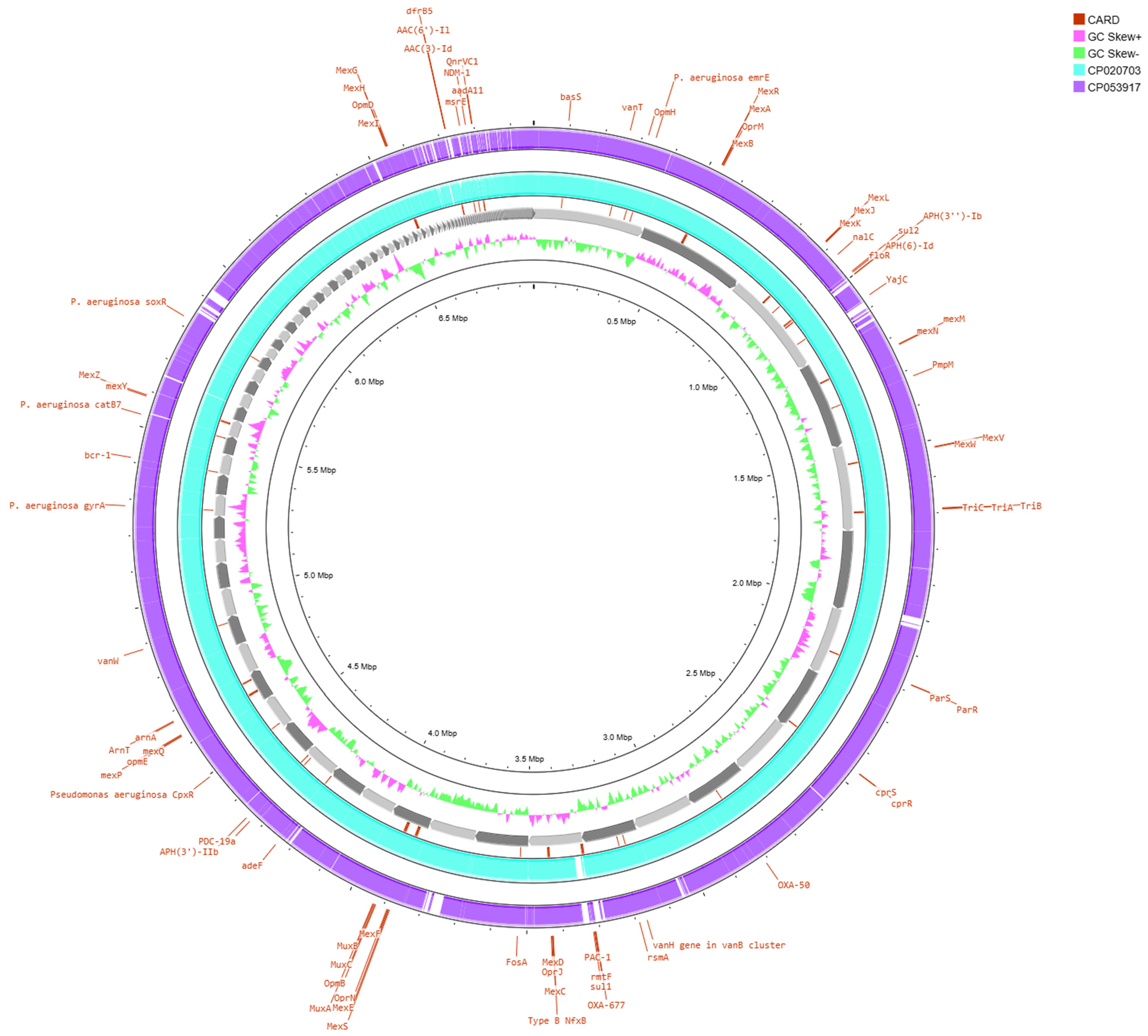
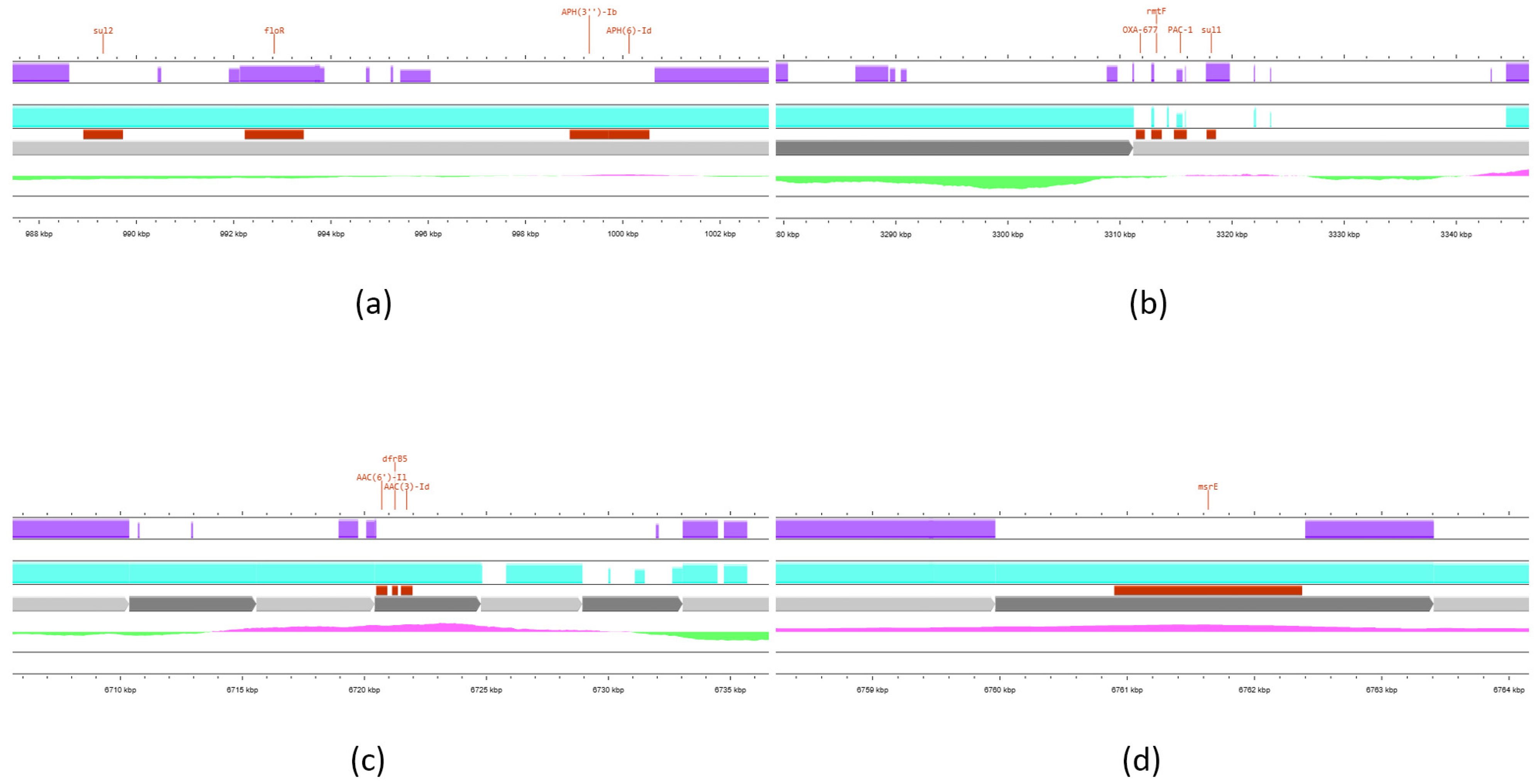
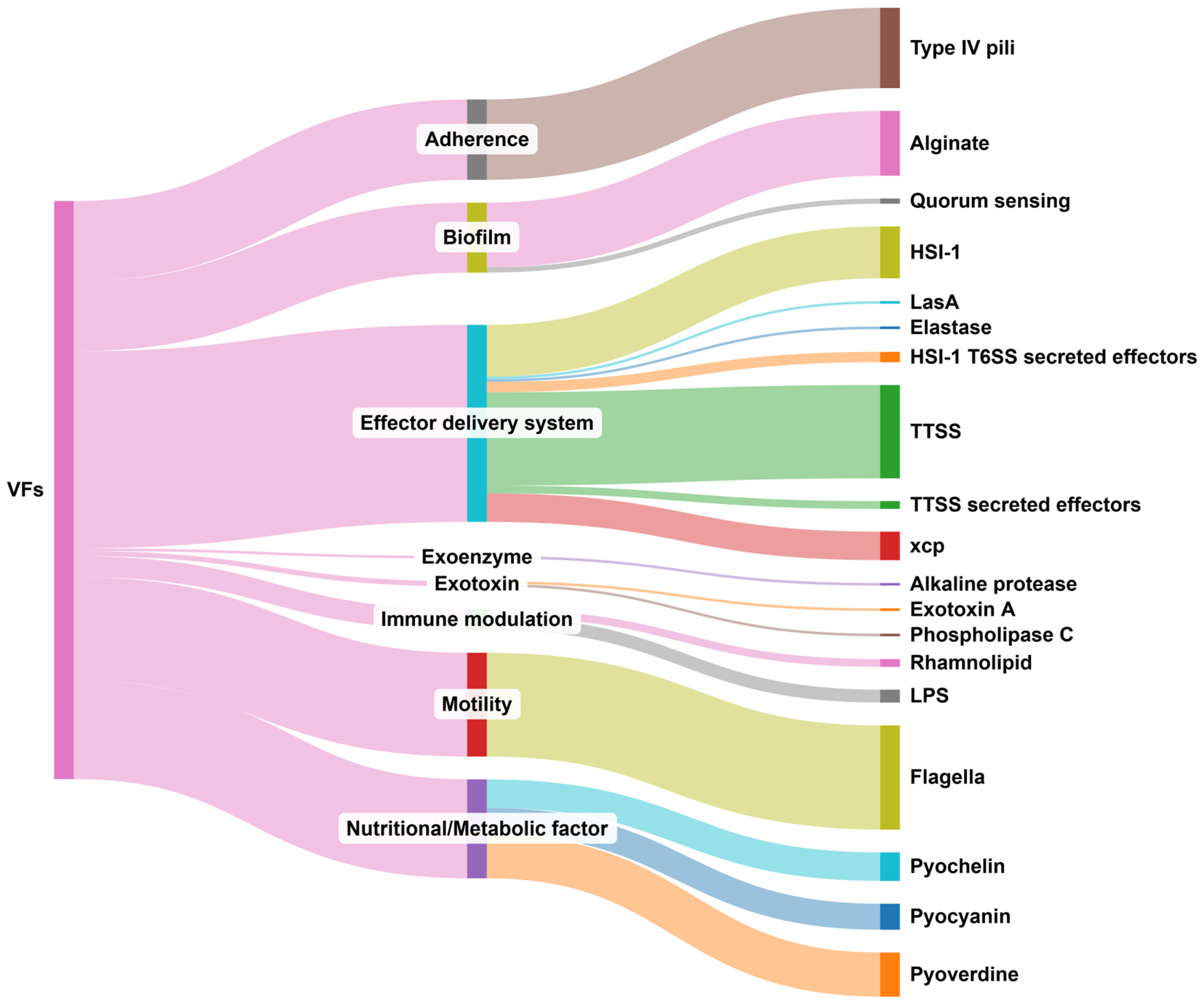
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Bacterial Priority Pathogens List 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance, 1st ed.; World Health Organization: Geneva, Switzerland, 2024; ISBN 978-92-4-009346-1. [Google Scholar]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, T.; Hébraud, M.; Dapkevicius, M.L.N.E.; Maltez, L.; Pereira, J.E.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 12892. [Google Scholar] [CrossRef]
- Breidenstein, E.B.M.; De La Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-Producing Pseudomonas aeruginosa—An Emerging Challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Hong, D.J.; Bae, I.K.; Jang, I.-H.; Jeong, S.H.; Kang, H.-K.; Lee, K. Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Infect. Chemother. 2015, 47, 81. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Verdial, C.; Serrano, I.; Tavares, L.; Gil, S.; Oliveira, M. Mechanisms of Antibiotic and Biocide Resistance That Contribute to Pseudomonas aeruginosa Persistence in the Hospital Environment. Biomedicines 2023, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Pottenger, S.; Timofte, D.; Moore, M.; Wright, L.; Kukavica-Ibrulj, I.; Jeukens, J.; Levesque, R.C.; Freschi, L.; Pinchbeck, G.L.; et al. Reservoirs of Resistance: Polymyxin Resistance in Veterinary-associated Companion Animal Isolates of Pseudomonas aeruginosa. Vet. Rec. 2019, 185, 206. [Google Scholar] [CrossRef]
- Pereira, P.F.V.; Reway, A.P.; Félix, A.; Beutemmüller, E.A.; Pretto-Giordano, L.G.; Alfieri, A.A.; Lisbôa, J.A.N.; Müller, E.E. Mammary Gland Health of Santa Inês Ewes at the Drying and Puerperium and Evaluation of a Dry-off Terapy with Gentamicin. Pesq. Vet. Bras. 2018, 38, 2194–2200. [Google Scholar] [CrossRef]
- Darwich, L.; Seminati, C.; Burballa, A.; Nieto, A.; Durán, I.; Tarradas, N.; Molina-López, R.A. Antimicrobial Susceptibility of Bacterial Isolates from Urinary Tract Infections in Companion Animals in Spain. Vet. Rec. 2021, 188, e60. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Thomson, N.; Weill, F.-X.; Holt, K.E. Genomic Insights into the Emergence and Spread of Antimicrobial-Resistant Bacterial Pathogens. Science 2018, 360, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef]
- Blanc, D.S.; Magalhães, B.; Koenig, I.; Senn, L.; Grandbastien, B. Comparison of Whole Genome (Wg-) and Core Genome (Cg-) MLST (BioNumerics™) Versus SNP Variant Calling for Epidemiological Investigation of Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 1729. [Google Scholar] [CrossRef] [PubMed]
- Pelegrin, A.C.; Palmieri, M.; Mirande, C.; Oliver, A.; Moons, P.; Goossens, H.; van Belkum, A. Pseudomonas aeruginosa: A Clinical and Genomics Update. FEMS Microbiol. Rev. 2021, 45, fuab026. [Google Scholar] [CrossRef]
- Tsilipounidaki, K.; Gkountinoudis, C.-G.; Florou, Z.; Fthenakis, G.C.; Miriagou, V.; Petinaki, E. First Detection and Molecular Characterization of Pseudomonas aeruginosa blaNDM-1 ST308 in Greece. Microorganisms 2023, 11, 2159. [Google Scholar] [CrossRef]
- Pappa, O.; Louka, C.; Karadimas, K.; Maikousi, E.; Tzoukmani, A.; Polemis, M.; Panopoulou, A.-D.; Daniil, I.; Chryssou, S.; Mellou, K.; et al. Emergence of NDM-1-Producing Pseudomonas aeruginosa Nosocomial Isolates in Attica Region of Greece. Microorganisms 2024, 12, 1753. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Lubbers, B.V.; Diaz-Campos, D.V.; Schwarz, S.; Sweeney, M.T.; Burbick, C.R.; Govendir, M.; Harris, B.; Holliday, N.M.; Hayes, J.; Lawhon, S.D.; et al. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; VET01S ed7; CLSI: Malvern, PA, USA, 2024. [Google Scholar]
- Lewis, J.S.; Mathers, A.J.; Bobenchik, A.M.; Bryson, A.M.; Campeau, S.; Cullen, S.K.; Dingle, T.; Esparza, G.; Humphires, R.M.; Kirn, T.J.; et al. Performance Standards for Antimicrobial Susceptibility Testing; M100ed35e; CLSI: Malvern, PA, USA, 2025. [Google Scholar]
- The Galaxy Community; Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; et al. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 February 2025).
- Joshi, N.A.; Fass, J.N. Sickle: A Windowed Adaptive Trimming Tool for FASTQ Files Using Quality. 2011. Available online: https://Github.Com/Najoshi/Sickle (accessed on 10 February 2025).
- Prjibelski, A.D.; Vasilinetc, I.; Bankevich, A.; Gurevich, A.; Krivosheeva, T.; Nurk, S.; Pham, S.; Korobeynikov, A.; Lapidus, A.; Pevzner, P.A. ExSPAnder: A Universal Repeat Resolver for DNA Fragment Assembly. Bioinformatics 2014, 30, i293–i301. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An Algorithm for Hybrid Assembly of Short and Long Reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Vasilinetc, I.; Prjibelski, A.D.; Gurevich, A.; Korobeynikov, A.; Pevzner, P.A. Assembling Short Reads from Jumping Libraries with Large Insert Sizes. Bioinformatics 2015, 31, 3262–3268. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Tseemann. MLST: Scan Contig Files against PubMLST Typing Schemes. 2016. Available online: https://github.com/Tseemann/Mlst (accessed on 10 February 2025).
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella Enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antiobiotic Resistance Genes. 2016. Available online: https://github.com/Tseemann/Abricate (accessed on 10 February 2025).
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification: Find out More about Bakta, the Motivation, Challenges and Applications, Here. Microb. Genom. 2021, 7, 685. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- Chew, K.L.; Octavia, S.; Ng, O.T.; Marimuthu, K.; Venkatachalam, I.; Cheng, B.; Lin, R.T.P.; Teo, J.W.P. Challenge of Drug Resistance in Pseudomonas aeruginosa: Clonal Spread of NDM-1-Positive ST-308 within a Tertiary Hospital. J. Antimicrob. Chemother. 2019, 74, 2220–2224. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Song, W.; Park, M.-J.; Jeong, S.; Lee, N.; Jeong, S.H. Molecular Characterization of the First Emerged NDM-1-Producing Pseudomonas aeruginosa Isolates in South Korea. Microb. Drug Resist. 2021, 27, 1063–1070. [Google Scholar] [CrossRef]
- VFDB—Virulence Factors of Bacterial Pathogens (RRID:SCR_007969). Available online: https://www.mgc.ac.cn/VFs/main.htm (accessed on 16 February 2025).
- Abdullahi, I.N.; Mejri, S.; Okwume, C.C.; Lawal, N.A.; Olusegun, O.A.; Sallem, R.B.; Slama, K.B. Global Epidemiology of High Priority and Pandemic Pseudomonas aeruginosa in Pets, Livestock, Wild, and Aquatic Animals: A Systematic Review and Meta-Analysis. Lett. Appl. Microbiol. 2025, 78, ovaf028. [Google Scholar] [CrossRef]
- Murphy, K.M. A Review of Techniques for the Investigation of Otitis Externa and Otitis Media. Clin. Tech. Small Anim. Pract. 2001, 16, 236–241. [Google Scholar] [CrossRef]
- Rosser, E.J. Causes of Otitis Externa. Vet. Clin. N. Am. Small Anim. Pract. 2004, 34, 459–468. [Google Scholar] [CrossRef]
- Khan, I.; Miskeen, S.; Khalil, A.T.; Phull, A.-R.; Kim, S.J.; Oh, D.-H. Foodborne Pathogens: Staphylococcus Aureus and Listeria Monocytogenes an Unsolved Problem of the Food Industry. Pak. J. Nutr. 2016, 15, 505–514. [Google Scholar] [CrossRef]
- Jeanvoine, A.; Meunier, A.; Puja, H.; Bertrand, X.; Valot, B.; Hocquet, D. Contamination of a Hospital Plumbing System by Persister Cells of a Copper-Tolerant High-Risk Clone of Pseudomonas aeruginosa. Water Res. 2019, 157, 579–586. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa Epidemic High-Risk Clones and Their Association with Horizontally-Acquired β-Lactamases: 2020 Update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.M.; Rajasekaram, G.; Puthucheary, S.D.; Chua, K.H. Detection of VIM-2-, IMP-1- and NDM-1-Producing Multidrug-Resistant Pseudomonas aeruginosa in Malaysia. J. Glob. Antimicrob. Resist. 2018, 13, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Schwarz, S.; Zhang, R.; Lei, L.; Liu, X.; Lin, D.; Shen, J. IMP-45-Producing Multidrug-Resistant Pseudomonas aeruginosa of Canine Origin. J. Antimicrob. Chemother. 2014, 69, 2579–2581. [Google Scholar] [CrossRef]
- Division of Epidemiology, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, India; Pruthvishree, B.S.; Vinodh Kumar, O.R.; Sivakumar, M.; Tamta, S.; Sunitha, R.; Sinha, D.K.; Singh, B.R. Molecular Characterization of Extensively Drug Resistant (XDR), Extended Spectrum Beta-Lactamases (ESBL) and New Delhi Metallo Beta-Lactamase-1 (blaNDM1) Producing Escherichia coli Isolated from a Male Dog—A Case Report. Vet. Arhiv 2018, 88, 139–148. [Google Scholar] [CrossRef]
- Lysitsas, M.; Triantafillou, E.; Chatzipanagiotidou, I.; Antoniou, K.; Valiakos, G. Antimicrobial Susceptibility Profiles of Acinetobacter Baumannii Strains, Isolated from Clinical Cases of Companion Animals in Greece. Vet. Sci. 2023, 10, 635. [Google Scholar] [CrossRef]
- Da Silva, L.C.A.; Do Nascimento Pessoa, D.A.; Maia, L.Â.; Matos, R.A.T.; Macêdo, M.M.D.S. Systemic Infection by Pseudomonas aeruginosa in a Dog. Acta Sci. Vet. 2016, 44, 5. [Google Scholar] [CrossRef]
- Hayashi, W.; Izumi, K.; Yoshida, S.; Takizawa, S.; Sakaguchi, K.; Iyori, K.; Minoshima, K.; Takano, S.; Kitagawa, M.; Nagano, Y.; et al. Antimicrobial Resistance and Type III Secretion System Virulotypes of Pseudomonas aeruginosa Isolates from Dogs and Cats in Primary Veterinary Hospitals in Japan: Identification of the International High-Risk Clone Sequence Type 235. Microbiol. Spectr. 2021, 9, e00408-21. [Google Scholar] [CrossRef]
- Leal-Vélez, L.; Quevedo-Caraballo, S.; Scarpellini, R.; García, M.E.; Blanco, J.L. Integrative Phenotypic and Genomic Analysis of Extended-Spectrum Beta-Lactamase (ESBL) and Carbapenemase Genes in Enterobacteriaceae and Pseudomonaceae Strains Isolated from Animals in a Spanish Veterinary Teaching Hospital. Res. Vet. Sci. 2025, 185, 105529. [Google Scholar] [CrossRef]
- Badis, A.; Heleili, N.; Merradi, M.; Ayachi, A.; Martino, P.A.; Meroni, G.; Soggiu, A. Outbreak of Carbapenem-Resistant High-Risk Clone ST244 of Pseudomonas aeruginosa in Dogs and Cats in Algeria. Antibiotics 2025, 14, 230. [Google Scholar] [CrossRef]
- Hyun, J.; Chung, T.; Hwang, C. Identification of VIM-2 Metallo-β-lactamase-producing Pseudomonas aeruginosa Isolated from Dogs with Pyoderma and Otitis in Korea. Vet. Dermatol. 2018, 29, 186. [Google Scholar] [CrossRef] [PubMed]
- Jangsangthong, A.; Lugsomya, K.; Apiratwarrasakul, S.; Phumthanakorn, N. Distribution of Sequence Types and Antimicrobial Resistance of Clinical Pseudomonas aeruginosa Isolates from Dogs and Cats Visiting a Veterinary Teaching Hospital in Thailand. BMC Vet. Res. 2024, 20, 234. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, T.; Garcês, A.; Silva, A.; Lopes, R.; Alegria, N.; Hébraud, M.; Igrejas, G.; Poeta, P. The Impact of the Virulence of Pseudomonas aeruginosa Isolated from Dogs. Vet. Sci. 2023, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lei, L.; Lv, Y.; Zhang, R.; Liu, X.; Li, M.; Zhang, F.; Wang, Y. Bla NDM-1 -Producing Multidrug-Resistant Escherichia coli Isolated from a Companion Dog in China. J. Glob. Antimicrob. Resist. 2018, 13, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.E.; Phan, H.T.T.; George, S.; Hubbard, A.T.M.; Stoesser, N.; Maciuca, I.E.; Crook, D.W.; Timofte, D. Occurrence and Characterization of Escherichia coli ST410 Co-Harbouring blaNDM-5, blaCMY-42 and blaTEM-190 in a Dog from the UK. J. Antimicrob. Chemother. 2019, 74, 1207–1211. [Google Scholar] [CrossRef]
- Nittayasut, N.; Yindee, J.; Boonkham, P.; Yata, T.; Suanpairintr, N.; Chanchaithong, P. Multiple and High-Risk Clones of Extended-Spectrum Cephalosporin-Resistant and blaNDM-5-Harbouring Uropathogenic Escherichia Coli from Cats and Dogs in Thailand. Antibiotics 2021, 10, 1374. [Google Scholar] [CrossRef]
- Ding, L.; Sun, Y.; Zhang, Y.; Shen, S.; Hu, F. In Vivo Development of Aztreonam Resistance in Meropenem-Resistant Pseudomonas aeruginosa Owing to Overexpression of the BlaPDC-16. Microbiol. Spectr. 2023, 11, e03080-22. [Google Scholar] [CrossRef]
- Karthik, M.; Kacha, S.; Rajendran, S.; Bakthavatchalam, Y.D.; Lal, B.; Walia, K.; Veeraraghavan, B. Genetic Characteristics and Diversity of PDC Variants of Pseudomonas aeruginosa and Its Clinical Relevance. Infect. Genet. Evol. 2024, 126, 105701. [Google Scholar] [CrossRef]
- Teo, J.Q.-M.; Lim, J.C.; Tang, C.Y.; Lee, S.J.-Y.; Tan, S.H.; Sim, J.H.-C.; Ong, R.T.-H.; Kwa, A.L.-H. Ceftolozane/Tazobactam Resistance and Mechanisms in Carbapenem-Nonsusceptible Pseudomonas aeruginosa. mSphere 2021, 6, e01026-20. [Google Scholar] [CrossRef]
- Mack, A.R.; Hujer, A.M.; Mojica, M.F.; Taracila, M.A.; Feldgarden, M.; Haft, D.H.; Klimke, W.; Prasad, A.B.; Bonomo, R.A. β-Lactamase Diversity in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2025, 69, e0078524. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Arima, S.; Niina, A.; Kataoka, Y.; Takahashi, T. Characterization of Pseudomonas aeruginosa Isolates from Dogs and Cats in Japan: Current Status of Antimicrobial Resistance and Prevailing Resistance Mechanisms. Microbiol. Immunol. 2012, 56, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; De Sousa, T.; Silva, C.; Igrejas, G.; Poeta, P. Impact of Antimicrobial Resistance of Pseudomonas aeruginosa in Urine of Small Companion Animals in Global Context: Comprehensive Analysis. Vet. Sci. 2025, 12, 157. [Google Scholar] [CrossRef]
- Feßler, A.T.; Scholtzek, A.D.; Schug, A.R.; Kohn, B.; Weingart, C.; Hanke, D.; Schink, A.-K.; Bethe, A.; Lübke-Becker, A.; Schwarz, S. Antimicrobial and Biocide Resistance among Canine and Feline Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii Isolates from Diagnostic Submissions. Antibiotics 2022, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Newstead, L.; Smith-Zaitlik, T.; Kelly, C.; Roberts, E.; Street, S.; Paterson, G.K. Genomic Characterization of Pseudomonas aeruginosa from Canine Otitis Highlights the Need for a One Health Approach to This Opportunistic Pathogen. Microb. Genom. 2025, 11, 001407. [Google Scholar] [CrossRef] [PubMed]
- Kung, V.L.; Ozer, E.A.; Hauser, A.R. The Accessory Genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2010, 74, 621–641. [Google Scholar] [CrossRef]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tümmler, B. Pseudomonas aeruginosa Genomic Structure and Diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef]
- Javanmardi, F.; Emami, A.; Pirbonyeh, N.; Keshavarzi, A.; Rajaee, M. A Systematic Review and Meta-Analysis on Exo-Toxins Prevalence in Hospital Acquired Pseudomonas aeruginosa Isolates. Infect. Genet. Evol. 2019, 75, 104037. [Google Scholar] [CrossRef]
- Recio, R.; Villa, J.; Viedma, E.; Orellana, M.Á.; Lora-Tamayo, J.; Chaves, F. Bacteraemia Due to Extensively Drug-Resistant Pseudomonas aeruginosa Sequence Type 235 High-Risk Clone: Facing the Perfect Storm. Int. J. Antimicrob. Agents 2018, 52, 172–179. [Google Scholar] [CrossRef]
- Morales-Espinosa, R.; Delgado, G.; Espinosa-Camacho, F.; Flores-Alanis, A.; Rodriguez, C.; Mendez, J.L.; Gonzalez-Pedraza, A.; Cravioto, A. Pseudomonas aeruginosa Strains Isolated from Animal with High Virulence Genes Content and Highly Sensitive to Antimicrobials. J. Glob. Antimicrob. Resist. 2024, 37, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Płókarz, D.; Czopowicz, M.; Bierowiec, K.; Rypuła, K. Virulence Genes as Markers for Pseudomonas aeruginosa Biofilm Formation in Dogs and Cats. Animals 2022, 12, 422. [Google Scholar] [CrossRef] [PubMed]
- Foksiński, P.; Blank, A.; Kaczorek-Łukowska, E.; Małaczewska, J.; Wróbel, M.; Wójcik, E.A.; Sowińska, P.; Pietrzyk, N.; Matusiak, R.; Wójcik, R. Does Every Strain of Pseudomonas aeruginosa Attack the Same? Results of a Study of the Prevalence of Virulence Factors of Strains Obtained from Different Animal Species in Northeastern Poland. Pathogens 2024, 13, 979. [Google Scholar] [CrossRef]
- Secker, B.; Shaw, S.; Atterbury, R.J. Pseudomonas Spp. in Canine Otitis Externa. Microorganisms 2023, 11, 2650. [Google Scholar] [CrossRef] [PubMed]
- Cotter, C.S.; Avidano, M.A.; Stringer, S.P.; Schultz, G.S. Inhibition of Proteases in Pseudomonas Otitis Media in Chinchillas. Otolaryngol.-Head Neck Surg. 1996, 115, 342–351. [Google Scholar] [CrossRef]
- Sánchez-Jiménez, A.; Llamas, M.A.; Marcos-Torres, F.J. Transcriptional Regulators Controlling Virulence in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2023, 24, 11895. [Google Scholar] [CrossRef] [PubMed]
- Abdolghanizadeh, S.; Salmeh, E.; Mirzakhani, F.; Soroush, E.; Siadat, S.D.; Tarashi, S. Microbiota Insights into Pet Ownership and Human Health. Res. Vet. Sci. 2024, 171, 105220. [Google Scholar] [CrossRef]
- Jin, M.; Osman, M.; Green, B.A.; Yang, Y.; Ahuja, A.; Lu, Z.; Cazer, C.L. Evidence for the Transmission of Antimicrobial Resistant Bacteria between Humans and Companion Animals: A Scoping Review. One Health 2023, 17, 100593. [Google Scholar] [CrossRef]
- Mauri, C.; Maraolo, A.E.; Di Bella, S.; Luzzaro, F.; Principe, L. The Revival of Aztreonam in Combination with Avibactam against Metallo-β-Lactamase-Producing Gram-Negatives: A Systematic Review of In Vitro Studies and Clinical Cases. Antibiotics 2021, 10, 1012. [Google Scholar] [CrossRef]
- Sebola, D.C.; Oguttu, J.W.; Kock, M.M.; Qekwana, D.N. Hospital-Acquired and Zoonotic Bacteria from a Veterinary Hospital and Their Associated Antimicrobial-Susceptibility Profiles: A Systematic Review. Front. Vet. Sci. 2023, 9, 1087052. [Google Scholar] [CrossRef]
| Antibacterial Agent | Disk Content (μg) | Breakpoints | AST Results | ||
|---|---|---|---|---|---|
| Inhibition Zone (mm) | MIC μg/mL | DD | MIC | ||
| Ceftazidime | 30 | S: ≥18 I: 15–17, R: ≤14 | S: ≤8, I: 16, R: ≥32 | R | R |
| Cefepime | 30 | S: ≥18 I: 15–17, R: ≤14 | S: ≤8, I: 16, R: ≥32 | R | R |
| Piperacillin–tazobactam | 100/10 | S: ≥22 I: 18–21, R: ≤17 | S: ≤16/4, I: 32/4, R: ≥64/4 | R | R |
| Ticarcillin–clavulanate | 75/10 | S: ≥24 I: 16–23, R: ≤15 | - | R | NT |
| Aztreonam | 30 | S: ≥22 I: 16–21, R: ≤15 | S: ≤8, I: 16, R: ≥32 | R | I |
| Imipenem | 10 | S: ≥19 I: 16–18, R: ≤15 | S: ≤2, I: 4, R: ≥8 | R | R |
| Meropenem | 10 | S: ≥19 I: 16–18, R: ≤15 | S: ≤2, I: 4, R: ≥8 | R | R |
| Ceftazidime–avibactam | - | - | S: ≤8/4, R: ≥16/4 | NT | R |
| Ceftolozane–tazobactam | - | - | S: ≤4/4, I: 8/4, R: ≥16/4 | NT | R |
| Imipenem–relabactam | - | - | S: ≤2/4, I: 4/4, R: ≥8/4 | NT | R |
| Meropenem + vaborbactam | - | - | S: ≤16/4, I: 32/4, R: ≥64/4 | NT | R |
| Amikacin | 30 | S: ≥17 I: 15–16, R: ≤14 | S: ≤16, I: 32, R: ≥64 | R | R |
| Gentamicin | 10 | S: ≥16 I: 13–14, R: ≤12 | S: ≤2, I: 4, R: ≥8 | R | R |
| Tobramycin | 10 | S: ≥19 I: 13–18, R: ≤12 | S: ≤1, I: 2, R: ≥4 | R | R |
| Ciprofloxacin | 5 | S: ≥25 I: 19–24, R: ≤18 | S: ≤0.5, I: 1, R: ≥2 | R | R |
| Enrofloxacin | 5 | S: ≥23 I: 17–22, R: ≤16 | S: ≤0.5, I: 1–2, R: ≥4 | R | R |
| Fosfomycin 1 | 200 | - | - | R | R |
| Colistin | - | - | S: ≤2, R: ≥4 | NT | S |
| Resistance Gene | Predicted Phenotype |
|---|---|
| aac(6′)-Il, aadA11, aph(3″)-Ib, aph(3′)-IIb, aph(6)-Id, EmrE | amikacin, gentamicin, kanamycin, neomycin, spectinomycin, streptomycin, tobramycin |
| blaNDM-1, blaPDC-19a, blaPAC-1, blaOXA-677, blaOXA-50 | all beta-lactams except monobactams |
| catB7 | chloramphenicol |
| floR, mexM, mexN | florfenicol |
| qnrVC1 | ciprofloxacin |
| fosA | fosfomycin |
| msrE | erythromycin, azithromycin |
| dfrB5 | trimethoprim |
| sul1, sul2 | sulfamethoxazole |
| cprR, cprS, basS, arnA | peptide antibiotic |
| MexA, OprM, YajC, rsmA, MexE, MexF, OpmB, MuxC, MuxB, CpxR, MexB, MexK, MexJ, MexL, PmpM, MexV, MexW, MexC, MexD, OprJ, OprN, MuxA, opmE, mexQ, mexP, MexG, MexH, MexI, OpmD, nalC, Type A NfxB, MexS, MexZ, ParS, ParR, soxR, MexR | aminocoumarins, aminoglycosides, beta-lactams, diaminopyrimidines, fluoroquinolones, macrolides, tetracyclines, oxazolidinones, peptide antibiotics, phenicols, rifamycin, sulfonamides, disinfecting agents and antiseptics (multidrug efflux pumps) |
| TriA, TriB, TriC, OpmH | disinfecting agents and antiseptics |
| Metric | CP053917_vs_Sequenced_Strain | CP020703_vs_Sequenced_Strain |
|---|---|---|
| Average Nucleotide Identity (ANI) | 99.094 | 99.9328 |
| Query Sequence Fragments | 2308 | 2297 |
| Orthologous Matches | 2064 | 2188 |
| % orthology | 89.4 | 95.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lysitsas, M.; Triantafillou, E.; Chatzipanagiotidou, I.; Triantafillou, A.; Agorou, G.; Filippitzi, M.E.; Giakountis, A.; Valiakos, G. Companion Animals as Reservoirs of Multidrug Resistance—A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece. Vet. Sci. 2025, 12, 576. https://doi.org/10.3390/vetsci12060576
Lysitsas M, Triantafillou E, Chatzipanagiotidou I, Triantafillou A, Agorou G, Filippitzi ME, Giakountis A, Valiakos G. Companion Animals as Reservoirs of Multidrug Resistance—A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece. Veterinary Sciences. 2025; 12(6):576. https://doi.org/10.3390/vetsci12060576
Chicago/Turabian StyleLysitsas, Marios, Eleftherios Triantafillou, Irene Chatzipanagiotidou, Anastasios Triantafillou, Georgia Agorou, Maria Eleni Filippitzi, Antonis Giakountis, and George Valiakos. 2025. "Companion Animals as Reservoirs of Multidrug Resistance—A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece" Veterinary Sciences 12, no. 6: 576. https://doi.org/10.3390/vetsci12060576
APA StyleLysitsas, M., Triantafillou, E., Chatzipanagiotidou, I., Triantafillou, A., Agorou, G., Filippitzi, M. E., Giakountis, A., & Valiakos, G. (2025). Companion Animals as Reservoirs of Multidrug Resistance—A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece. Veterinary Sciences, 12(6), 576. https://doi.org/10.3390/vetsci12060576







