Establishment of a One–Pot RAA–CRISPR/Cas13a Assay-Based TGEV S Gene Detection
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Samples
2.2. Nucleic Acid Extraction
2.3. Construction of Standard Recombinant Plasmid Containing Target Sequence
2.4. Quantitative Real–Time PCR (qPCR) Assay
2.5. RAA Primers, ssRNA Reporter, and crRNA Preparation
2.6. CRISPR/Cas13a Complex Cleavage System
2.7. Establishment of TGEV Assay Based on RAA–CRISPR/Cas13a System
2.8. Optimization of TGEV Assay System Based on RAA–CRISPR/Cas13a
2.9. Specificity Analysis Test of RAA–CRISPR/Cas13a Assay
2.10. Sensitivity Analysis Test of RAA–CRISPR/Cas13a Assay
2.11. Stability Analysis Test of RAA–CRISPR/Cas13a Assay
2.12. Effectiveness Evaluation of Testing Clinical Samples
2.13. Statistical Analysis
3. Results
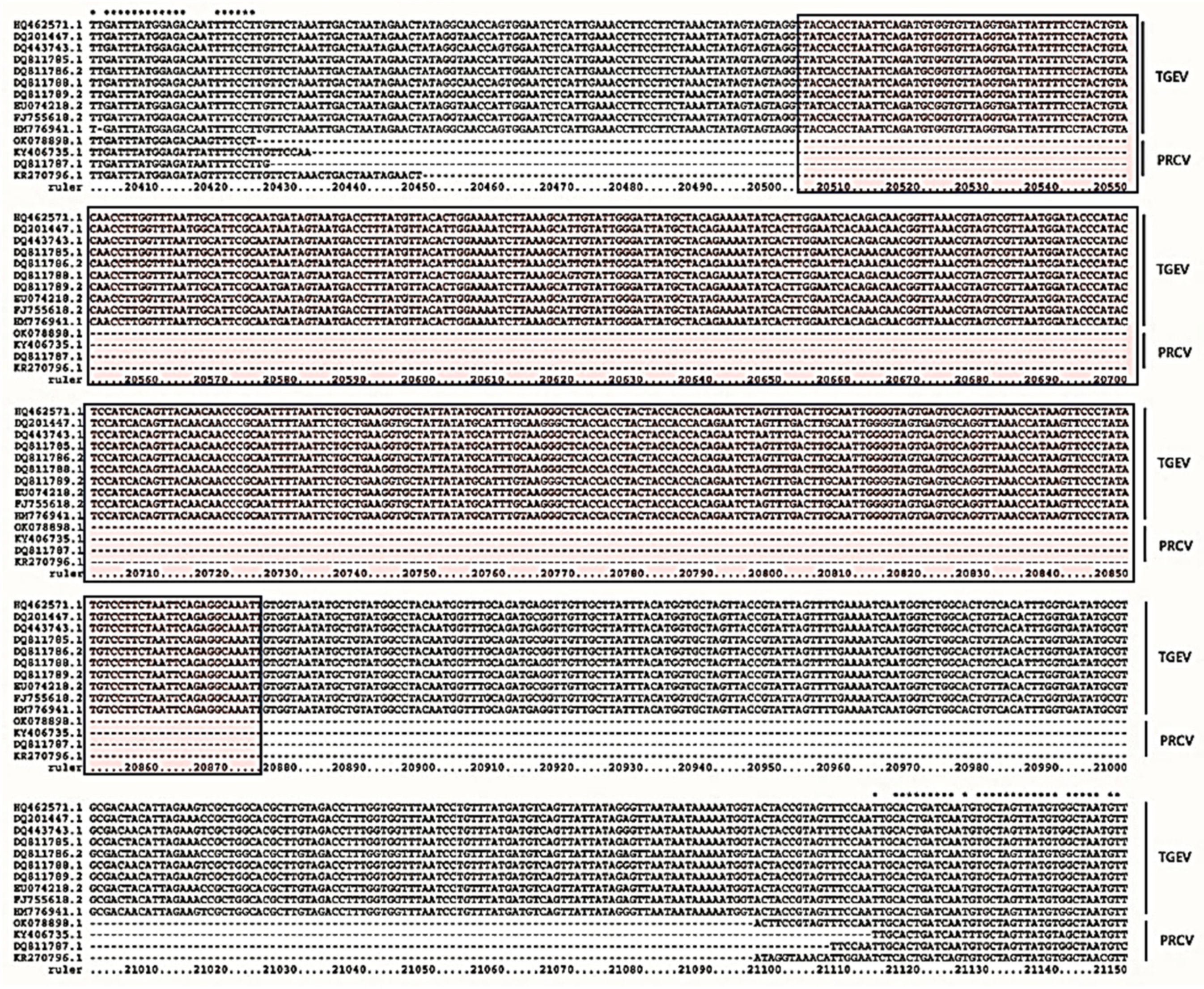
3.1. Standard Plasmid Containing Target Sequence
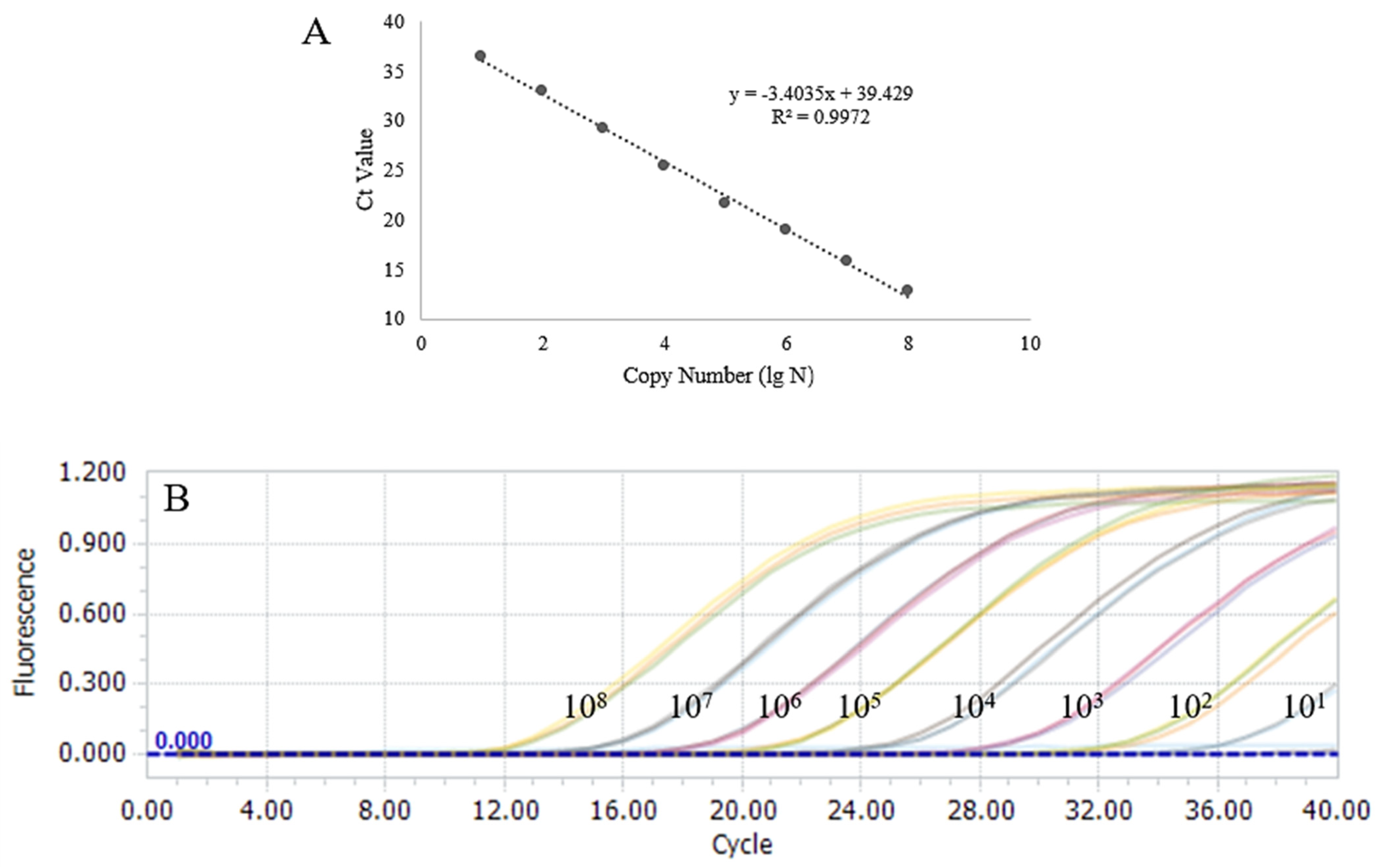
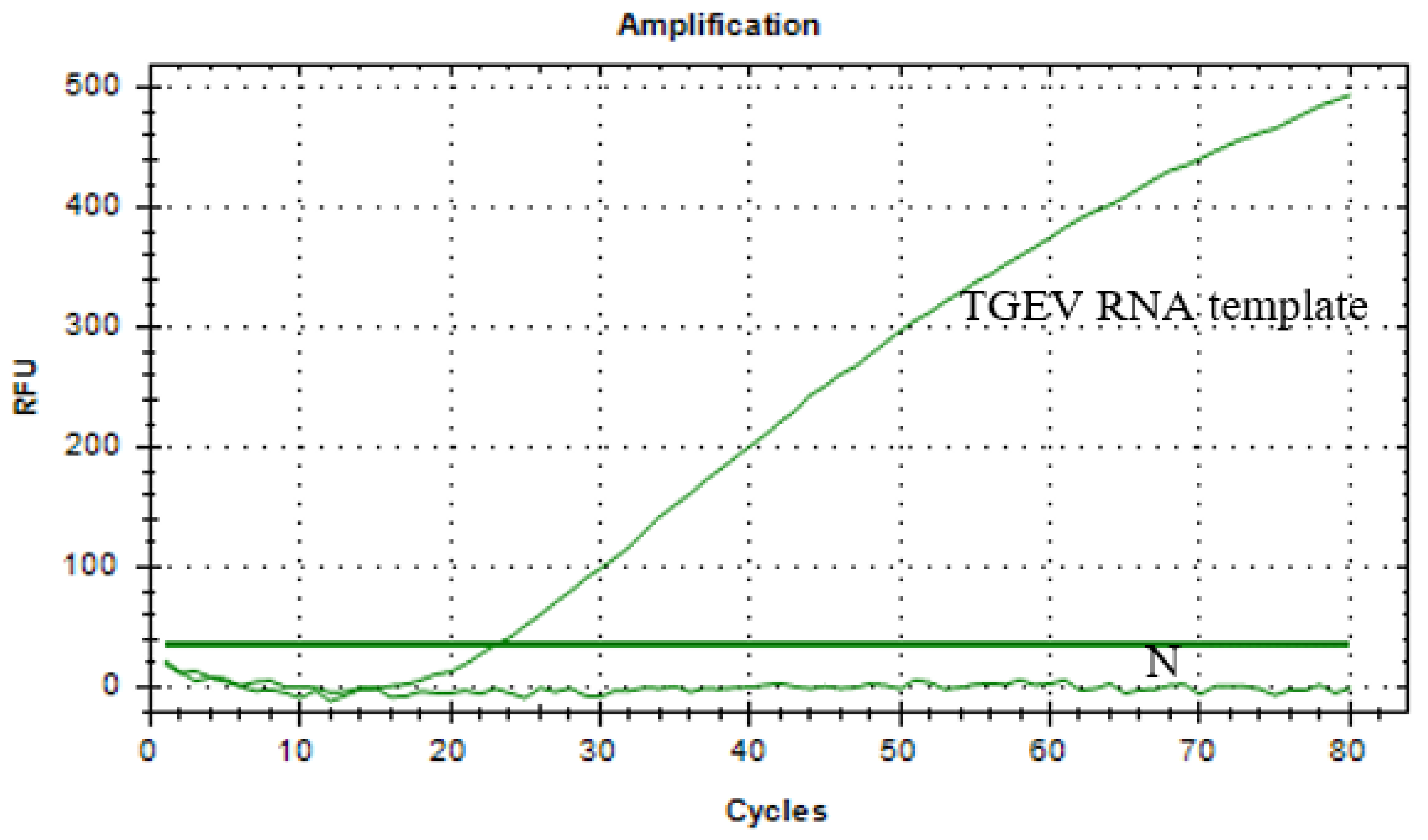
3.2. Sensitivity and Specificity Validation of qPCR
3.3. Optimal Primer Screening for RAA
3.4. Detection of CRISPR/Cas13a Complex Cleavage
3.5. Orthogonal Test Optimization and Establishment of RAA–CRISPR/Cas13a Assay
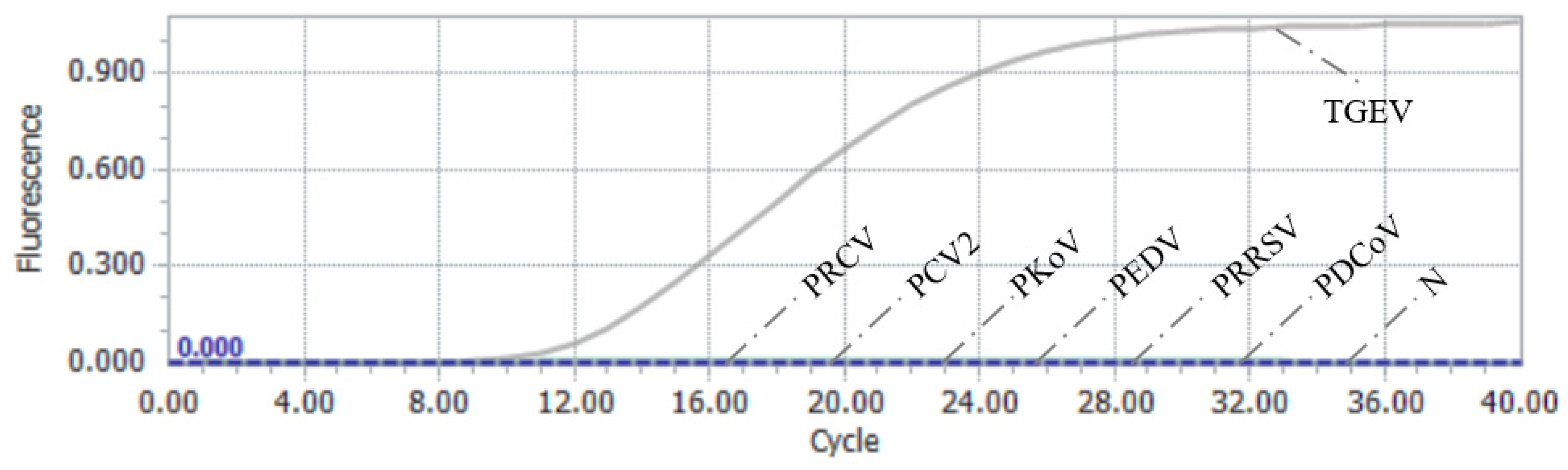
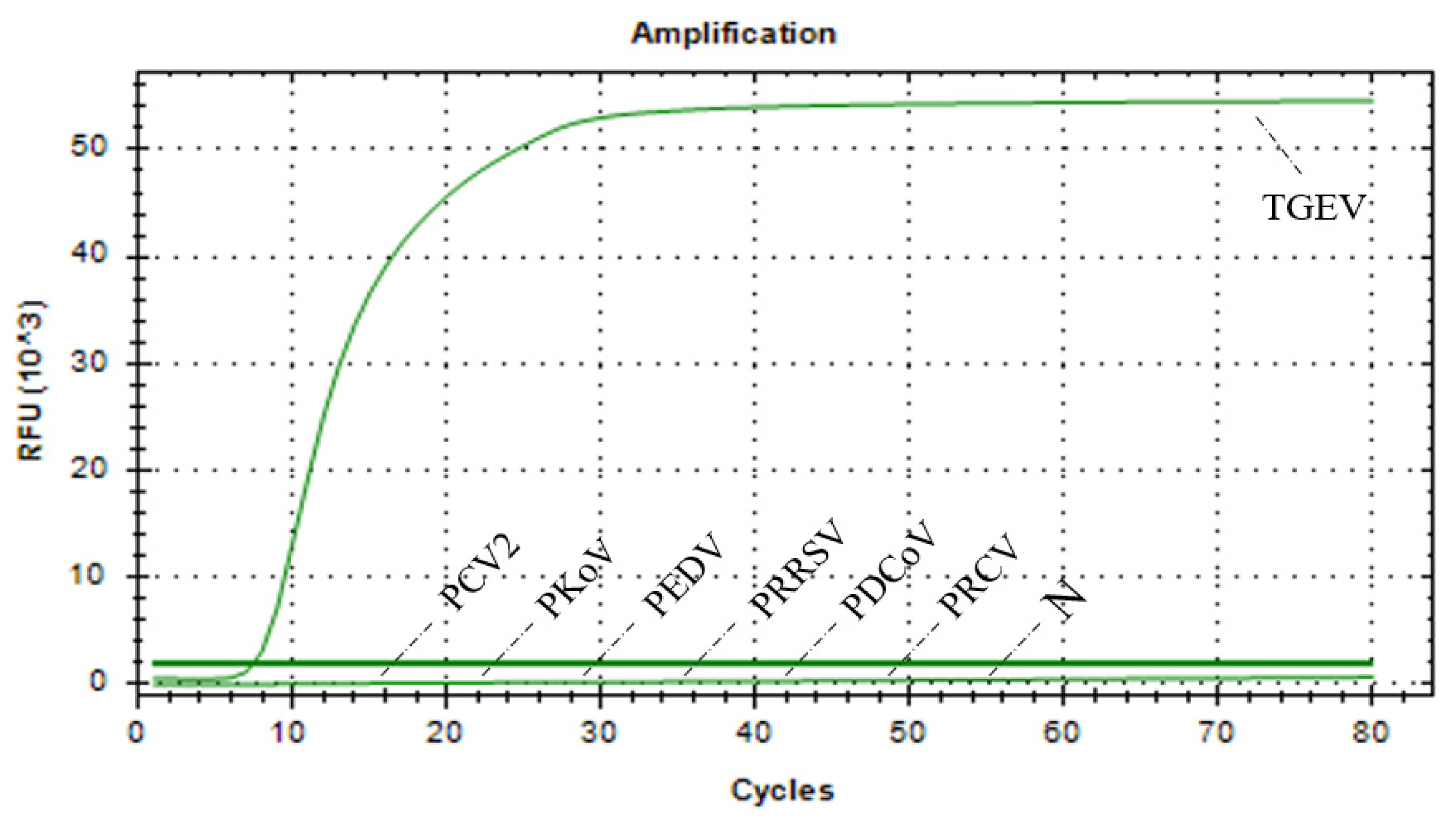
3.6. Results of Specificity Assessment of RAA–CRISPR/Cas13a Assay
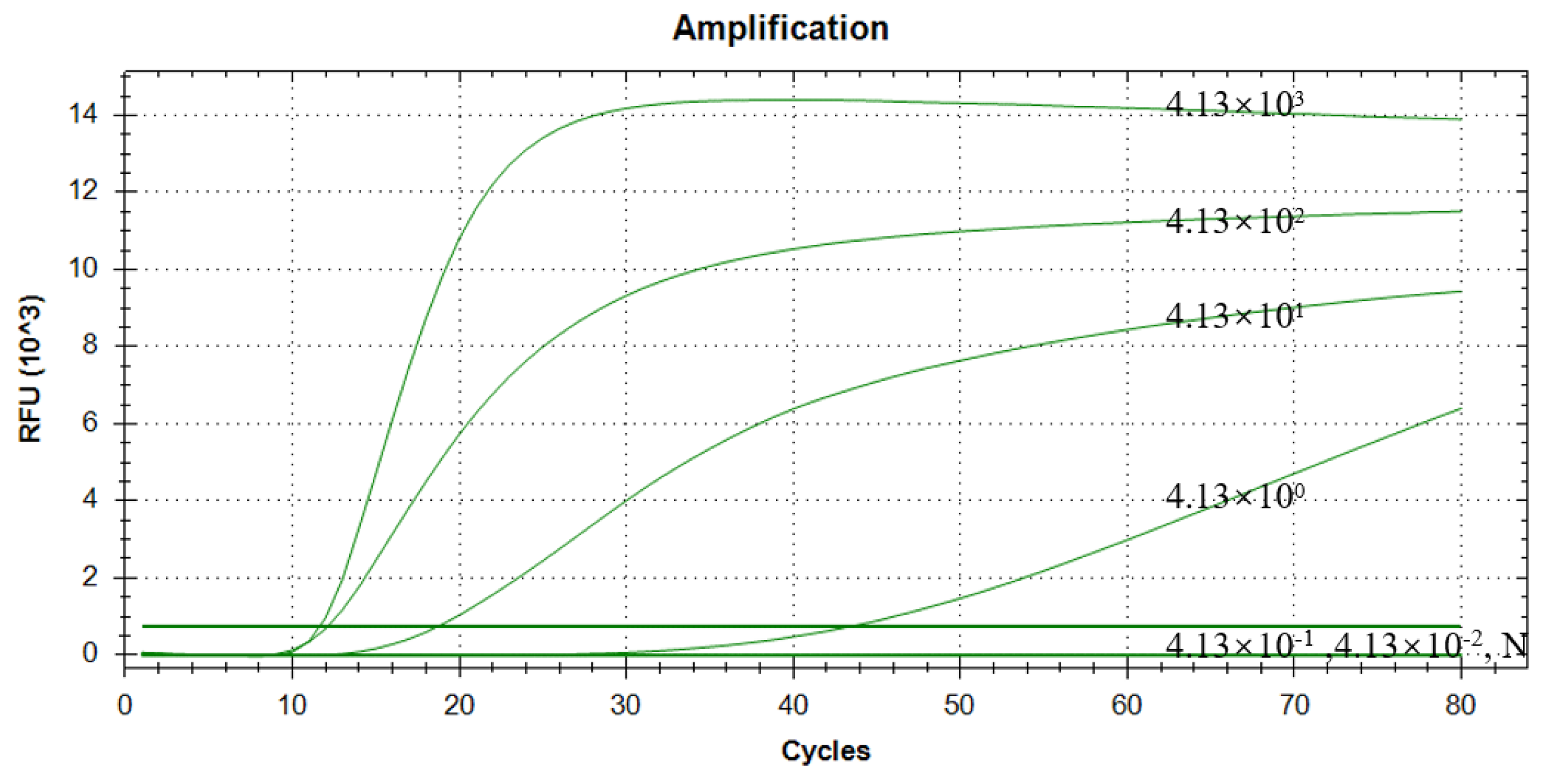
3.7. Results of Sensitivity Assessment of RAA–CRISPR/Cas13a Assay
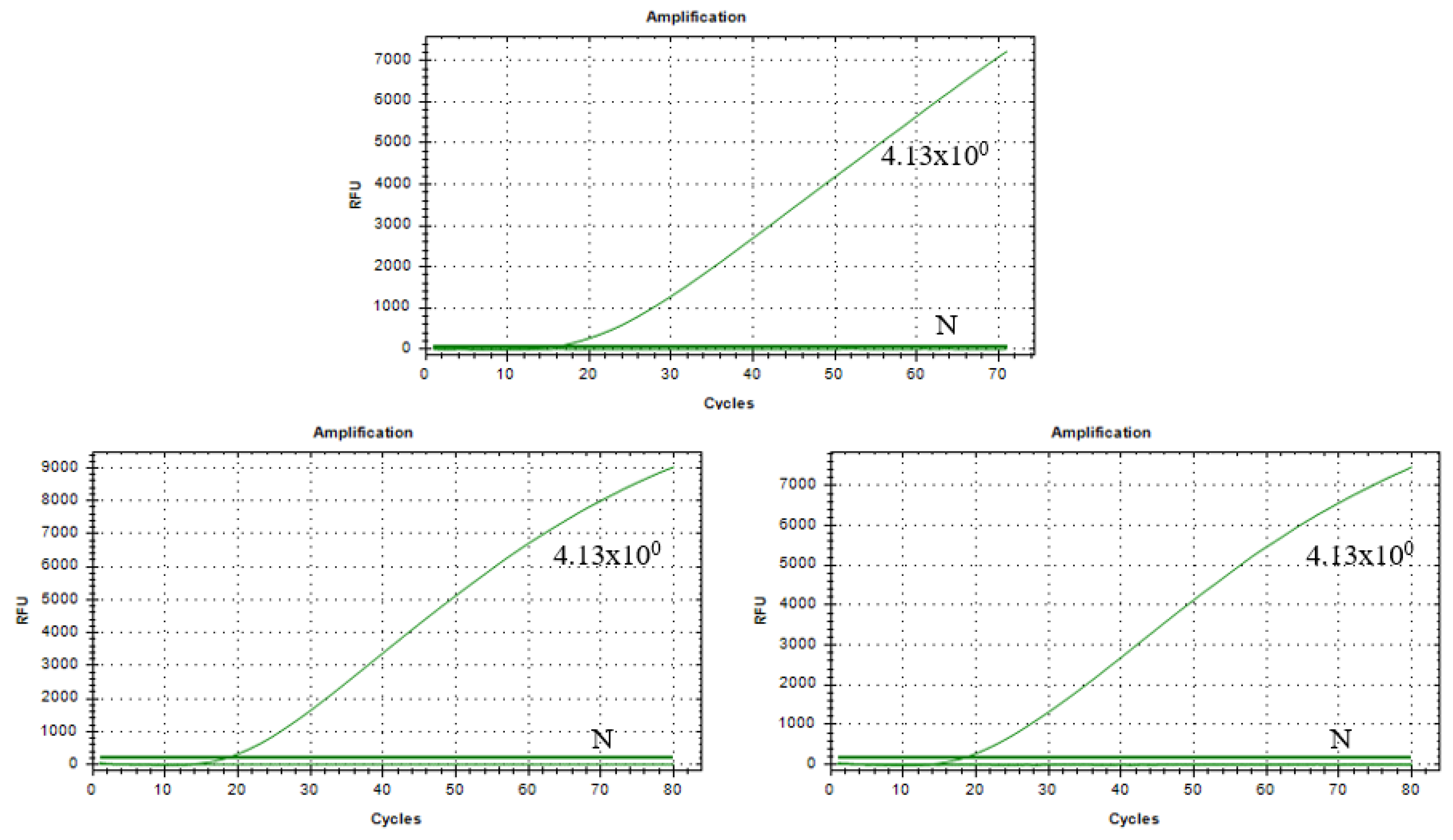
3.8. Results of Stability Assessment of RAA–CRISPR/Cas13a Assay
3.9. Clinical Sample Detection of RAA–CRISPR/Cas13a Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| ASFV | African swine fever virus |
| CRISPR | Clustered regularly interspaced short palindromic repeat |
| Cas | CRISPR–associated proteins |
| CV | Coefficient of variation |
| CSFV | Classical swine fever virus |
| PCV2 | Porcine circovirus 2 |
| PDCoV | Porcine delta coronavirus |
| PEDV | Porcine epidemic diarrhea virus |
| PKoV | Porcine kobuvirus |
| PRCV | Porcine respiratory coronavirus |
| PRRSV | Porcine reproductive and respiratory syndrome virus |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| TGEV | Porcine transmissible gastroenteritis virus |
| RAA | Recombinase–aided amplification |
References
- Kim, L.; Chang, K.O.; Sestak, K.; Parwani, A.; Saif, L.J. Development of a reverse transcription–nested polymerase chain reaction assay for differential diagnosis of transmissible gastroenteritis virus and porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J. Vet. Diagn. Investig. 2000, 12, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef] [PubMed]
- Garwes, D.J. Transmissible gastroenteritis. Vet. Rec. 1988, 122, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Sestak, K.; Hodgins, D.C.; Shoup, D.I.; Ward, L.A.; Jackwood, D.J.; Saif, L.J. Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (Tgev) and recombinant Tgev spike protein vaccines and protection of their suckling pigs against virulent Tgev challenge exposure. Am. J. Vet. Res. 1998, 59, 1002–1008. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Wang, X.; Zhou, J.; Ma, L.; Li, J.; Yang, L.; Ouyang, H.; Yuan, H.; Pang, D. Transmissible Gastroenteritis Virus: An Update Review and Perspective. Viruses 2023, 15, 359. [Google Scholar] [CrossRef]
- Magtoto, R.; Poonsuk, K.; Baum, D.; Zhang, J.; Chen, Q.; Ji, J.; Piñeyro, P.; Zimmerman, J.; Giménez–Lirola, L.G. Evaluation of the Serologic Cross–Reactivity between Transmissible Gastroenteritis Coronavirus and Porcine Respiratory Coronavirus Using Commercial Blocking Enzyme–Linked Immunosorbent Assay Kits. mSphere 2019, 4, e00017-19. [Google Scholar] [CrossRef]
- Woods, R.D.; Wesley, R.D.; Kapke, P.A. Neutralization of porcine transmissible gastroenteritis virus by complement–dependent monoclonal antibodies. Am. J. Vet. Res. 1988, 49, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Comparative pathogenesis of enteric viral infections of swine. Adv. Exp. Med. Biol. 1999, 473, 47–59. [Google Scholar] [CrossRef]
- Saif, L.J.; Jung, K. Comparative Pathogenesis of Bovine and Porcine Respiratory Coronaviruses in the Animal Host Species and SARS-CoV-2 in Humans. J. Clin. Microbiol. 2020, 58, e01355-20. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with Crispr–Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single–component programmable RNA–guided RNA–targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA–Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726.e10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Zhang, Y.; Qin, G.; Sun, W.; Wang, A.; Wang, Y.; Zhang, G.; Zhao, J. On–site detection and differentiation of African swine fever virus variants using an orthogonal CRISPR–Cas12b/Cas13a–based assay. iScience 2024, 27, 109050. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yin, D.D.; Shao, Y.; Song, X.; Wang, Z.; Pan, X.; Tu, J.; He, C.; Zhu, L.; Qi, K.; et al. Establishment and preliminary application of RAA–CRISPR/Cas13a assay for porcine epidemic diarrhea virus. J. Anim. Husb. Vet. Sci. 2023, 54, 3991–3997. [Google Scholar]
- Wang, L.; Zhou, J.; Wang, Q.; Wang, Y.; Kang, C. Rapid design and development of CRISPR–Cas13a targeting SARS-CoV-2 spike protein. Theranostics 2021, 11, 649–664. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Z.H.; Li, P.; Lin, X.; Jia, H.; Jiang, Y.T.; Wang, X.J.; Hou, S.H. Application of portable real–time recombinase–aided amplification (rt–RAA) assay in the clinical diagnosis of ASFV and prospective DIVA diagnosis. Appl. Microbiol. Biotechnol. 2021, 105, 3249–3264. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Li, F.; Shen, X.X.; Fu, S.H.; He, Y.; Lei, W.W.; Liang, G.D.; Wang, H.Y.; Ma, X.J. A Reverse–transcription Recombinase–aided Amplification Assay for the Rapid Detection of the Far–Eastern Subtype of Tick–borne Encephalitis Virus. Biomed. Environ. Sci. 2019, 32, 357–362. [Google Scholar] [CrossRef]
- Lu, Q.; Zhao, Z.Y.; Yin, D.W.; Liu, Y.; Wang, W.; Zheng, M.; Hu, S.; Zhao, C.; Zhang, X.; Lei, X.; et al. Establishment and preliminary application of fluorescent RT–RAA method for porcine infectious gastroenteritis virus. J. Anim. Sci. Vet. Med. 2023, 54, 2208–2214. [Google Scholar]
- Lu, L.D.; Mou, H.; Hu, X.; Liu, M.; Li, S.; Li, X.; Song, Z.; Yang, L. Establishment and preliminary application of RAA detection method for S gene of porcine transmissible gastroenteritis virus. Chin. J. Anim. Husb. Vet. Med. 2024, 55, 3590–3599. [Google Scholar]
- Turlewicz, P.H.; Pomorska, M.M. Porcine Coronaviruses: Overview of the State of the Art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef]
- Chen, F.; Knutson, T.P.; Rossow, S.; Saif, L.J.; Marthaler, D.G. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci. Rep. 2019, 9, 3953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013, 158, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Huang, F.Q.; Zuo, X.L.; Liang, J.; Liang, K.; Shan, J.; Li, Z.; Yu, J.; Luo, L.; Xuan, Z.; et al. Establishment of an elasmobranch virus detection method for largemouth bass based on CRISPR/Cas13a system. J. Aquat. 2024, 48, 283–291. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Wang, S.M.; Duan, H.Y.; Zhao, K.; Dong, S.; Gao, F.; Li, L. Establishment and application of TaqMan real–time PCR for detection of porcine infectious gastroenteritis virus. China Vet. Sci. 2024, 4, 479–484. [Google Scholar] [CrossRef]
- Boyle, D.S.; Lehman, D.A.; Lillis, L.; Peterson, D.; Singhal, M.; Armes, N.; Parker, M.; Piepenburg, O.; Overbaugh, J. Rapid detection of HIV–1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. MBio 2013, 4, e00135-13. [Google Scholar] [CrossRef]
- Yu, J.X.; Wei, S.S.; Qin, S.M.; Wu, J.; Yang, L.; Chen, F.; Xu, L.; Qin, S.; Hua, J.; Wei, J.; et al. Establishment of a Rapid Detection Method for Porcine Reproductive and Respiratory Syndrome Virus Based on CRISPR/Cas12a–RT–RAA. China Anim. Husb. Vet. Med. 2024, 51, 3237–3246. [Google Scholar] [CrossRef]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra–sensitive and high–throughput CRISPR–p owered COVID–19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR–Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR–Cas12b–assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Das, D.; Lin, C. –W.; Chuang, H.–S. LAMP–Based Point–of–Care Biosensors for Rapid Pathogen Detection. Biosensors 2022, 12, 1068. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Li, J.; Chen, K.; Yu, X.; Sun, C.; Zhang, M. Multiplex Recombinase Polymerase Amplification Assay for the Simultaneous Detection of Three Foodborne Pathogens in Seafood. Foods 2020, 9, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Jiang, W.; Wu, Y.; Fang, R.; Deng, F.; Yang, D. Advances in CRISPR/Cas13a–based biosensors for non–coding RNA detection. Talanta 2025, 294, 128223. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Cui, J.; Zheng, H.; Guo, T.; Wei, R.; Sha, Z.; Gu, S.; Ni, B. Implementation of RT–RAA and CRISPR/Cas13a for an NiV Point–of–Care Test: A Promising Tool for Disease Control. Viruses 2025, 17, 483. [Google Scholar] [CrossRef]
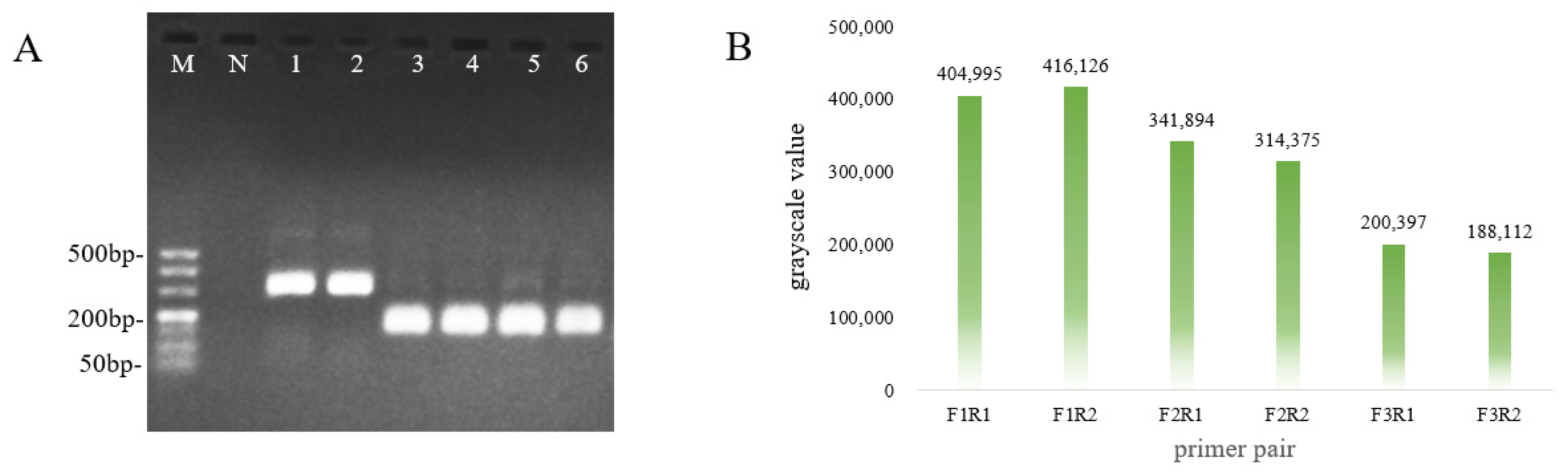
| Number | Primer Sequences | Primer Pairing | Product Size, bp |
|---|---|---|---|
| F1 | 5′-GGTGTTAGGTGATTATTTTCCTACTGTACA-3′ | F1/R1 | 308 |
| F2 | 5′-ACGGTTAAACGTAGTCGTTAATGGATACCC-3′ | F2/R1 | 164 |
| F3 | 5′-CGGTTAAACGTAGTCGTTAATGGATACCCA-3′ | F3/R1 | 163 |
| R1 | 5′-ACCTGCACTCACTACCCCAATTGCAAGTCA-3′ | F1/R2 | 309 |
| R2 | 5′-AACCTGCACTCACTACCCCAATTGCAAGTC-3′ | F2/R2 | 165 |
| F3/ R2 | 164 |
| Test No. | Experimental Condition Parameters | |||||
|---|---|---|---|---|---|---|
| A (Reporter RNA Concentration, μM) | B (Primer Concentration, μM) | C (crRNA Concentration, μM) | ||||
| 1 | 1 | 8 | 1 | 8 | 1 | 2.5 |
| 2 | 1 | 8 | 2 | 10 | 3 | 10 |
| 3 | 1 | 8 | 3 | 12 | 2 | 5 |
| 4 | 2 | 10 | 1 | 8 | 3 | 10 |
| 5 | 2 | 10 | 2 | 10 | 2 | 5 |
| 6 | 2 | 10 | 3 | 12 | 1 | 2.5 |
| 7 | 3 | 12 | 1 | 8 | 2 | 5 |
| 8 | 3 | 12 | 2 | 10 | 1 | 2.5 |
| 9 | 3 | 12 | 3 | 12 | 3 | 10 |
| Intra–Repetitive Assay | Inter–Repetitive Assay | ||||
|---|---|---|---|---|---|
| Standard Deviation (s) | Coefficient of Variation (CV/%) | Standard Deviation (s) | Coefficient of Variation (CV/%) | ||
| 35.58 | 0.44 | 1.2 | 36.01 | 0.52 | 1.4 |
| 35.86 | 0.41 | 1.1 | |||
| 36.60 | 0.42 | 1.1 | |||
| Test No. | Experimental Condition Parameters | |||
|---|---|---|---|---|
| A (Probe) | B (Primer) | C (crRNA) | Ct Value | |
| 1 | 1 | 1 | 1 | 2.89 |
| 2 | 1 | 2 | 3 | 2.49 |
| 3 | 1 | 3 | 2 | 2.56 |
| 4 | 2 | 1 | 3 | 1.84 |
| 5 | 2 | 2 | 2 | 1.98 |
| 6 | 2 | 3 | 1 | 2.01 |
| 7 | 3 | 1 | 2 | 1.88 |
| 8 | 3 | 2 | 1 | 1.84 |
| 9 | 3 | 3 | 3 | 1.88 |
| Results of Orthogonal Test | |||
|---|---|---|---|
| A (Probe) | B (Primer) | C (crRNA) | |
| K1 | 7.93325 | 6.60910 | 6.74525 |
| K2 | 5.83047 | 6.31189 | 6.41953 |
| K3 | 5.60377 | 6.44651 | 6.20272 |
| k1 | 2.64442 | 2.20303 | 2.24842 |
| k2 | 1.94349 | 2.10396 | 2.13984 |
| k3 | 1.86792 | 2.14884 | 2.06757 |
| R | 0.77649 | 0.09907 | 0.18084 |
| Optimal conditions | A3 | B2 | C3 |
| Source of Variation | SS | df | MS | F | p |
|---|---|---|---|---|---|
| Intercept | 41.689 | 1 | 41.689 | 1801.233 | 0.001 ** |
| A (ssRNA reporter concentration, μM) | 1.109 | 2 | 0.554 | 23.957 | 0.040 * |
| B (primer concentration, μM) | 0.015 | 2 | 0.008 | 0.325 | 0.755 |
| C (crRNA concentration, μM) | 0.047 | 2 | 0.024 | 1.026 | 0.494 |
| Residual | 0.046 | 2 | 0.023 |
| No. | Region | Time | Quantity | Methods | ||
|---|---|---|---|---|---|---|
| RAA–CRISPR/Cas13a Detection Positive Results | RT–qPCR Detection Positive Results | RT–PCR Detection Positive Results | ||||
| 1 | Wanzhou | 19 February 2023 | 70 | 5/70 | 5/70 | 5/70 |
| 2 | Luzhou | 09 March 2023 | 4 | 1/4 | 1/4 | 1/4 |
| 3 | Wulong | 04 November 2023 | 20 | 2/20 | 2/20 | 2/20 |
| 4 | Changshou | 02 December 2023 | 8 | 0/8 | 0/8 | 0/8 |
| 5 | Banan | 17 December 2023 | 11 | 0/11 | 0/11 | 0/11 |
| 6 | Rongchang | 23 February 2024 | 16 | 0/16 | 0/16 | 0/16 |
| 7 | Dazhou | 21 November 2024 | 7 | 0/7 | 0/7 | 0/7 |
| 8 | Longchang | 24 December 2024 | 4 | 0/4 | 0/4 | 0/4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, L.; Mu, H.; Li, S.; Gao, J.; Liu, M.; Niu, S.; Xu, G.; Fu, L.; Song, Z.; Yang, L. Establishment of a One–Pot RAA–CRISPR/Cas13a Assay-Based TGEV S Gene Detection. Vet. Sci. 2025, 12, 464. https://doi.org/10.3390/vetsci12050464
Lv L, Mu H, Li S, Gao J, Liu M, Niu S, Xu G, Fu L, Song Z, Yang L. Establishment of a One–Pot RAA–CRISPR/Cas13a Assay-Based TGEV S Gene Detection. Veterinary Sciences. 2025; 12(5):464. https://doi.org/10.3390/vetsci12050464
Chicago/Turabian StyleLv, Lindan, Hao Mu, Shaomei Li, Jieqi Gao, Mingni Liu, Shuizhu Niu, Guoyang Xu, Lizhi Fu, Zhenhui Song, and Liu Yang. 2025. "Establishment of a One–Pot RAA–CRISPR/Cas13a Assay-Based TGEV S Gene Detection" Veterinary Sciences 12, no. 5: 464. https://doi.org/10.3390/vetsci12050464
APA StyleLv, L., Mu, H., Li, S., Gao, J., Liu, M., Niu, S., Xu, G., Fu, L., Song, Z., & Yang, L. (2025). Establishment of a One–Pot RAA–CRISPR/Cas13a Assay-Based TGEV S Gene Detection. Veterinary Sciences, 12(5), 464. https://doi.org/10.3390/vetsci12050464







