Distinguished Loop-Mediated Isothermal Amplification Assay to Detect Porcine Epidemic Diarrhea Virus Genotypes I and II
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA and RNA Extraction
2.3. LAMP Primer Design
2.4. Plasmid Standard Construction
2.5. Conventional RT-PCR
2.6. RT-LAMP
2.7. Sensitivity and Specificity
2.8. Reproducibility
2.9. Detection of PEDV in Clinical Samples by Conventional RT-PCR and RT-LAMP
3. Results
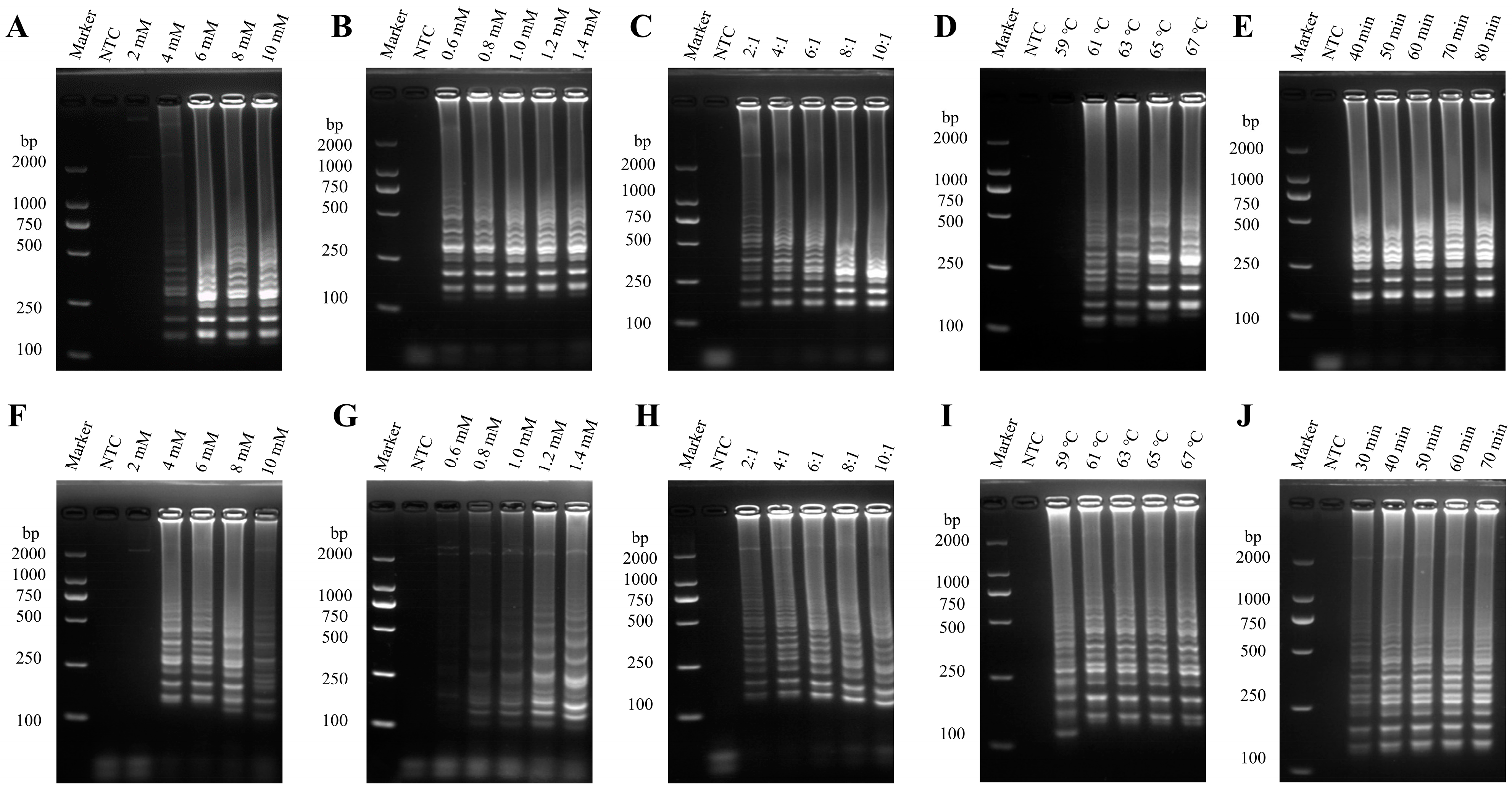
3.1. Optimization of the PEDV RT-LAMP Method
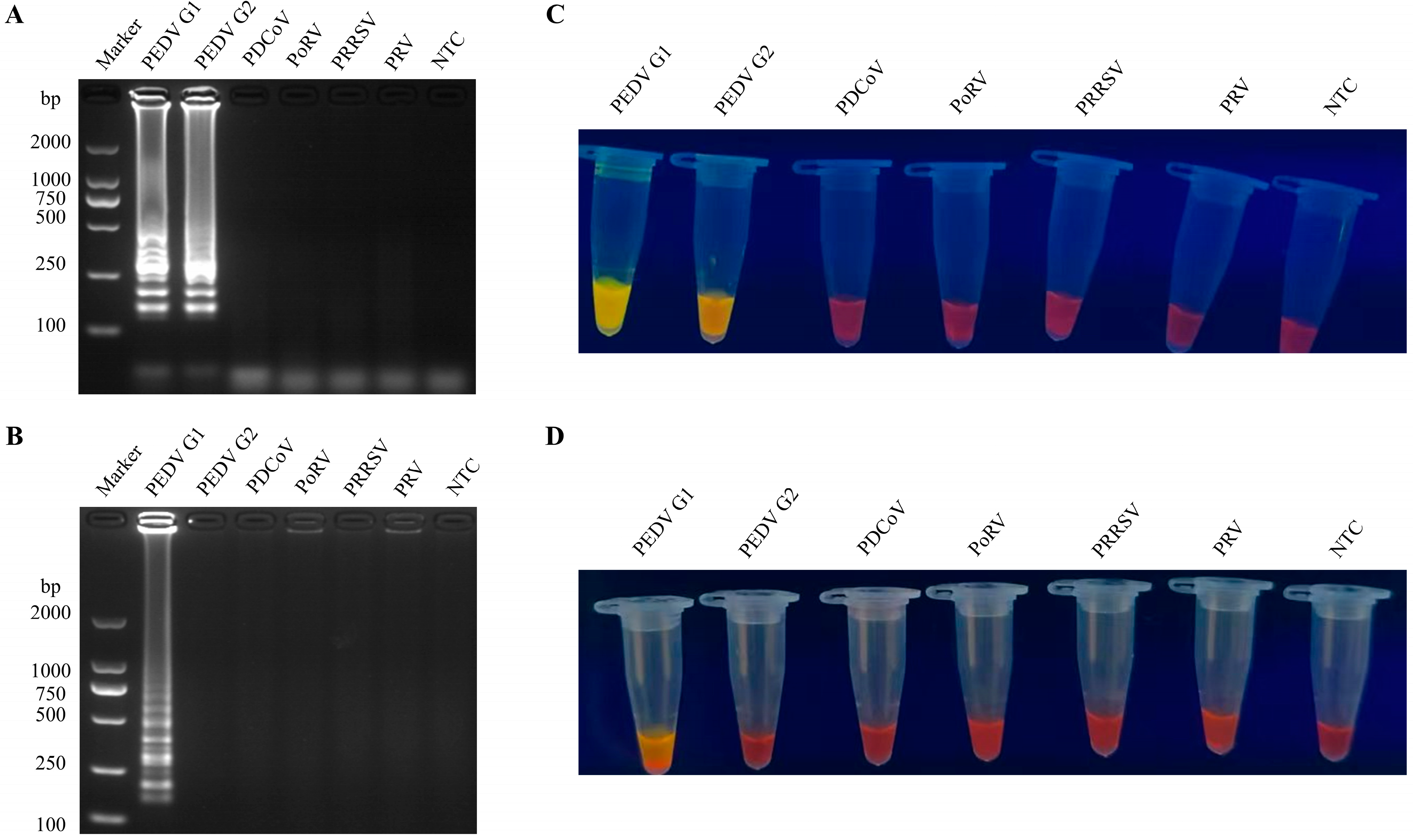
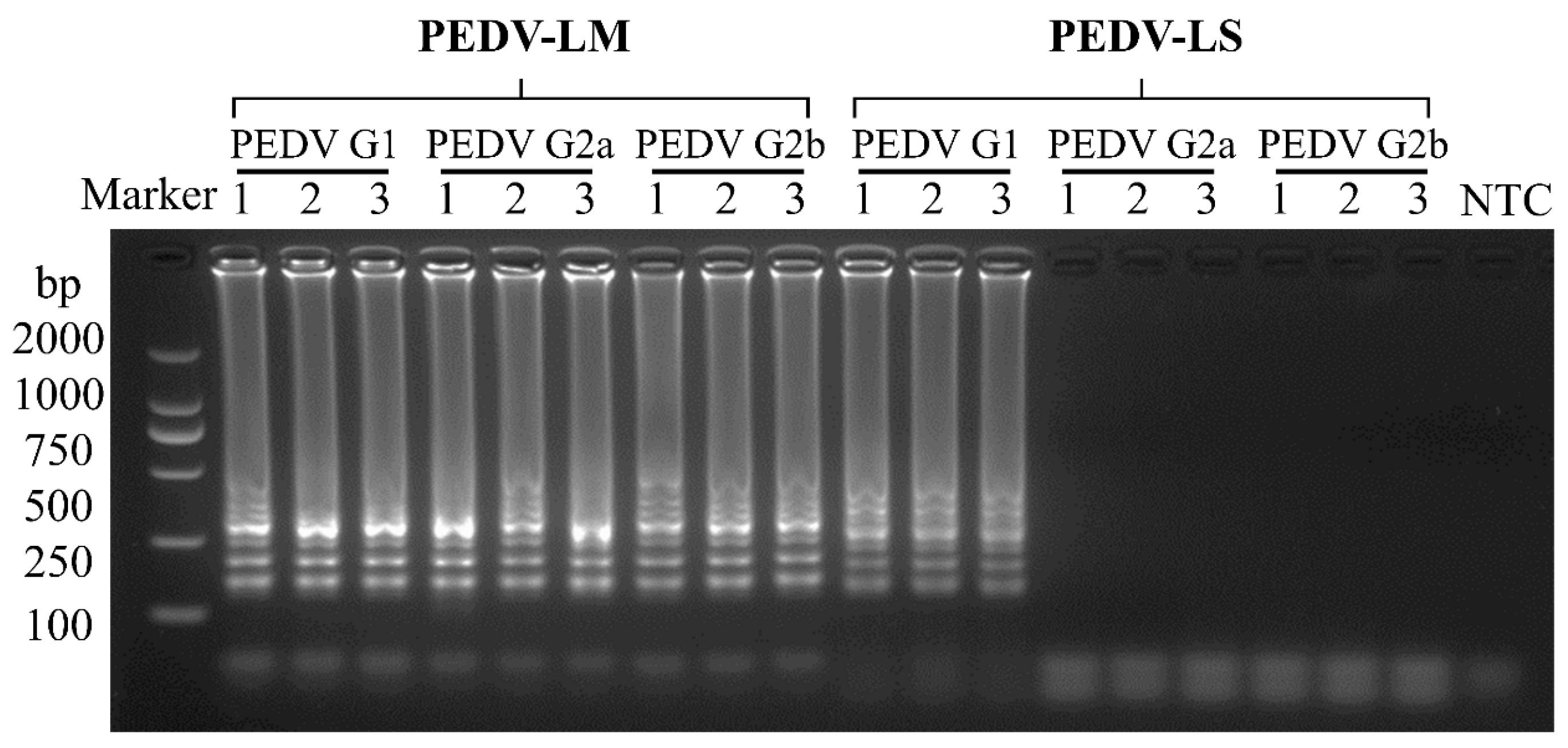
3.2. Specificity of the RT-LAMP Assay
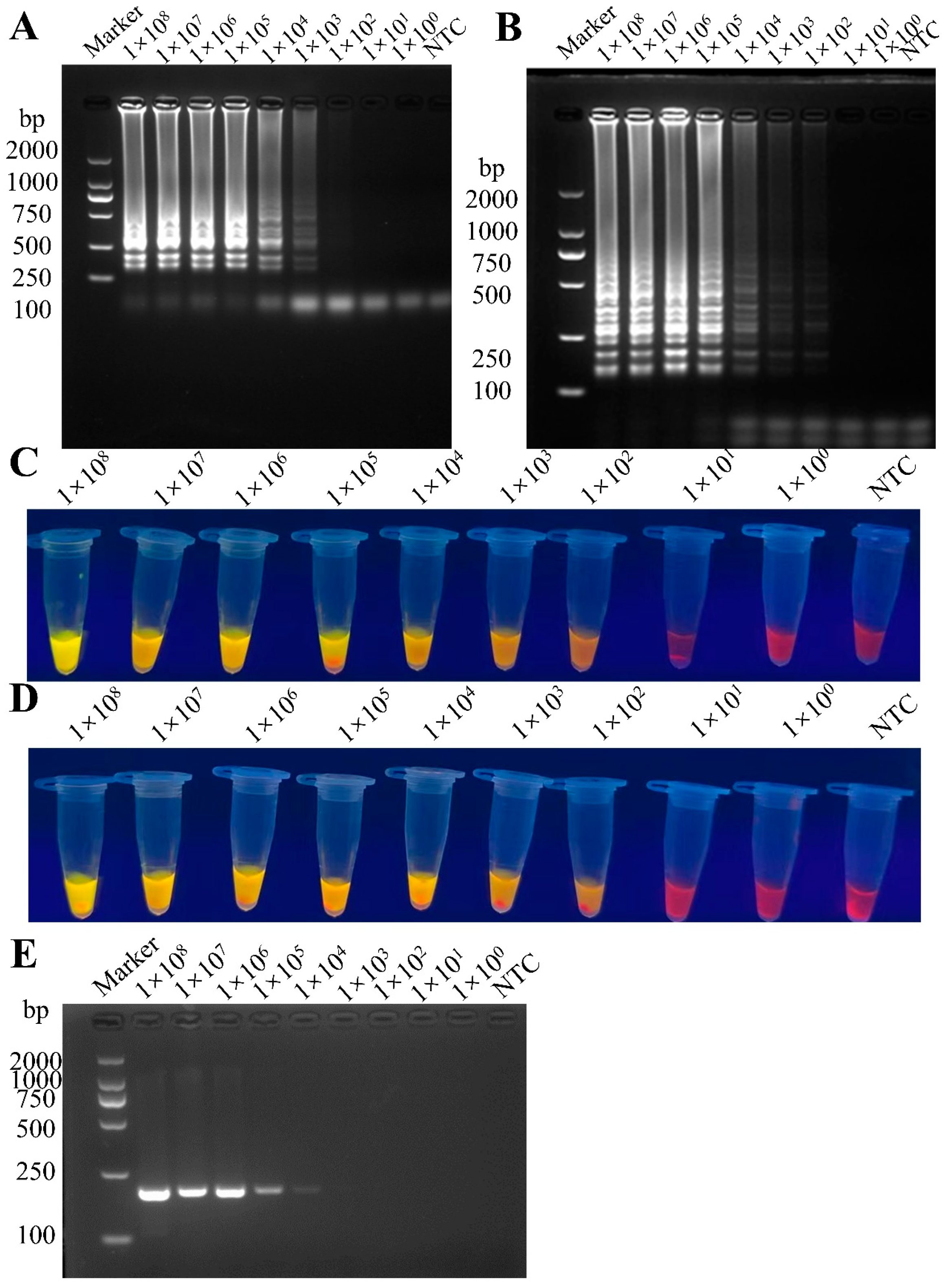
3.3. Sensitivity Comparison Between RT-LAMP and RT-PCR
3.4. Reproducibility of the RT-LAMP Assay
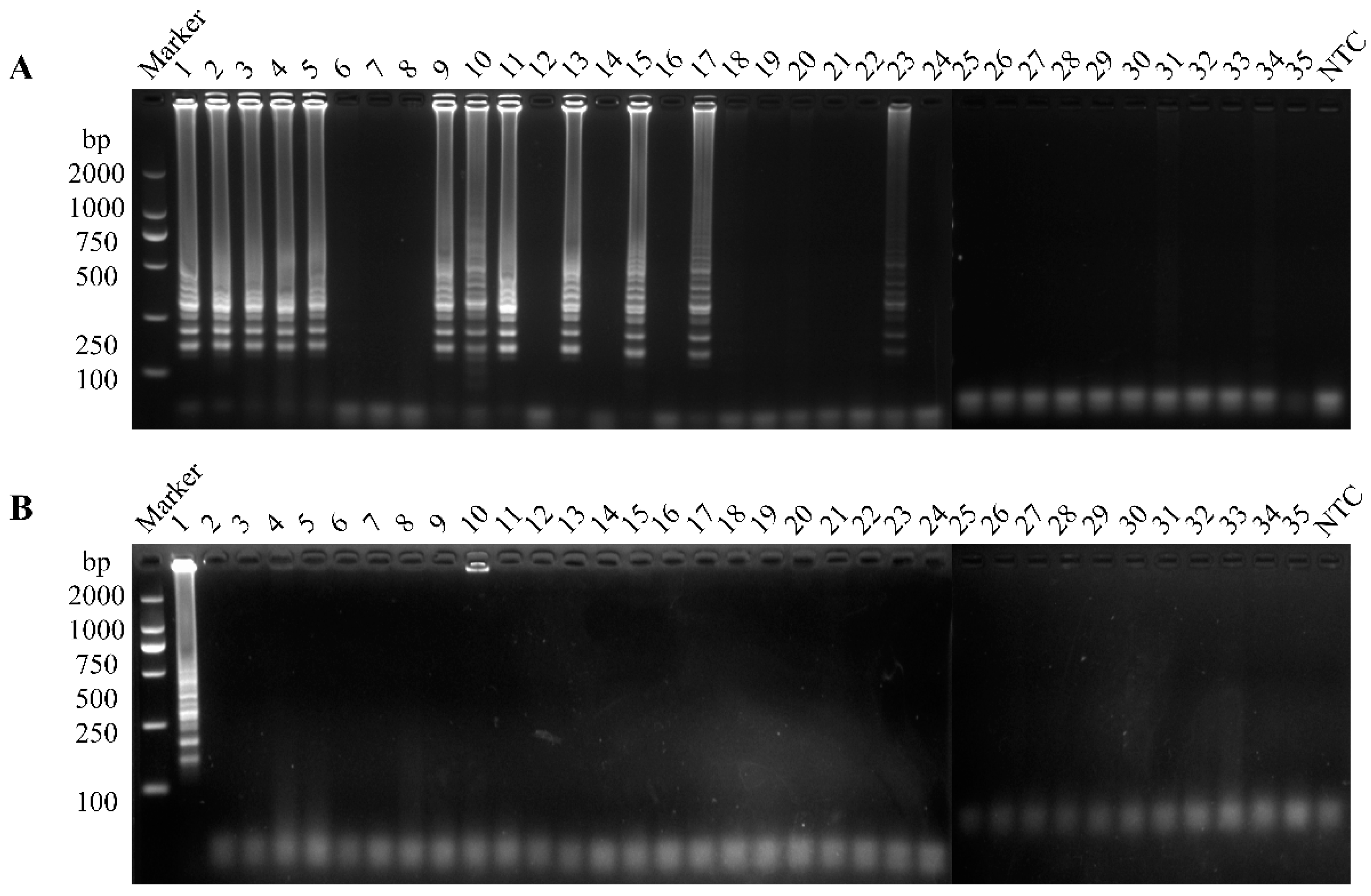
3.5. Clinical Sample Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, D.; Huang, D.; Peng, Q.; Huang, T.; Chen, Y.; Zhang, T.; Nie, X.; He, H.; Wang, P.; Liu, Q.; et al. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea viruses associated with outbreaks of severe diarrhea in piglets in Jiangxi, China 2013. PLoS ONE 2015, 10, e0120310. [Google Scholar] [CrossRef] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 204, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, Y.; Lin, C.; Tan, M.; Wan, P.; Xie, B.; Xiong, L.; Ji, H. Epidemiological monitoring and genetic variation analysis of pathogens associated with porcine viral diarrhea in southern China from 2021 to 2023. Front. Microbiol. 2024, 15, 1303915. [Google Scholar] [CrossRef]
- Sun, D.; Wang, X.; Wei, S.; Chen, J.; Feng, L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: A mini-review. J. Vet. Med. Sci. 2016, 78, 355–363. [Google Scholar] [CrossRef]
- Bai, J.; Du, C.; Lu, Y.; Wang, R.; Su, X.; Yu, K.; Qin, Q.; Chen, Y.; Wei, Z.; Huang, W.; et al. Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022. Animals 2023, 13, 1215. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef]
- Park, J.; Lee, C. Emergence and evolution of novel G2b-like porcine epidemic diarrhea virus inter-subgroup G1b recombinants. Arch. Virol. 2020, 165, 2471–2478. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.A.-O.; Lee, S.A.-O.; Lee, C.A.-O. Assessing the risk of recurrence of porcine epidemic diarrhea virus in affected farms on Jeju Island, South Korea. J. Vet. Sci. 2021, 22, e48. [Google Scholar] [CrossRef]
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 2001, 23, 137–144. [Google Scholar] [CrossRef]
- Lin, C.M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhang, H.; Li, L.; Yang, Y.; Zou, X.; Chen, J.; Tang, X. PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses 2022, 14, 1744. [Google Scholar] [CrossRef]
- Zhou, S.; Han, S.; Shi, J.; Wu, J.; Yuan, X.; Cong, X.; Xu, S.; Wu, X.; Li, J.; Wang, J. Loop-mediated isothermal amplification for detection of porcine circovirus type 2. Virol. J. 2011, 8, 497. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Li, C.; Liang, J.; Yang, D.; Zhang, Q.; Miao, D.; He, X.; Du, Y.; Zhang, W.; Ni, J.; Zhao, K. Visual and Rapid Detection of Porcine Epidemic Diarrhea Virus (PEDV) Using Reverse Transcription Loop-Mediated Isothermal Amplification Method. Animals 2022, 12, 2712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ye, Y.; Song, D.; Guo, N.; Peng, Q.; Li, A.; Zhou, X.; Chen, Y.; Zhang, M.; Huang, D.; et al. A simple and rapid identification method for newly emerged porcine Deltacoronavirus with loop-mediated isothermal amplification. Biol. Res. 2017, 50, 30. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Lee, E.; Park, S.; Lim, C.S.; Jang, W.S. Enhanced Point-of-Care SARS-CoV-2 Detection: Integrating RT-LAMP with Microscanning. Biosensors 2024, 14, 348. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Li, T.; Zhang, G.; Cao, J.; Jin, Y.; Xing, G.; Liao, M.; Zhou, J. Development of a loop-mediated isothermal amplification method to rapidly detect porcine circovirus genotypes 2a and 2b. Virol. J. 2012, 9, 318. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, Z.; Zhou, Z.; Sun, J.; Yan, S.; Gao, W.; Shao, Y.; Bai, Y.; Wu, Y.; Yan, Z.; et al. A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses 2022, 14, 1819. [Google Scholar] [CrossRef]
- Yu, K.; Liu, X.; Lu, Y.; Long, M.; Bai, J.; Qin, Q.; Su, X.; He, G.; Mi, X.; Yang, C.; et al. Biological Characteristics and Pathogenicity Analysis of a Low Virulence G2a Porcine Epidemic Diarrhea Virus. Microbiol. Spectr. 2023, 11, e0453522. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Y.; Chen, Y.; Xing, G.; Hao, H.; Wei, Q.; Liang, Y.; Xie, W.; Li, D.; Huang, H.; et al. A novel duplex TaqMan probe-based real-time RT-qPCR for detecting and differentiating classical and variant porcine epidemic diarrhea viruses. Mol. Cell. Probes 2018, 37, 6–11. [Google Scholar] [CrossRef]
- Gill, P.; Hadian Amree, A. AS-LAMP: A New and Alternative Method for Genotyping. Avicenna J. Med. Biotechnol. 2020, 12, 2–8. [Google Scholar]
- Badolo, A.; Bando, H.; Traoré, A.; Ko-Ketsu, M.; Guelbeogo, W.M.; Kanuka, H.; Ranson, H.; Sagnon, N.; Fukumoto, S. Detection of G119S ace-1 (R) mutation in field-collected Anopheles gambiae mosquitoes using allele-specific loop-mediated isothermal amplification (AS-LAMP) method. Malar. J. 2015, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Kampeera, J.; Chareanchim, W.; Rattanajak, R.; Pornthanakasem, W.; Kiatpathomchai, W.; Kongkasuri-yachai, D. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol. Int. 2017, 66, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Li, B.; Shi, Y.; Feng, S.; Yin, Y.; Long, F.; Pan, Y.; Wei, Y. Phylogenetic and Evolutionary Analysis of Porcine Epidemic Diarrhea Virus in Guangxi Province, China, during 2020 and 2024. Viruses 2024, 16, 1126. [Google Scholar] [CrossRef]
- Zhao, P.D.; Bai, J.; Jiang, P.; Tang, T.S.; Li, Y.; Tan, C.; Shi, X. Development of a multiplex TaqMan probe-based real-time PCR for discrimination of variant and classical porcine epidemic diarrhea virus. J. Virol. Methods 2014, 206, 150–155. [Google Scholar] [CrossRef]
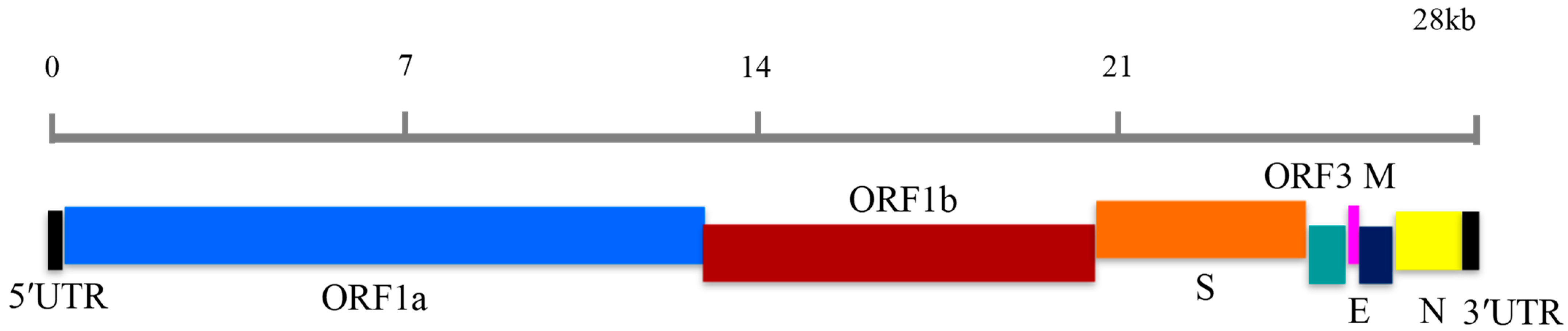
| Primer Set | Primer ID | Sequence (5′-3′) |
|---|---|---|
| PEDV-LM | PEDV-LM-FIP | CCTGTACGCCAGTAGCAACCTTGAGCACCAACTGGTGTAACG |
| PEDV-LM-BIP | ATTTCGTCACAGTCGCCAAGGCGACTGAACGACCAACACGT | |
| PEDV-LM-F3 | TCTGTGATGGGCCGACAG | |
| PEDV-LM-B3 | CCAGTGCCAGATGAAGCATT | |
| PEDV-LS | PEDV-LS-FIP | ACTAGCAGTTTCAATGCCTGTGGTTACCTACCTAGTATGAACTCT |
| PEDV-LS-BIP | GGCGTTCATGGTATTTTTCTCAGCCTAGGATCAAACGGCTCTTG | |
| PEDV-LS-F3 | AAATTTAATGTTCAGGCACCT | |
| PEDV-LS-B3 | GTGGCCTTATGTAAATAAAGCT | |
| qPCR [19] | PEDV-GI-F | TGTTTTGGGTGGTTATCTACCTA |
| PEDV-GI-R | AGCTGGTAACCACTAGGAT | |
| PEDV-GII-F | CCAGTACTTTCAACACTTAGCCTA | |
| PEDV-GII-R | GCCACTAGCAGTTGGATG |
| PEDV Genotype | Sample No. | qPCR | RT-LAMP | Positive Rate | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | qPCR | RT-LAMP | ||
| GI | 35 | 1 | 23 | 1 | 23 | 34.3% | 34.3% |
| GII | 11 | 11 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, L.; Fang, M.; Wei, X.; Li, J.; Wu, Q.; Yang, X.; Ye, Y.; Wan, G.; Huang, D.; et al. Distinguished Loop-Mediated Isothermal Amplification Assay to Detect Porcine Epidemic Diarrhea Virus Genotypes I and II. Vet. Sci. 2025, 12, 399. https://doi.org/10.3390/vetsci12050399
Liu Z, Li L, Fang M, Wei X, Li J, Wu Q, Yang X, Ye Y, Wan G, Huang D, et al. Distinguished Loop-Mediated Isothermal Amplification Assay to Detect Porcine Epidemic Diarrhea Virus Genotypes I and II. Veterinary Sciences. 2025; 12(5):399. https://doi.org/10.3390/vetsci12050399
Chicago/Turabian StyleLiu, Zhong, Lanlan Li, Mengtao Fang, Xiaoqing Wei, Jieqiong Li, Qi Wu, Xiaoxue Yang, Yu Ye, Gen Wan, Dongyan Huang, and et al. 2025. "Distinguished Loop-Mediated Isothermal Amplification Assay to Detect Porcine Epidemic Diarrhea Virus Genotypes I and II" Veterinary Sciences 12, no. 5: 399. https://doi.org/10.3390/vetsci12050399
APA StyleLiu, Z., Li, L., Fang, M., Wei, X., Li, J., Wu, Q., Yang, X., Ye, Y., Wan, G., Huang, D., & Song, D. (2025). Distinguished Loop-Mediated Isothermal Amplification Assay to Detect Porcine Epidemic Diarrhea Virus Genotypes I and II. Veterinary Sciences, 12(5), 399. https://doi.org/10.3390/vetsci12050399






