Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 in Henan, China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. DNA Extraction and PCR Detection of PCV2
2.3. Amplification and Sequencing of Complete Genome of PCV2 Strains
2.4. Sequence Comparison and Phylogenetic Analysis
2.5. Rates of Substitution
3. Results
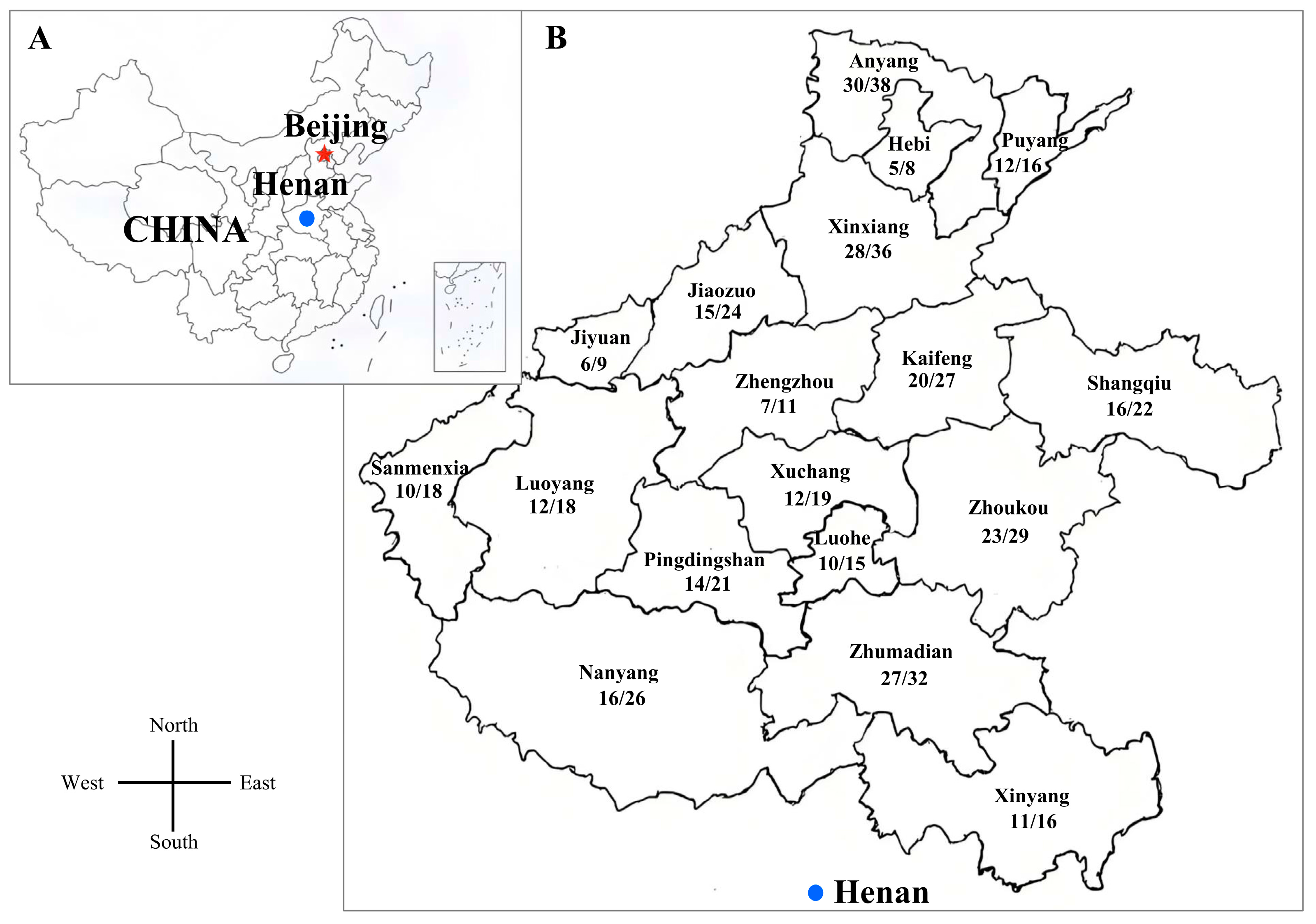
3.1. The Frequency of PCV2 in Different Regions During 2020 and 2023
3.2. Genome Sequence Analysis of PCV2
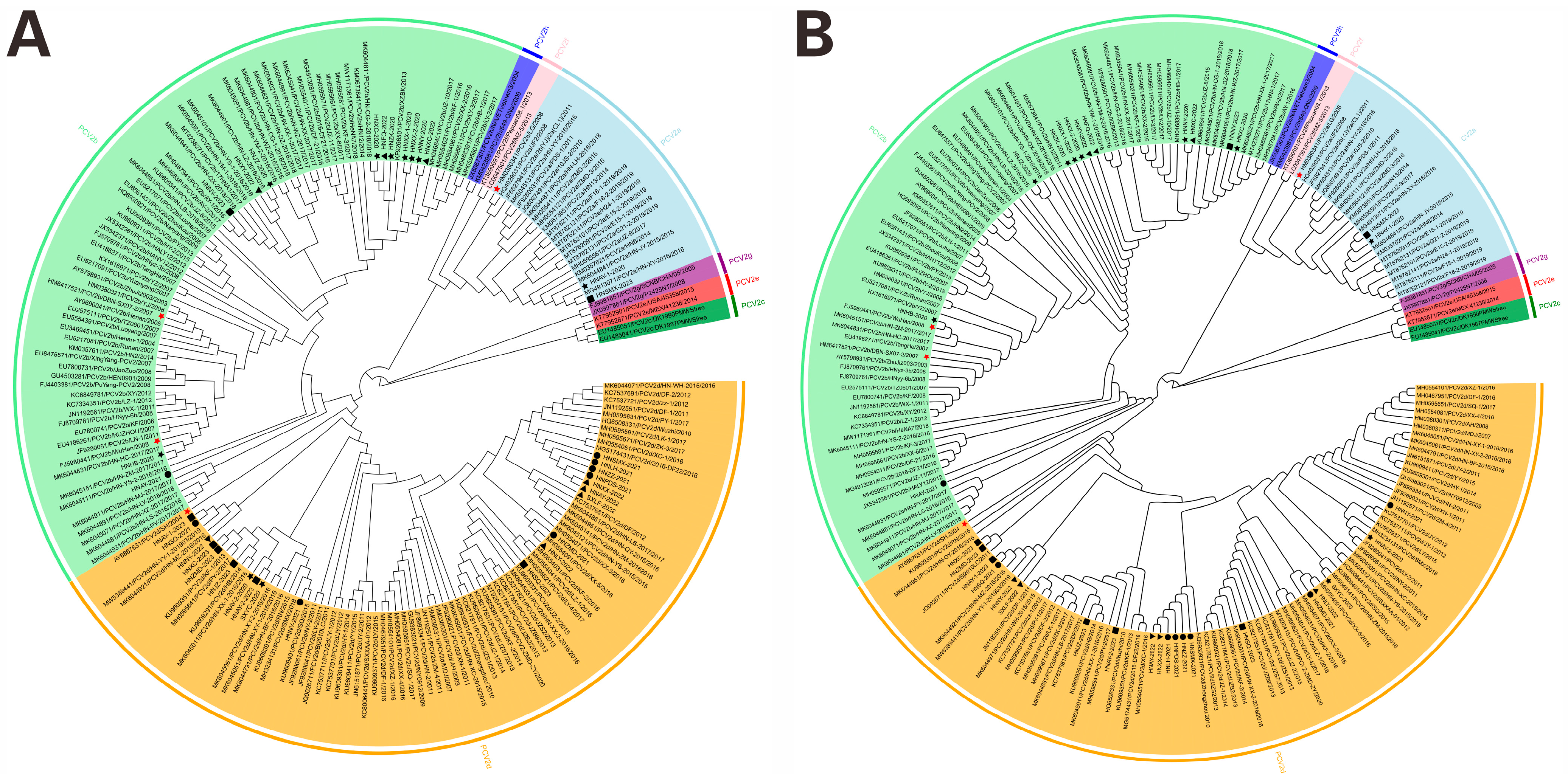
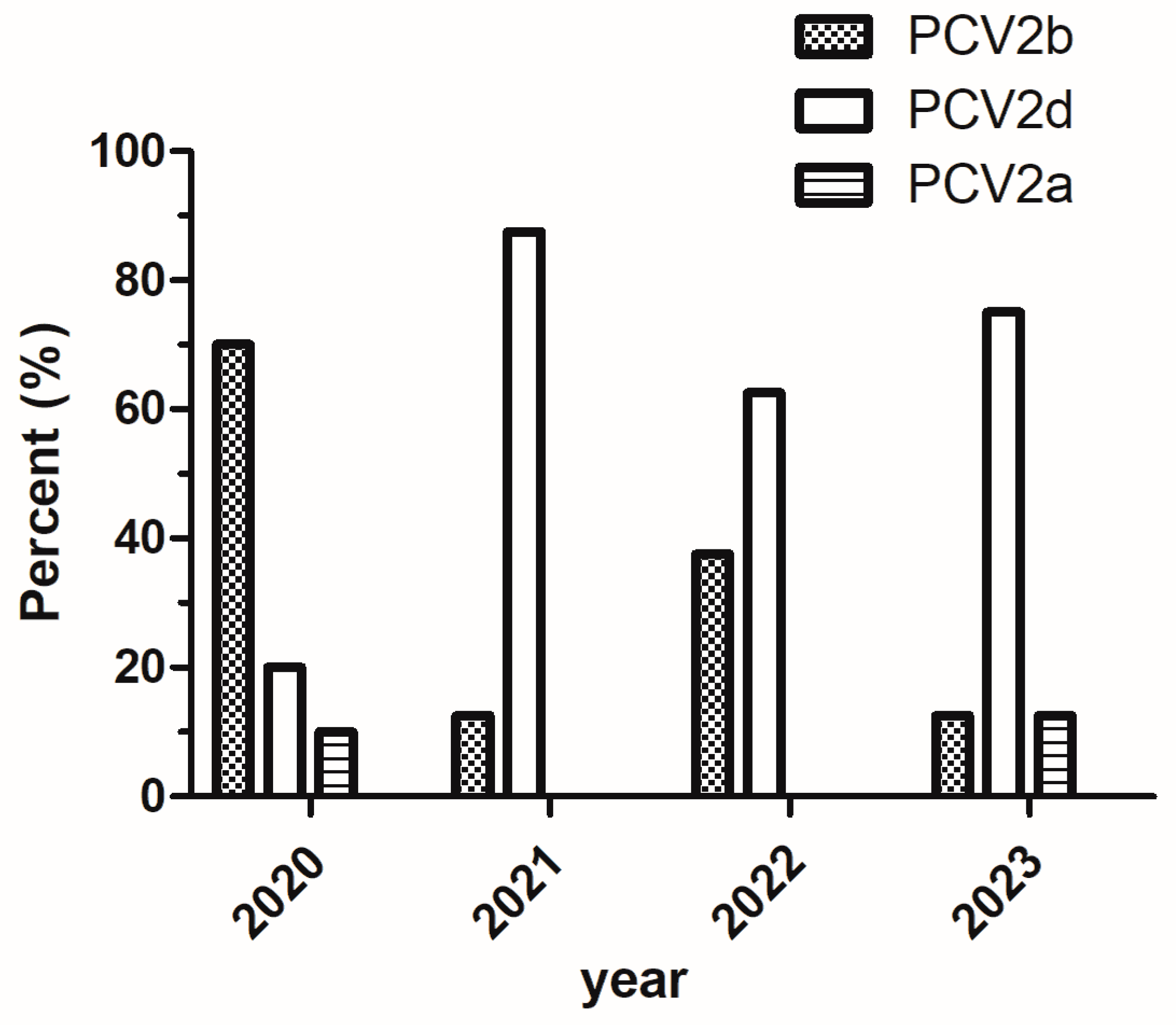
3.3. Phylogenetic Analysis of PCV2 Strains
3.4. Mutation Analysis of Cap Proteins
3.5. Rates of Substitution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Xu, T.; Wen, J.; Yang, L.; Lai, S.; Sun, X.; Xu, Z.; Zhu, L. Prevalence and phylogenetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) in the Southwest of China during 2020–2022. Front. Vet. Sci. 2022, 9, 1042792. [Google Scholar] [CrossRef]
- Kang, L.; Wahaab, A.; Shi, K.; Mustafa, B.E.; Zhang, Y.; Zhang, J.J.; Li, Z.J.; QIU, Y.F. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China. Viruses 2022, 14, 289. [Google Scholar] [CrossRef]
- Li, N.; Liu, J.; Qi, J.; Hao, F.; Xu, L.; Guo, K. Genetic Diversity and Prevalence of Porcine Circovirus Type 2 in China During 2000–2019. Front. Vet. Sci. 2021, 8, 788172. [Google Scholar] [CrossRef]
- Tegegne, D.; Tsegaye, G.; Aman, S.; Faustini, G.; Franzo, G. Molecular Epidemiology and Genetic Characterization of PCV2 Circulating in Wild Boars in Southwestern Ethiopia. J. Trop. Med. 2022, 2022, 5185247. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, Q.L.; Nie, L.B.; Wang, Q.; Ge, G.Y.; Li, D.L.; Ma, B.Y.; Sheng, C.Y.; Su, N.; Zong, Y.; et al. Prevalence of porcine circovirus 2 throughout China in 2015-2019: A systematic review and meta-analysis. Microb. Pathog. 2020, 149, 104490. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016-2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef]
- de Sousa Moreira, A.; Santos-Silva, S.; Mega, J.; Palmeira, J.D.; Torres, R.T.; Mesquita, J.R. Epidemiology of Porcine Circovirus Type 2 Circulating in Wild Boars of Portugal during the 2018-2020 Hunting Seasons Suggests the Emergence of Genotype 2d. Animals 2022, 12, 451. [Google Scholar] [CrossRef]
- Ouyang, T.; Zhang, X.W.; Liu, X.H.; Ren, L.Z. Co-Infection of Swine with Porcine Circovirus Type 2 and Other Swine Viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Harding, J.C. The clinical expression and emergence of porcine circovirus 2. Vet. Microbiol. 2004, 98, 131–135. [Google Scholar] [CrossRef]
- Rose, N.; Opriessnig, T.; Grasland, B.; Jestin, A. Epidemiology and transmission of porcine circovirus type 2 (PCV2). Virus Res. 2012, 164, 78–89. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, M.; Liu, Z.; Meng, F.; Wang, H.; Cao, L.; Li, Y.; Jiao, Q.; Han, Z.; Liu, S. Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong province in China from 2015 to 2018. BMC Vet. Res. 2021, 17, 17. [Google Scholar] [CrossRef]
- Chae, C. Porcine respiratory disease complex: Interaction of vaccination and porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, and Mycoplasma hyopneumoniae. Vet. J. 2016, 212, 1–6. [Google Scholar] [CrossRef]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef]
- Franzo, G.; Cortey, M.; de Castro, A.M.; Piovezan, U.; Szabo, M.P.; Drigo, M.; Segalés, J.; Richtzenhain, L.J. Genetic characterisation of Porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Vet. Microbiol. 2015, 178, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J. Gen. Virol. 2015, 96, 1830–1841. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; He, D.S.; Qi, W.B.; Chen, S.N.; Deng, S.F.; Hu, J.; Li, X.P.; Li, L.; Chen, R.A.; Luo, M.L.; et al. Complete genome characterization and phylogenetic analysis of three distinct buffalo-origin PCV2 isolates from China. Infect. Genet. Evol. 2014, 28, 278–282. [Google Scholar] [CrossRef]
- Guo, L.J.; Lu, Y.H.; Wei, Y.W.; Huang, L.P.; Liu, C.M. Porcine circovirus type 2 (PCV2): Genetic variation and newly emerging genotypes in China. Virol. J. 2010, 7, 273. [Google Scholar] [CrossRef]
- Jantafong, T.; Boonsoongnern, A.; Poolperm, P.; Urairong, K.; Lekcharoensuk, C.; Lekcharoensuk, P. Genetic characterization of porcine circovirus type 2 in piglets from PMWS-affected and -negative farms in Thailand. Virol. J. 2011, 8, 88. [Google Scholar] [CrossRef]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef]
- Yang, X.; Chen, F.; Cao, Y.; Pang, D.; Ouyang, H.; Ren, L. Complete genome sequence of porcine circovirus 2b strain CC1. J. Virol. 2012, 86, 9536. [Google Scholar] [CrossRef]
- Mankertz, A.; Mankertz, J.; Wolf, K.; Buhk, H.J. Identification of a protein essential for replication of porcine circovirus. J. Gen. Virol. 1998, 79, 381–384. [Google Scholar] [CrossRef]
- Finsterbusch, T.; Mankertz, A. Porcine circoviruses--small but powerful. Virus Res. 2009, 143, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef]
- Shi, R.; Hou, L.; Liu, J. Host immune response to infection with porcine circoviruses. Anim. Dis. 2021, 1, 23. [Google Scholar] [CrossRef]
- Olvera, A.; Cortey, M.; Segalés, J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, Y.H.; Tian, R.B.; Hou, C.Y.; Li, X.S.; Zheng, L.L.; Wang, L.Q.; Chen, H.Y. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in central China. Infect. Genet. Evol. 2021, 94, 105016. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, P.; Rushton, J.; Wieland, B. Cost of post-weaning multi-systemic wasting syndrome and porcine circovirus type-2 subclinical infection in England—An economic disease model. Prev. Vet. Med. 2013, 110, 88–102. [Google Scholar] [CrossRef]
- Lang, H.W.; Zhang, G.C.; Wu, F.Q.; Zhang, C.G. Detection of serum antibody against postweaning multisystemic wasting syndrome in pigs. Chin. J. Vet. Sci. Technol. 2000, 30, 3–5. [Google Scholar]
- Wang, S.Y.; Sun, Y.F.; Wang, Q.; Yu, L.X.; Zhu, S.Q.; Liu, X.M.; Yao, Y.; Wang, J.; Shan, T.L.; Zheng, H.; et al. An epidemiological investigation of porcine circovirus type 2 and porcine circovirus type 3 infections in Tianjin, North China. PeerJ 2020, 8, e9735. [Google Scholar] [CrossRef]
- Zheng, G.; Lu, Q.; Wang, F.; Xing, G.; Feng, H.; Jin, Q.; Guo, Z.; Teng, M.; Hao, H.; Li, D.; et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet. Res. 2020, 16, 6. [Google Scholar] [CrossRef]
- Jia, Y.; Zhu, Q.; Xu, T.; Chen, X.; Li, H.; Ma, M.; Zhang, Y.; He, Z.; Chen, H. Detection and genetic characteristics of porcine circovirus type 2 and 3 in Henan province of China. Mol. Cell Probes 2022, 61, 101790. [Google Scholar] [CrossRef]
- Han, L.; Yuan, G.F.; Chen, S.J.; Dai, F.; Hou, L.S.; Fan, J.H.; Zuo, Y.Z. Porcine circovirus type 2 (PCV2) infection in Hebei Province from 2016 to 2019: A retrospective study. Arch. Virol. 2021, 166, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Cortey, M.; Segalés, J.; Hughes, J.; Drigo, M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenetics Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef]
- Xiao, Q.; Wen, L.; Yin, L.; Zhang, F.; Li, T.; Banma, Z.; He, K.; Suolang, S.; Nan, W.; Wu, J.; et al. Prevalence and genetic diversity of porcine circovirus type 2 in northern Guangdong Province during 2016–2021. Front. Vet. Sci. 2022, 9, 932612. [Google Scholar] [CrossRef]
- Xu, G.; Sarkar, A.; Qian, L.; Shuxia, Z.; Rahman, M.A.; Yongfeng, T. The impact of the epidemic experience on the recovery of production of pig farmers after the outbreak-Evidence from the impact of African swine fever (ASF) in Chinese pig farming. Prev. Vet. Med. 2022, 199, 105568. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.L.; Zhao, Y.; Zheng, H.H.; Tian, R.B.; Han, H.Y.; Chen, H.Y.; Zheng, L.L. Analysis of genetic variation of porcine circovirus type 2 within pig populations in central China. Arch. Virol. 2019, 164, 1445–1451. [Google Scholar] [CrossRef]
- Hu, Y.; Zhan, Y.; Wang, D.; Xie, X.; Liu, T.; Liu, W.; Wang, N.; Deng, Z.; Lei, H.; Yang, Y.; et al. Evidence of natural co-infection with PCV2b subtypes in vivo. Arch. Virol. 2017, 162, 2015–2020. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Hill, V.; Baele, G. Bayesian Estimation of Past Population Dynamics in BEAST 1.10 Using the Skygrid Coalescent Model. Mol. Biol. Evol. 2019, 36, 2620–2628. [Google Scholar] [CrossRef]
- Suh, J.; Oh, T.; Park, K.; Yang, S.; Cho, H.; Chae, C. A Comparison of Virulence of Three Porcine Circovirus Type 2 (PCV2) Genotypes (a, b, and d) in Pigs Singularly Inoculated with PCV2 and Dually Inoculated with PCV2 and Porcine Reproductive and Respiratory Syndrome Virus. Pathogens 2021, 10, 891. [Google Scholar] [CrossRef]
- Constans, M.; Ssemadaali, M.; Kolyvushko, O.; Ramamoorthy, S. Antigenic Determinants of Possible Vaccine Escape by Porcine Circovirus Subtype 2b Viruses. Bioinform. Biol. Insights 2015, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Wang, H.; Feng, Y.; Song, M.; Li, Q.; Li, J. Genetic variation and phylogenetic analysis of Porcine circovirus type 2 in China from 2016 to 2018. Acta Virol. 2019, 63, 459–468. [Google Scholar] [CrossRef]
- Yue, W.; Li, Y.; Zhang, X.; He, J.; Ma, H. Prevalence of Porcine Circoviruses in Slaughterhouses in Central Shanxi Province, China. Front. Vet. Sci. 2022, 9, 820914. [Google Scholar] [CrossRef]
- Jang, G.; Yoo, H.; Kim, Y.; Yang, K.; Lee, C. Genetic and phylogenetic analysis of porcine circovirus type 2 on Jeju Island, South Korea, 2019-2020: Evidence of a novel intergenotypic recombinant. Arch. Virol. 2021, 166, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Lv, N.; Zhu, L.; Li, W.; Li, Z.; Qian, Q.; Zhang, T.; Liu, L.; Hong, J.; Zheng, X.; Wang, Y.; et al. Molecular epidemiology and genetic variation analyses of porcine circovirus type 2 isolated from Yunnan Province in China from 2016–2019. TBMC Vet. Res. 2020, 16, 96. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Long, Y.; Yang, L.; Song, W.; Liu, J.; Li, Q.; Liang, G.; Yu, D.; Huang, C.; et al. Epidemiological Analysis From 2018 to 2020 in China and Prevention Strategy of Porcine Circovirus Type 2. Front. Vet. Sci. 2021, 8, 753297. [Google Scholar] [CrossRef]
- Xu, T.; Hou, C.Y.; Zhang, Y.H.; Li, H.X.; Chen, X.M.; Pan, J.J.; Chen, H.Y. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 2022, 808, 145991. [Google Scholar] [CrossRef]
- Zhan, Y.; Yu, W.; Cai, X.; Lei, X.; Lei, H.; Wang, A.; Sun, Y.; Wang, N.; Deng, Z.; Yang, Y. The Carboxyl Terminus of the Porcine Circovirus Type 2 Capsid Protein Is Critical to Virus-Like Particle Assembly, Cell Entry, and Propagation. J. Virol. 2020, 94, e00042-00020. [Google Scholar] [CrossRef]
- Hou, Q.; Hou, S.; Chen, Q.; Jia, H.; Xin, T.; Jiang, Y.; Guo, X.; Zhu, H. Nuclear localization signal regulates porcine circovirus type 2 capsid protein nuclear export through phosphorylation. Virus Res. 2018, 246, 12–22. [Google Scholar] [CrossRef]
- Lekcharoensuk, P.; Morozov, I.; Paul, P.S.; Thangthumniyom, N.; Wajjawalku, W.; Meng, X.J. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 2004, 78, 8135–8145. [Google Scholar] [CrossRef]
- Tan, C.Y.; Thanawongnuwech, R.; Arshad, S.S.; Hassan, L.; Fong, M.W.C.; Ooi, P.T. Genotype Shift of Malaysian Porcine Circovirus 2 (PCV2) from PCV2b to PCV2d within a Decade. Animals 2022, 12, 1849. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Mahé, D.; Blanchard, P.; Truong, C.; Arnauld, C.; Le Cann, P.; Cariolet, R.; Madec, F.; Albina, E.; Jestin, A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 2000, 81, 1815–1824. [Google Scholar] [CrossRef]
- Larochelle, R.; Magar, R.; D’Allaire, S. Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 2002, 90, 101–112. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Sequence (5′−3′) | Length | Annealing Temperature | Purpose |
|---|---|---|---|---|
| PCV2-F | CTGTTTTCGAACGCAGTGCC | 486 bp | 56 °C | Detection of PCV2 [36] |
| PCV2-R | GCATCTTCAACACCCGCCT | |||
| PCV2-C-F | CGGGGTACCACTGAGTCTTTTTTATCACTTCG | 1767–1768 bp | 58 °C | PCV2 whole-genome amplification [37] |
| PCV2-C-R | CCCAAGCTTAAGACTCAGTAATTTATTTCATATGG |
| Region | No. of Tested Samples | No. of PCV2 Positive Samples | Frequency |
|---|---|---|---|
| Anyang | 38 | 30 | 78.95% |
| Puyang | 16 | 12 | 75.00% |
| Hebi | 8 | 5 | 62.50% |
| Xinxiang | 36 | 28 | 77.78% |
| Jiaozuo | 24 | 15 | 62.50% |
| Jiyuan | 9 | 6 | 66.67% |
| Zhenzhou | 11 | 7 | 63.64% |
| Kaifeng | 27 | 20 | 74.07% |
| Shangqiu | 22 | 16 | 72.73% |
| Zhoukou | 29 | 23 | 79.31% |
| Xuchang | 19 | 12 | 63.16% |
| Luohe | 15 | 10 | 66.67% |
| Zhumadian | 32 | 27 | 84.38% |
| Pingdingshan | 21 | 14 | 66.67% |
| Nanyang | 26 | 16 | 61.54% |
| Luoyang | 18 | 12 | 66.67% |
| Sanmenxia | 18 | 10 | 55.56% |
| Xinyang | 16 | 11 | 68.75% |
| Total | 385 | 274 | 71.17% |
| Strain | Collection Year | Isolation Origin | Genotype | Accession No. | Length (nt) |
|---|---|---|---|---|---|
| HNZK-2020 | 2020 | Zhoukou, Henan | PCV2b | OQ749408 | 1767 |
| HNXX-1-2020 | 2020 | Xinxiang, Henan | PCV2b | OQ749409 | 1767 |
| HNNY-2020 | 2020 | Nanyang, Henan | PCV2b | OQ749410 | 1767 |
| HNJZ-2020 | 2020 | Jiaozuo, Henan | PCV2b | OQ749411 | 1767 |
| HNXC-2020 | 2020 | Xuchang, Henan | PCV2b | OQ749412 | 1767 |
| HNAY-1-2020 | 2020 | Anyang, Henan | PCV2a | OQ749413 | 1767 |
| HNXX-2-2020 | 2020 | Xinxiang, Henan | PCV2b | OQ749414 | 1767 |
| HNHB-2020 | 2020 | Hebi, Henan | PCV2b | OQ749415 | 1767 |
| HNAY-2-2020 | 2020 | Anyang, Henan | PCV2d | OQ749416 | 1767 |
| HNJY-2020 | 2020 | Jiyuan, Henan | PCV2d | OQ749417 | 1767 |
| HNAY-2021 | 2021 | Anyang, Henan | PCV2b | OQ749418 | 1767 |
| HNZZ-2021 | 2021 | Zhengzhou, Henan | PCV2d | OQ749419 | 1767 |
| HNPDS-2021 | 2021 | Pingdingshan, Henan | PCV2d | OQ749420 | 1767 |
| HNSQ-2021 | 2021 | Shangqiu, Henan | PCV2d | OQ749421 | 1767 |
| HNLH-2021 | 2021 | Luohe, Henan | PCV2d | OQ749422 | 1767 |
| HNNY-2021 | 2021 | Nanyang, Henan | PCV2d | OQ749423 | 1767 |
| HNZMD-2021 | 2021 | Zhumadian, Henan | PCV2d | OQ749424 | 1767 |
| HNSMX-2021 | 2021 | Sanmenxia, Henan | PCV2d | OQ749425 | 1767 |
| HNXX-2022 | 2022 | Xinxiang, Henan | PCV2d | OQ749426 | 1767 |
| HNAY-2022 | 2022 | Anyang, Henan | PCV2d | OQ749427 | 1767 |
| HNXY-2022 | 2022 | Xinyang, Henan | PCV2b | OQ749428 | 1767 |
| HNXC-2022 | 2022 | Xuchang, Henan | PCV2b | OQ749429 | 1767 |
| HNLY-2022 | 2022 | Luoyang, Henan | PCV2d | OQ749430 | 1767 |
| HNNY-2022 | 2022 | Nanyang, Henan | PCV2d | OQ749431 | 1767 |
| HNFQ-2022 | 2022 | Xinxiang, Henan | PCV2b | OQ749432 | 1767 |
| HNLY-2022 | 2022 | Luoyang, Henan | PCV2d | OQ749433 | 1767 |
| HNJZ-2023 | 2023 | Jiaozuo, Henan | PCV2d | OR114683 | 1767 |
| HNZMD-2023 | 2023 | Zhumadian, Henan | PCV2d | OR114684 | 1767 |
| HNXC-2023 | 2023 | Xuchang, Henan | PCV2d | OR114685 | 1767 |
| HNSQ-2023 | 2023 | Shangqiu, Henan | PCV2d | OR114686 | 1767 |
| HNNY-2023 | 2023 | Nanyang, Henan | PCV2b | OR114687 | 1767 |
| HNAY-1-2023 | 2023 | Anyang, Henan | PCV2d | OR114688 | 1767 |
| HNSMX-2023 | 2023 | Sanmenxia, Henan | PCV2a | OR114689 | 1767 |
| HNAY-2-2023 | 2023 | Anyang, Henan | PCV2d | OR114690 | 1767 |
| Position | PCV2a | PCV2b | PCV2d | Position | PCV2a | PCV2b | PCV2d |
|---|---|---|---|---|---|---|---|
| 8 | F | F(1/12/Y(11/12) | F(16/20)/Y(4/20) | 121 | S | S | T |
| 47 | A | T | T | 131 | M | T | T |
| 53 | F | F(11/12)/I(1/12) | I | 133 | V | A | A |
| 57 | V | I | V | 134 | P | T | N |
| 59 | A | K(11/12)/R(1/12) | K | 136 | Q | L | L |
| 63 | S | K(1/12)/R(11/12) | R | 151 | P | T | T |
| 68 | S | A | N | 169 | S | S(11/12)/C(1/12) | R(3/20)/G(17/20) |
| 73 | L | M | M | 185 | M | L | L |
| 77 | L | I | I | 190 | S | A(1/12)/T(11/12) | T |
| 78 | D | N | N | 191 | R | G | G |
| 80 | V | L | L | 206 | T/K | I | I |
| 86 | T | S | S | 210 | D | E | D |
| 88 | K | P | P | 215 | V | V | I |
| 89 | I | R(11/12)/L(1/12) | L | 232 | N | N | N |
| 90 | S | S | T | 234 | K | K(1/12)/(11/12) | K |
| 91 | I | V | V |
| PCV2 Genotype | Mean Rate of Substitution (Substitution/site/year) | 95% Highest Posterior Density | Effective Sample Size |
|---|---|---|---|
| PCV2a | 4.658 × 10−4 | 8.643 × 10−4 | 245,643 |
| PCV2b | 6.476 × 10−4 | 1.063 × 10−3 | 254,638 |
| PCV2d | 2.127 × 10−3 | 2.853 × 10−3 | 67,627 |
| Combined analysis for all genotypes | 1.098 × 10−3 | 1.381 × 10−3 | 81,829 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Lv, H.; Zhang, H.; Zhao, L.; Li, H.; He, Y.; Zhao, K.; Qiao, H.; Song, Y.; Bian, C. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 in Henan, China. Vet. Sci. 2025, 12, 343. https://doi.org/10.3390/vetsci12040343
Peng Z, Lv H, Zhang H, Zhao L, Li H, He Y, Zhao K, Qiao H, Song Y, Bian C. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 in Henan, China. Veterinary Sciences. 2025; 12(4):343. https://doi.org/10.3390/vetsci12040343
Chicago/Turabian StylePeng, Zhifeng, Huifang Lv, Han Zhang, Li Zhao, Huawei Li, Yanyu He, Kangdi Zhao, Hongxing Qiao, Yuzhen Song, and Chuanzhou Bian. 2025. "Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 in Henan, China" Veterinary Sciences 12, no. 4: 343. https://doi.org/10.3390/vetsci12040343
APA StylePeng, Z., Lv, H., Zhang, H., Zhao, L., Li, H., He, Y., Zhao, K., Qiao, H., Song, Y., & Bian, C. (2025). Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 in Henan, China. Veterinary Sciences, 12(4), 343. https://doi.org/10.3390/vetsci12040343







