Genomic Sequencing and Analysis of Enzootic Nasal Tumor Virus Type 2 Provides Evidence for Recombination within the Prevalent Chinese Strains
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. RNA Extraction
2.3. The Complete Genome Amplification of ENTV-2 CQ2
2.4. Sequence Assembly and Phylogenetic Analysis
2.5. Sequence Annotation and Comparative Analysis of ENTV-2 Whole Genome Sequences
2.6. Recombination Analysis
3. Results
3.1. Molecular Detection and Genomic Sequencing of ENTV-2
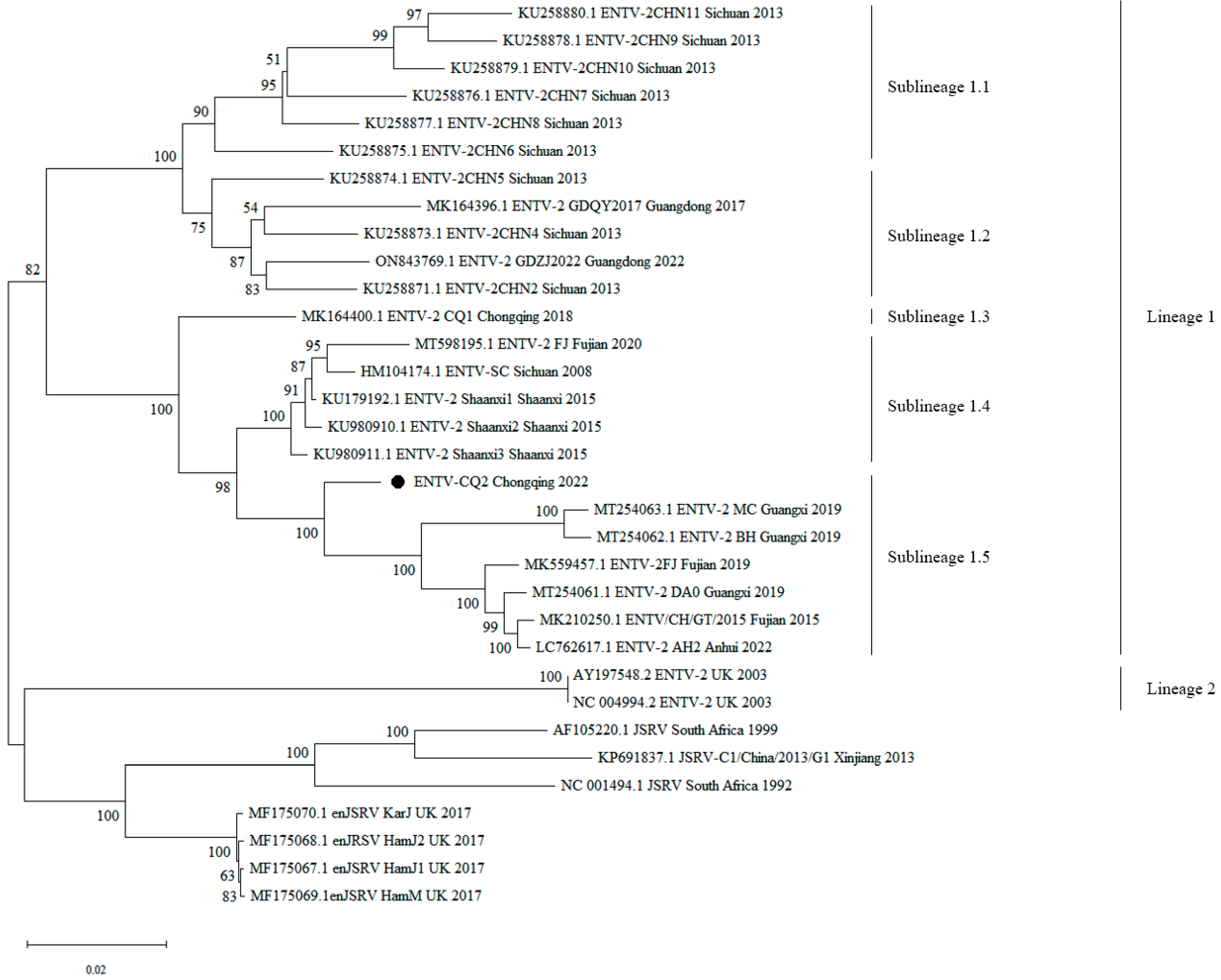
3.2. Phylogenetic Analysis of ENTV-2 CQ2
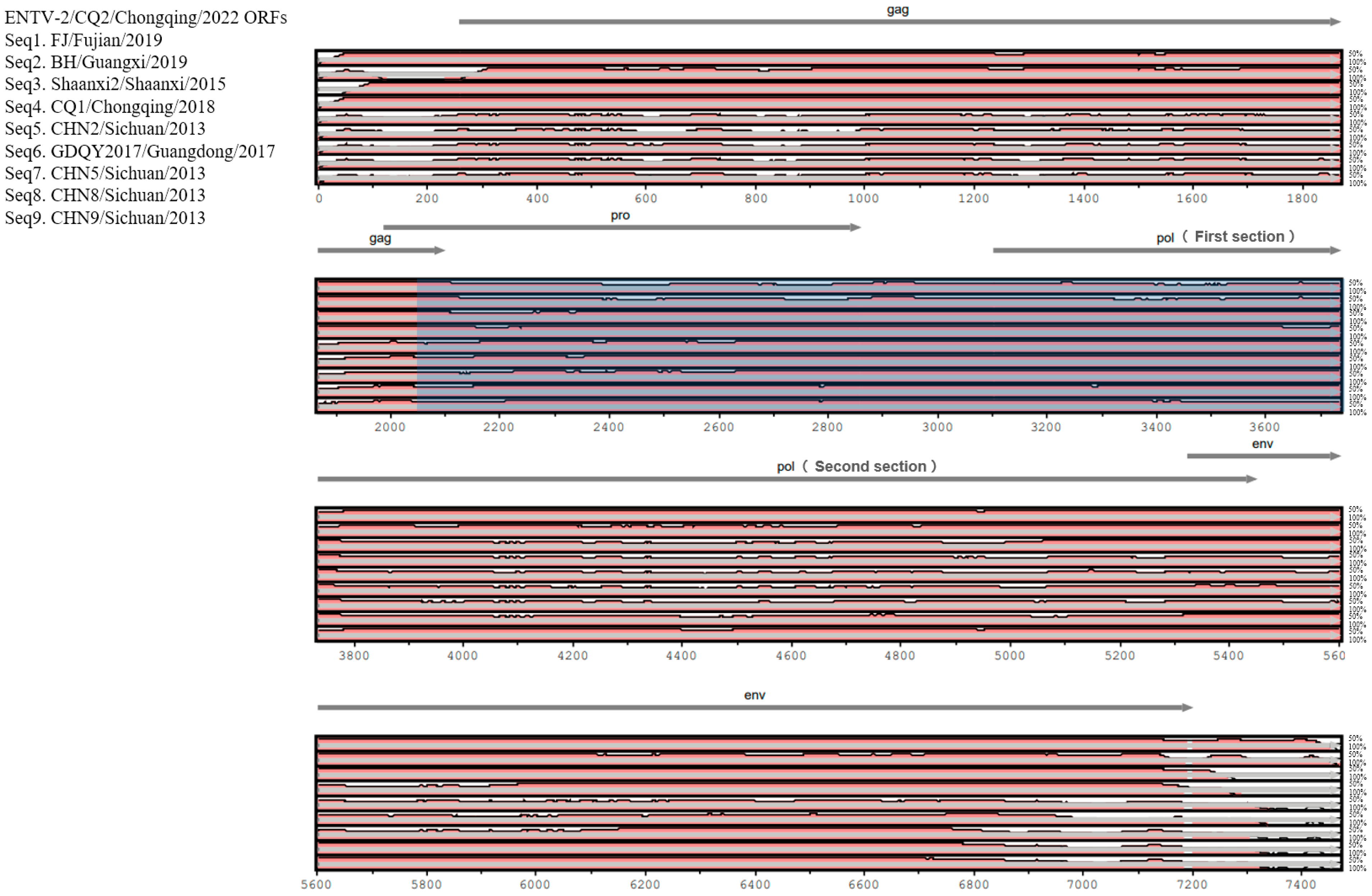
3.3. Comparative Genomic Analysis of ENTV-2 Strains
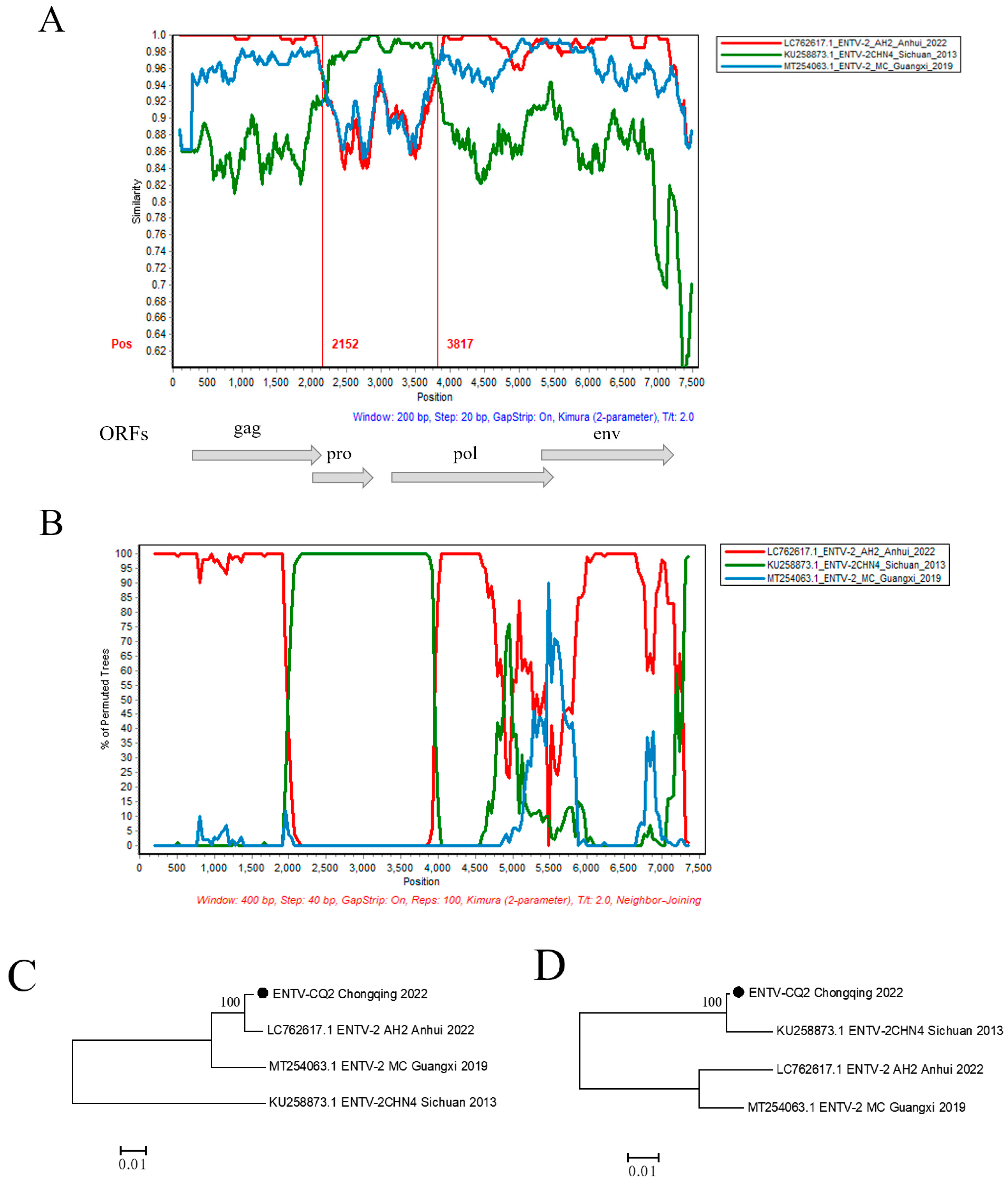
3.4. Recombination Analysis among ENTV-2 Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Las, H.M.; Ortin, A.; Cousens, C.; Minguijon, E.; Sharp, J.M. Enzootic nasal adenocarcinoma of sheep and goats. Curr. Top. Microbiol. 2003, 275, 201–223. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.R.; Linnerth-Petrik, N.M.; Laporte, A.N.; Menzies, P.I.; Foster, R.A.; Wootton, S.K. Full-length genome sequence analysis of enzootic nasal tumor virus reveals an unusually high degree of genetic stability. Virus Res. 2010, 151, 74–87. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Q.; Wang, J.; Zhou, M.; Fu, M.; Xu, X. Full-length genome sequence analysis of enzootic nasal tumor virus isolated from goats in China. Virol. J. 2017, 14, 141. [Google Scholar] [CrossRef]
- York, D.F.; Vigne, R.; Verwoerd, D.W.; Querat, G. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 1992, 66, 4930–4939. [Google Scholar] [CrossRef] [PubMed]
- Ortin, A.; Cousens, C.; Minguijon, E.; Pascual, Z.; Villarreal, M.P.; Sharp, J.M.; Heras, M.L. Characterization of enzootic nasal tumour virus of goats: Complete sequence and tissue distribution. J. Gen. Virol. 2003, 84, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Vitellozzi, G.; Mughetti, L.; Palmarini, M.; Mandara, M.T.; Mechelli, L.; Sharp, J.M.; Manocchio, I. Enzootic intranasal tumour of goats in Italy. Zentralbl. Veterinarmed. B 1993, 40, 459–468. [Google Scholar] [CrossRef]
- de Cecco, B.S.; Lorenzett, M.P.; Henker, L.C.; Weber, M.N.; Mosena, A.; Baumbach, L.; Canal, C.W.; Driemeier, D.; Pavarini, S.P.; Sonne, L. Detection of enzootic nasal tumor virus (ENTV) in a sheep flock in southern Brazil. Trop. Anim. Health PRO 2019, 51, 2095–2098. [Google Scholar] [CrossRef] [PubMed]
- De Las, H.M.; Borobia, M.; Ortin, A. Neoplasia-Associated Wasting Diseases with Economic Relevance in the Sheep Industry. Animals 2021, 11, 381. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Q.; Liu, W.; Chen, Y.; Jiang, J.; Wu, D.; Lin, Y.; Fang, Y.; Zheng, X.; Huang, S.; et al. Epidemiological Investigation on Goat Endemic Intranasal Tumor Virus Type 2 in Fuqing City of Fujian Province. China Anim. Health Insp. 2023, 40, 25–30. (In Chinese) [Google Scholar] [CrossRef]
- Ye, C.; Huang, Q.; Chen, T.; Jiang, J.; Hou, F.; Xu, D.; Peng, Y.; Fang, R.; Chen, J. First detection and genotypic analysis of goat enzootic nasal tumor virus 2 in Chongqing, China. Arch. Virol. 2019, 164, 1647–1650. [Google Scholar] [CrossRef]
- Wang, B.; Ye, N.; Cao, S.J.; Wen, X.T.; Huang, Y.; Yan, Q.G. Identification of novel and differentially expressed MicroRNAs in goat enzootic nasal adenocarcinoma. BMC Genom. 2016, 17, 896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Meng, J.; Li, Z.; He, Y.; Wang, D.; Li, N.; Sun, J.; Bai, F.; Wang, J. Detection and Sequence Analysis of Goat Enzootic Nasal Tumor Virus in Jiangcheng County, Yunnan Province. China Anim. Health Insp. 2022, 39, 48–52. (In Chinese) [Google Scholar] [CrossRef]
- Lei, H.; Su, M.; Ning, L.; Kang, Y.; Chen, K.; Zeng, Y. Investigation and diagnosis of Enzootic Nasal Tumor in Goats. Prog. Vet. Med. 2006, 2, 112–114. (In Chinese) [Google Scholar] [CrossRef]
- He, R.; Du, Y.; Gan, L.; Mohsin, M.A.; He, B.X. Development of a SYBR Green-based real-time quantitative polymerase chain reaction assay to detect enzootic nasal tumor virus in goats. Can. J. Vet. Res. 2021, 85, 145–150. [Google Scholar] [PubMed]
- Hou, H.; Zhu, D.; Zhang, D.; Hu, X.; Zhao, R.; Dai, Y. Cloning and Analysis of gag Gene of Enzootic Nasal Tumor Virus in Goats. China Herbiv. Sci. 2018, 38, 53–55. (In Chinese) [Google Scholar] [CrossRef]
- Zhai, S.L.; Lv, D.H.; Xu, Z.H.; Yu, J.S.; Wen, X.H.; Zhang, H.; Chen, Q.L.; Jia, C.L.; Zhou, X.R.; Zhai, Q.; et al. A Novel Enzootic Nasal Tumor Virus Circulating in Goats from Southern China. Viruses 2019, 11, 956. [Google Scholar] [CrossRef] [PubMed]
- Vuilleumier, S.; Bonhoeffer, S. Contribution of recombination to the evolutionary history of HIV. Curr. Opin. Hiv. Aids 2015, 10, 84–89. [Google Scholar] [CrossRef]
- Burke, D.S. Recombination in HIV: An important viral evolutionary strategy. Emerg. Infect. Dis. 1997, 3, 253–259. [Google Scholar] [CrossRef]
- Zhang, M.; Foley, B.; Schultz, A.K.; Macke, J.P.; Bulla, I.; Stanke, M.; Morgenstern, B.; Korber, B.; Leitner, T. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 2010, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.A.; van der Meer, F.; Checkley, S.; Joseph, T.; King, R.; Ravi, M.; Peters, D.; Fonseca, K.; Gagnon, C.A.; Provost, C.; et al. Analysis of Whole-Genome Sequences of Infectious laryngotracheitis Virus Isolates from Poultry Flocks in Canada: Evidence of Recombination. Viruses 2020, 12, 1302. [Google Scholar] [CrossRef]
- Yan, T.; Guo, L.; Jiang, X.; Wang, H.; Yao, Z.; Zhu, S.; Diao, Y.; Tang, Y. Discovery of a novel recombinant avian orthoreovirus in China. Vet. Microbiol. 2021, 260, 109094. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Ye, C.; Chen, T.; Jiang, J.; Peng, Y.; Chen, J.; Fang, R. EvaGreen-based real-time PCR assay for sensitive detection of enzootic nasal tumor virus 2. Mol. Cell. Probes 2019, 44, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.; Rheinstein, P.H. Mouse mammary tumor viral env sequences are not present in the human genome but are present in breast tumors and normal breast tissues. Virus Res. 2019, 266, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Swanstrom, R.; Wills, J.W. Synthesis, Assembly, and Processing of Viral Proteins-Retroviruses-NCBI Bookshelf; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Gifford, R.; Tristem, M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 2003, 26, 291–315. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.D. Identification of Hyal2 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr. Top. Microbiol. 2003, 275, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Monot, M.; Archer, F.; Gomes, M.; Mornex, J.F.; Leroux, C. Advances in the study of transmissible respiratory tumours in small ruminants. Vet. Microbiol. 2015, 181, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.K.; Duh, F.M.; Vigdorovich, V.; Danilkovitch-Miagkova, A.; Lerman, M.I.; Miller, A.D. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 2001, 98, 4443–4448. [Google Scholar] [CrossRef]
- Dirks, C.; Duh, F.M.; Rai, S.K.; Lerman, M.I.; Miller, A.D. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 2002, 76, 2141–2149. [Google Scholar] [CrossRef]
- Rosenberg, N.; Jolicoeur, P. Retroviral Pathogenesis-Retroviruses-NCBI Bookshelf; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Maeda, N.; Inoshima, Y.; De Las, H.M.; Maenaka, K. Enzootic nasal tumor virus type 2 envelope of goats acts as a retroviral oncogene in cell transformation. Virus Genes 2021, 57, 50–59. [Google Scholar] [CrossRef]
- White, K.A.; Enjuanes, L.; Berkhout, B. RNA virus replication, transcription and recombination. RNA Biol. 2011, 8, 182–183. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′-3′) | Position in CQ2 (OR682176) (bp) | Product Length (bp) |
|---|---|---|---|
| ENTV-2-1-F | ACAAGGCATCAGCCATTTTGGT | ||
| ENTV-2-1-R | AACCTCACCAAGTCGCTGC | 1–1002 | 1002 |
| ENTV-2-2-F | GGGGAACAAATTCGAACTCAT TATACT | ||
| ENTV-2-2-R | CCCATCTCAGGTGCTAGTATT GTAT | 433–2110 | 1678 |
| ENTV-2-3-F | GCAACCCTGCAGGTTCCAA | ||
| ENTV-2-3-R | TGTGGGTGCTCGAGGGGTA | 1999–3865 | 1867 |
| ENTV-2-4-F | ATTGCTGATGAAAAAATT | ||
| ENTV-2-4-R | TATTGAGGGAGAACAAA | 3497–4740 | 1244 |
| ENTV-2-5-F | GCAAATGATTGAAACTGT | ||
| ENTV-2-5-R | ACTATTGCCATGACCAAA | 4393–6152 | 1760 |
| ENTV-2-6-F | CTCCTTGGACTTTATGTCGAGC | ||
| ENTV-2-6-R | TGTTTTATTGTGTCATAGTATA | 5944–7468 | 1525 |
| Gene | Loci in the Genome of CQ2 | CQ2 | CHN4 | AH2 | MC |
|---|---|---|---|---|---|
| pro | 2273 | C a (K) b | C (K) | T (R) | T (R) |
| 2369 | A (E) | A (E) | T (D) | T (D) | |
| 2503–2504 | AA (E) | AA (E) | GT (G) | GT (G) | |
| 2608–2609 | GG (R) | GG (R) | CA (T) | CA (T) | |
| 2738 | A (E) | A (E) | T (D) | T (D) | |
| 2799 | A (I) | A (I) | G (V) | G (V) | |
| 2829–2831 | AGT (S) | AGT (S) | GAG (E) | GAG (E) | |
| pol | 3176 | A (I) | A (I) | G (V) | G (V) |
| 3294 | G (R) | G (R) | A (K) | A (K) | |
| 3356 | T (L) | T (L) | A (I) | A (I) | |
| 3461 | A (I) | A (I) | G (V) | G (V) | |
| 3494 | G (V) | G (V) | A (I) | A (I) | |
| 3555 | A (Y) | A (Y) | T (F) | T (F) | |
| 3564–3565 | TT (V) | TT (V) | CC (A) | CC (A) | |
| 3746 | T (S) | T (S) | C (P) | C (P) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Niu, J.; Liu, Y.; Dai, Y.; Ni, H.; Wang, J.; Fang, R.; Ye, C. Genomic Sequencing and Analysis of Enzootic Nasal Tumor Virus Type 2 Provides Evidence for Recombination within the Prevalent Chinese Strains. Vet. Sci. 2024, 11, 248. https://doi.org/10.3390/vetsci11060248
Li Y, Niu J, Liu Y, Dai Y, Ni H, Wang J, Fang R, Ye C. Genomic Sequencing and Analysis of Enzootic Nasal Tumor Virus Type 2 Provides Evidence for Recombination within the Prevalent Chinese Strains. Veterinary Sciences. 2024; 11(6):248. https://doi.org/10.3390/vetsci11060248
Chicago/Turabian StyleLi, Yixuan, Jingyi Niu, Yiyu Liu, Yu Dai, Hongbo Ni, Jinliang Wang, Rendong Fang, and Chao Ye. 2024. "Genomic Sequencing and Analysis of Enzootic Nasal Tumor Virus Type 2 Provides Evidence for Recombination within the Prevalent Chinese Strains" Veterinary Sciences 11, no. 6: 248. https://doi.org/10.3390/vetsci11060248
APA StyleLi, Y., Niu, J., Liu, Y., Dai, Y., Ni, H., Wang, J., Fang, R., & Ye, C. (2024). Genomic Sequencing and Analysis of Enzootic Nasal Tumor Virus Type 2 Provides Evidence for Recombination within the Prevalent Chinese Strains. Veterinary Sciences, 11(6), 248. https://doi.org/10.3390/vetsci11060248








