Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Genealogical Study
2.3. Animal Sampling for the Genomic Assays
2.4. Genotyping and Quality Control
2.5. Genomic Characterization
2.6. Identification of Selection Signatures and Genes Involved
3. Results
3.1. Genealogical Study
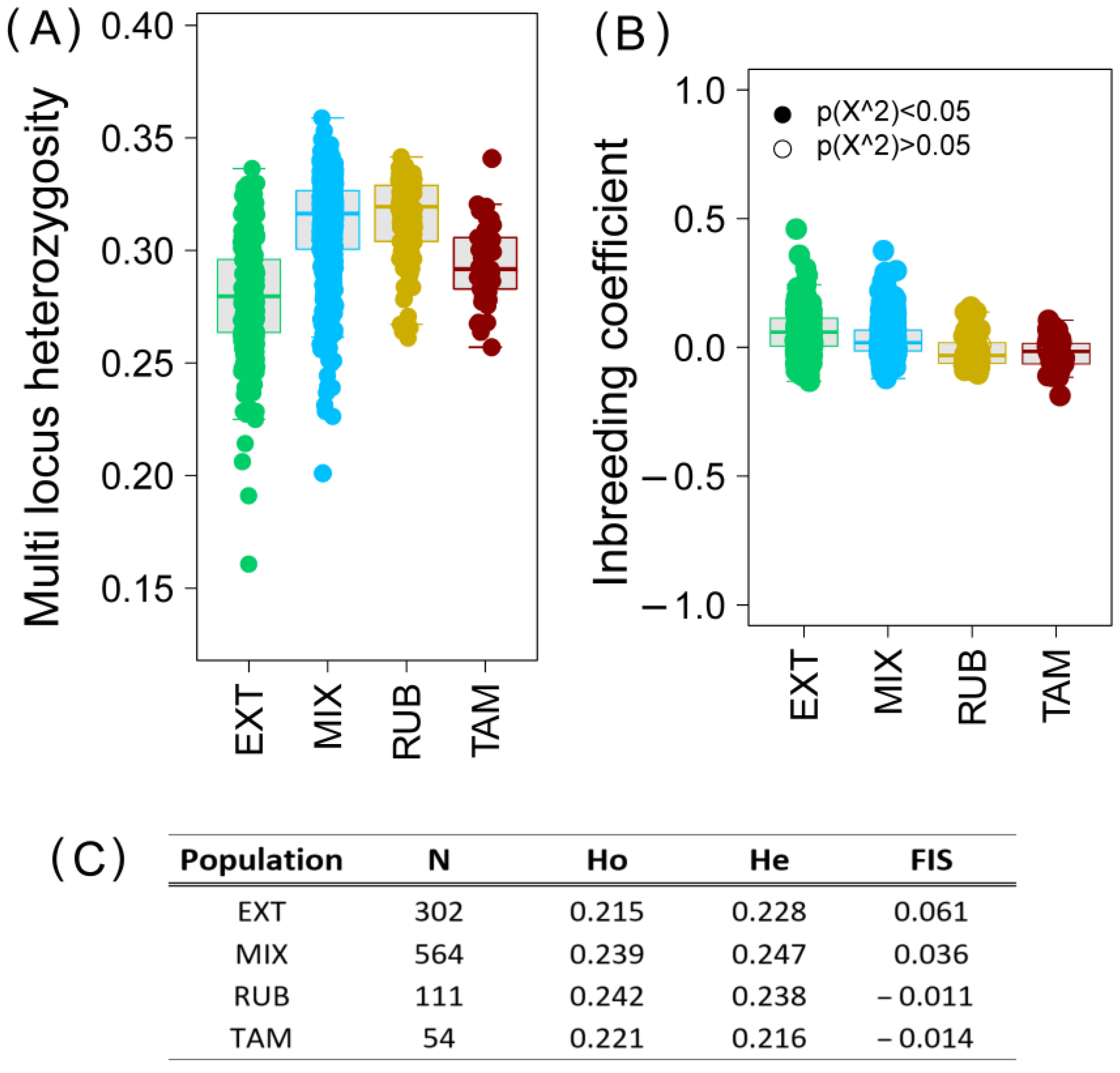
3.2. Genomic Diversity
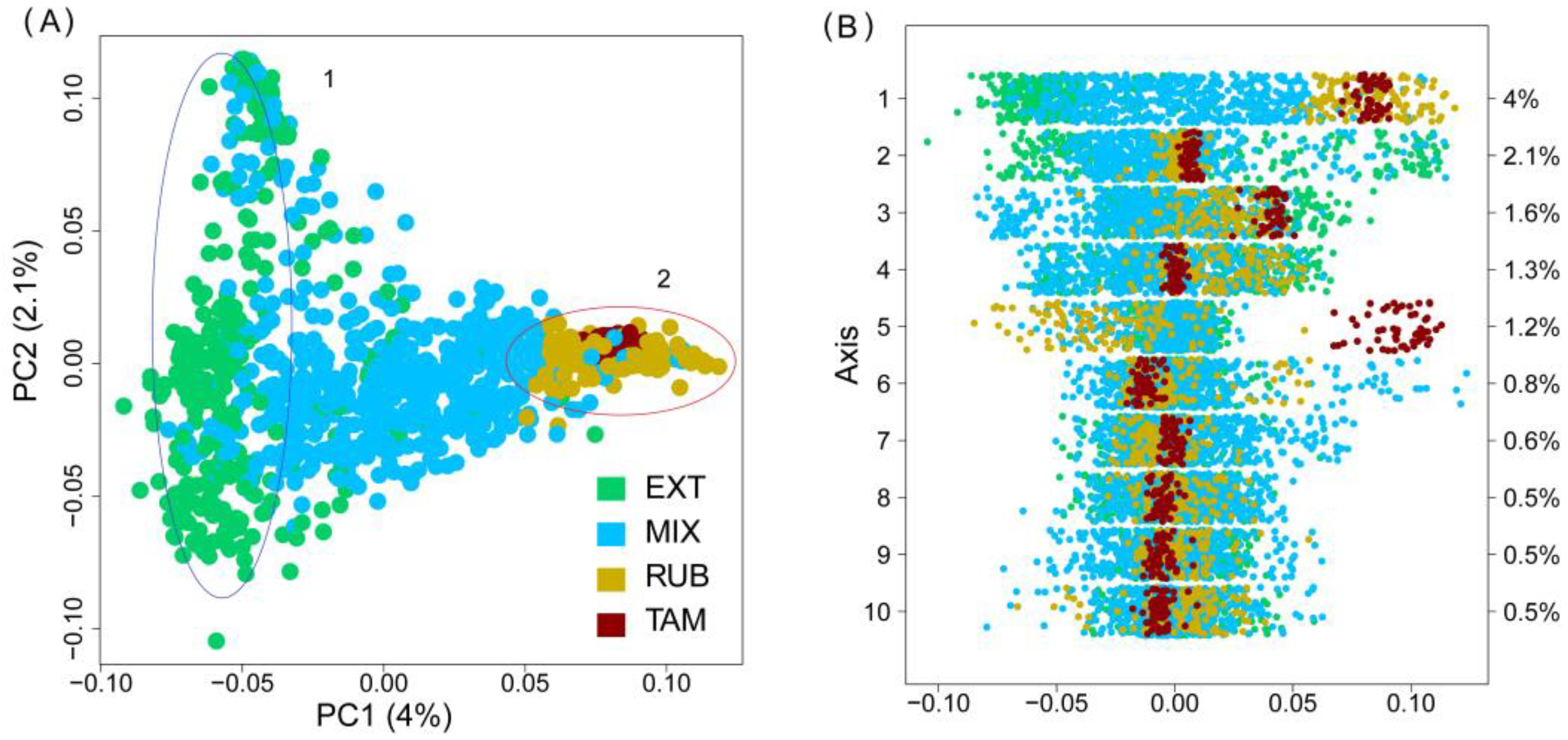
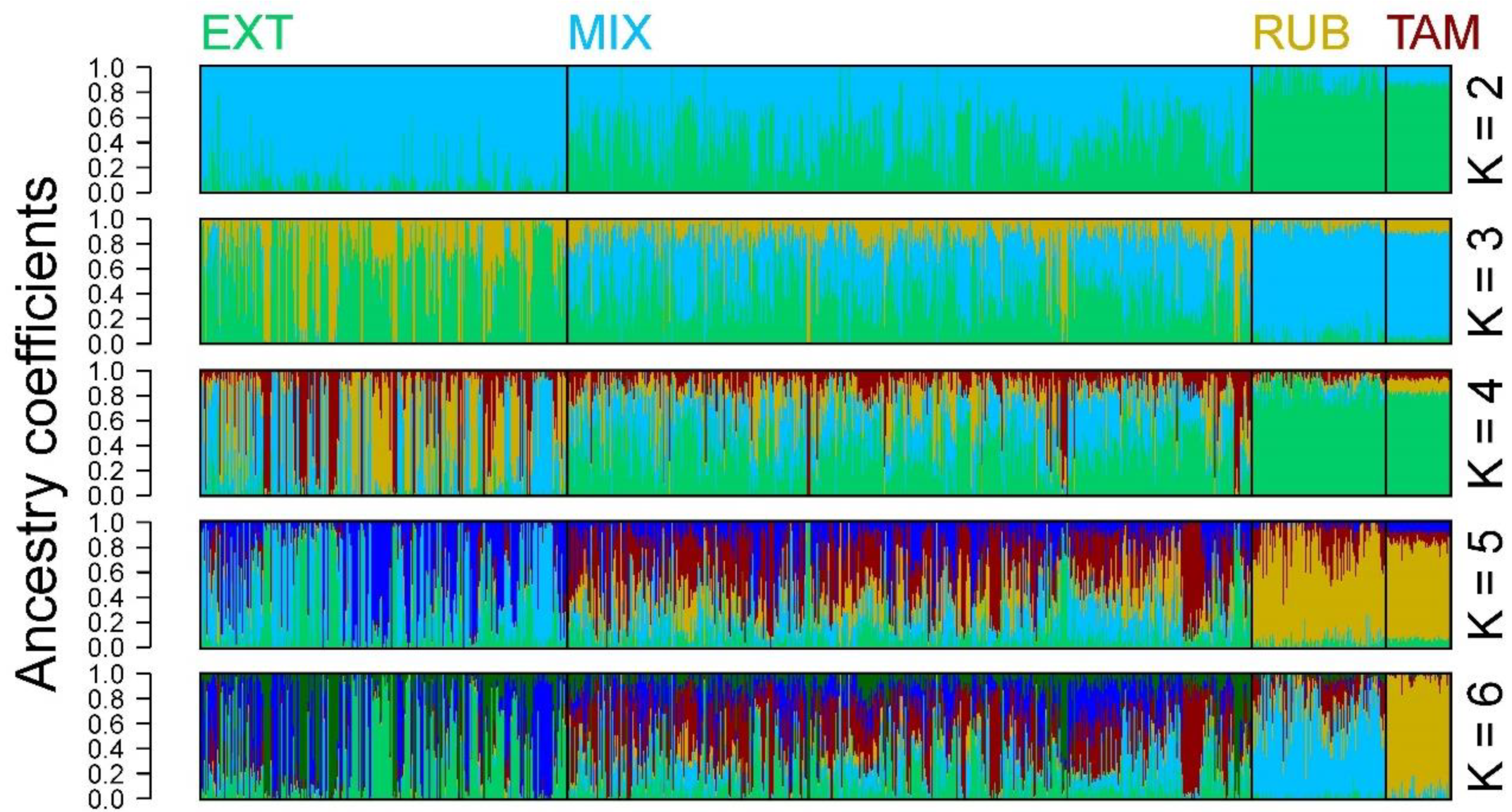
3.3. Population Structure and Genetic Differentiation
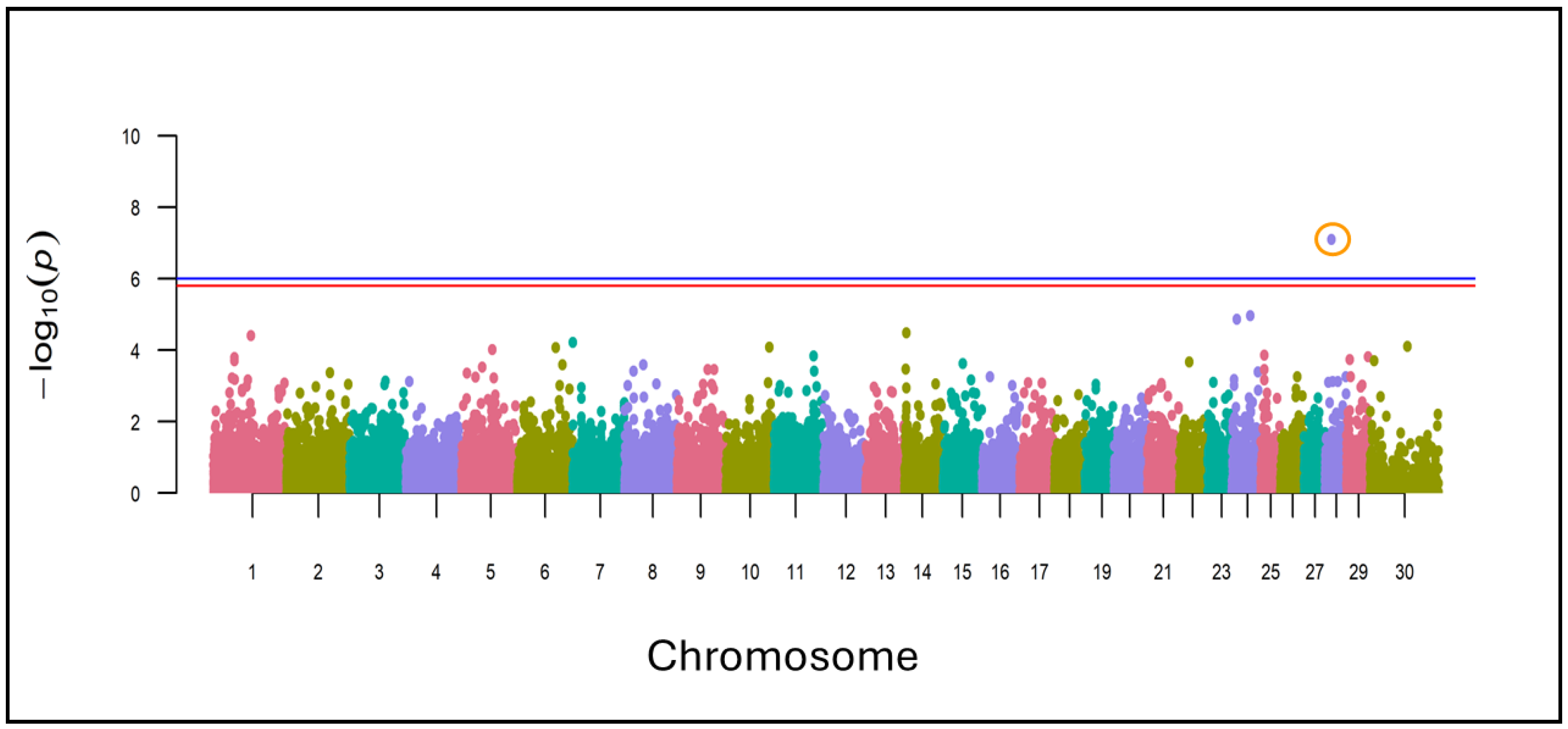
3.4. Identification of Selection Signatures and Genes Involved
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morales, R.; Menendez-Buxadera, A.; Aviles, C.; Molina, A. Direct and maternal genetic effects for preweaning growth in Retinta cattle estimated by a longitudinal approach throughout the calving trajectory of the cow. J. Anim. Breed. Genet. 2013, 130, 425–434. [Google Scholar] [CrossRef] [PubMed]
- ARCA. Datos Censales Raza Retinta. Available online: https://servicio.mapa.gob.es/arca/flujos.html?_flowId=datosCensalesRaza-flow&tipoOperacion=CONSULTA&formatoPagina=0&id=50218 (accessed on 1 January 2023).
- Pérez-González, J.; Frantz, A.C.; Torres-Porras, J.; Castillo, L.; Carranza, J. Population structure, habitat features and genetic structure of managed red deer populations. Eur. J. Wildl. Res. 2012, 58, 933–943. [Google Scholar] [CrossRef]
- Rodero-Serrano, E.; Demyda-Peyrás, S.; González-Martinez, A.; Rodero-Franganillo, A.; Moreno-Millán, M. The rob(1;29) chromosome translocation in endangered Andalusian cattle breeds. Livest. Sci. 2013, 158, 32–39. [Google Scholar] [CrossRef]
- Aparicio Sánchez, G. Zootecnia Especial Etnologia Compendiada; Moderna: Cambridge, MA, USA, 1960. [Google Scholar]
- Fuentes, F.; Sánchez, J.; Gonzalo, G. Tratado de Etnología Animal: Razas de Rumiantes y Monogástricos; Marín, D., Ed.; Diego Marín: Murcia, Spain, 2006. [Google Scholar]
- Peña Blanco, F.; Herrera García, M.; Rodero Serrano, E.; Gutiérrez Cabezas, M. Sobre el origen de la raza retinta. Arch. Zootec. 1995, 44, 99–110. [Google Scholar]
- Miretti, M.M.; Dunner, S.; Naves, M.; Contel, E.P.; Ferro, J.A. Predominant African-Derived mtDNA in Caribbean and Brazilian Creole Cattle is also Found in Spanish Cattle (Bos taurus). J. Hered. 2004, 95, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.M.A.G.; Moreno-Jiménez, S.; Jiménez, J.M.; Demyda-Peyrás, S.; Molina, A. Genomic and phenotypic differentiation of the Tamarona line of the Spanish Retinta beef cattle breed. In Proceedings of the 73rd Annual Congress of the European Federation of Animal Science (EAAP), Porto, Portugal, 5–8 September 2022. [Google Scholar]
- Gutiérrez García, J.P.; Goyache, F. A note on ENDOG: A computer program for analysing pedigree information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Enciso, M. Use of the uncertain relationship matrix to compute effective population size. J. Anim. Breed. Genet. 1995, 112, 327–332. [Google Scholar] [CrossRef]
- James, J. A note on selection differential and generation length when generations overlap. Anim. Sci. 1977, 24, 109–112. [Google Scholar] [CrossRef]
- FAO. 11th Session of the ITWG on Animal Genetic Resources for Food and Agriculture. Available online: https://www.fao.org/animal-genetics/events/events-detail/en/c/1369166/ (accessed on 1 March 2024).
- Thermo Fisher Scientific. Axiom CNV Summary Tool User Manual. Available online: https://tools.thermofisher.com/content/sfs/manuals/axiom_cnv_summary_tool_usermanual.pdf (accessed on 15 August 2020).
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef]
- de Jong, M.J.; de Jong, J.F.; Hoelzel, A.R.; Janke, A. SambaR: An R package for fast, easy and reproducible population-genetic analyses of biallelic SNP data sets. Mol. Ecol. Resour. 2021, 21, 1369–1379. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.J.; Niamir, A.; Wolf, M.; Kitchener, A.C.; Lecomte, N.; Seryodkin, I.V.; Fain, S.R.; Hagen, S.B.; Saarma, U.; Janke, A. Range-wide whole-genome resequencing of the brown bear reveals drivers of intraspecies divergence. Commun. Biol. 2023, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Turner, S. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Morales, R.M.; Menéndez-Buxadera, A.; Demyda-Peyrás, S.; Molina, A. Genetic effect of season on the preweaning growth of beef cattle: A first approach on retinta calves. Rev. Colomb. Cienc. Pecu. 2020, 33, 134–143. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Altarriba, J.; Díaz, C.; Quintanilla, R.; Cañón, J.; Piedrafita, J. Pedigree analysis of eight Spanish beef cattle breeds. Genet. Sel. Evol. 2003, 35, 43–63. [Google Scholar] [CrossRef] [PubMed]
- FAO. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/countryprofiles/webservices/en/?lang=en (accessed on 1 March 2024).
- Jiménez, J.M.; Morales, R.M.; Menéndez-Buxadera, A.; Demyda-Peyrás, S.; Laseca, N.; Molina, A. Estimation of the Genetic Components of (Co) variance and Preliminary Genome-Wide Association Study for Reproductive Efficiency in Retinta Beef Cattle. Animals 2023, 13, 501. [Google Scholar] [CrossRef]
- Damiran, D.; Larson, K.A.; Pearce, L.T.; Erickson, N.E.; Lardner, B.H. Effect of calving period on beef cow longevity and lifetime productivity in western Canada. Transl. Anim. Sci. 2018, 2, S61–S65. [Google Scholar] [CrossRef] [PubMed]
- Cañas-Álvarez, J.; González-Rodríguez, A.; Munilla, S.; Varona, L.; Díaz, C.; Baro, J.; Altarriba, J.; Molina, A.; Piedrafita, J. Genetic diversity and divergence among Spanish beef cattle breeds assessed by a bovine high-density SNP chip. J. Anim. Sci. 2015, 93, 5164–5174. [Google Scholar] [CrossRef] [PubMed]
- Mesner, L.D.; Ray, B.; Hsu, Y.-H.; Manichaikul, A.; Lum, E.; Bryda, E.C.; Rich, S.S.; Rosen, C.J.; Criqui, M.H.; Allison, M. Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. J. Clin. Investig. 2014, 124, 2736–2749. [Google Scholar] [CrossRef]
- Hepei, L.; Mingkun, X.; Li, W.; Jin, W. Assessment of BicC family RNA binding protein 1 and Ras protein specific guanine nucleotide releasing factor 1 as candidate genes for high myopia: A case–control study. Indian J. Ophthalmol. 2017, 65, 926–930. [Google Scholar] [PubMed]
- Li, Y.; Gao, Y.; Kim, Y.S.; Iqbal, A.; Kim, J.J. A whole genome association study to detect additive and dominant single nucleotide polymorphisms for growth and carcass traits in Korean native cattle, Hanwoo. Asian-Australas. J. Anim. Sci. 2017, 30, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Gotoh, M.; Kiyohara, K.; Akashima, T.; Iwasaki, H.; Kameyama, A.; Mochizuki, H.; Yada, T.; Inaba, N.; Togayachi, A. Differential Roles of TwoN-Acetylgalactosaminyltransferases, CSGalNAcT-1, and a Novel Enzyme, CSGalNAcT-2: Initiation and Elongation in Synthesis of Chondroitin Sulfate. J. Biol. Chem. 2003, 278, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ren, X.; Chen, Y.; Gao, Y.; Wang, N.; Lu, Z.; Gao, L.; Qin, L.; Wang, Y.; Gao, H. Chondroitin sulfate N-acetylgalactosaminyltransferase-2 contributes to the replication of infectious bursal disease virus via interaction with the capsid protein VP2. Viruses 2015, 7, 1474–1491. [Google Scholar] [CrossRef]
- Gasimli, L.; Hickey, A.M.; Yang, B.; Li, G.; dela Rosa, M.; Nairn, A.V.; Kulik, M.J.; Dordick, J.S.; Moremen, K.W.; Dalton, S.; et al. Changes in glycosaminoglycan structure on differentiation of human embryonic stem cells towards mesoderm and endoderm lineages. Biochim. Biophys. Acta BBA—Gen. Subj. 2014, 1840, 1993–2003. [Google Scholar] [CrossRef]
- Goodfellow, P.J.; Wells, S.A., Jr. RET gene and its implications for cancer. JNCI J. Natl. Cancer Inst. 1995, 87, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.; Grice, E.A.; Vinton, R.M.; Bessling, S.L.; McCallion, A.S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 2006, 312, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Huang, B.; Bai, F.; Wu, F.; Zhou, Z.; Lai, Z.; Li, S.; Qu, K.; Jia, Y.; Lei, C. Two novel SNPs in RET gene are associated with cattle body measurement traits. Animals 2019, 9, 836. [Google Scholar] [CrossRef] [PubMed]

| Population | Breeding Animal | Offspring | N | GI |
|---|---|---|---|---|
| Global | Bull | Male | 1497 | 5.85 |
| Female | 12,663 | 5.63 | ||
| Cow | Male | 1497 | 7.77 | |
| Female | 12,732 | 7.50 | ||
| EXT | Bull | Male | 425 | 5.62 |
| Female | 2932 | 5.42 | ||
| Cow | Male | 425 | 7.70 | |
| Female | 2933 | 7.38 | ||
| RUB | Bull | Male | 462 | 5.69 |
| Female | 2021 | 5.75 | ||
| Cow | Male | 462 | 7.24 | |
| Female | 2019 | 7.27 | ||
| TAM | Bull | Male | 6 | 5.79 |
| Female | 120 | 5.72 | ||
| Cow | Male | 6 | 8.27 | |
| Female | 120 | 8.72 |
| Global | EXT | RUB | TAM | |
|---|---|---|---|---|
| N (Ne) | 30,448 | 6169 | 2649 | 379 |
| Ne | 70.65 | 34.47 | 28.38 | 31.35 |
| N (F, AR) | 36,695 | 8341 | 4798 | 970 |
| F | 0.062 | 0.091 | 0.046 | 0.056 |
| AR | 0.020 | 0.057 | 0.027 | 0.109 |
| MG | 9.78 | 10.54 | 7.91 | 6.55 |
| CG | 3.22 | 3.43 | 2.97 | 3.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anaya, G.; Morales, R.; Demyda-Peyrás, S.; Moreno-Jiménez, S.; Jiménez, J.M.; Molina, A. Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages. Vet. Sci. 2024, 11, 247. https://doi.org/10.3390/vetsci11060247
Anaya G, Morales R, Demyda-Peyrás S, Moreno-Jiménez S, Jiménez JM, Molina A. Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages. Veterinary Sciences. 2024; 11(6):247. https://doi.org/10.3390/vetsci11060247
Chicago/Turabian StyleAnaya, Gabriel, Rosa Morales, Sebastián Demyda-Peyrás, Samuel Moreno-Jiménez, José María Jiménez, and Antonio Molina. 2024. "Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages" Veterinary Sciences 11, no. 6: 247. https://doi.org/10.3390/vetsci11060247
APA StyleAnaya, G., Morales, R., Demyda-Peyrás, S., Moreno-Jiménez, S., Jiménez, J. M., & Molina, A. (2024). Genomic Diversity of the Retinta Breed Derived from Two Ancestral Bovine Lineages. Veterinary Sciences, 11(6), 247. https://doi.org/10.3390/vetsci11060247





