Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Reproductive Performance Parameters
2.2. Detection of PPVs, PCV2, PCV3, PCV4, and PRRSV
2.3. Statistical Analysis
3. Results
3.1. Herd Management and Reproductive Performance Parameters
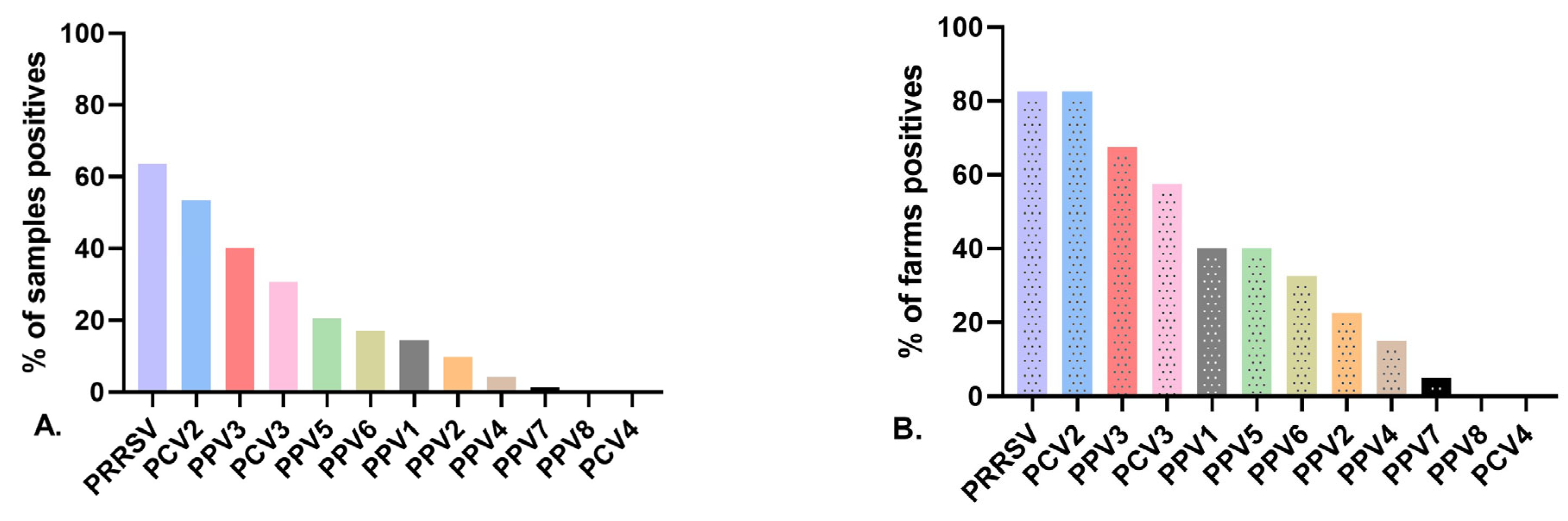
3.2. Detection Rates of Porcine Parvovirus in Gilts
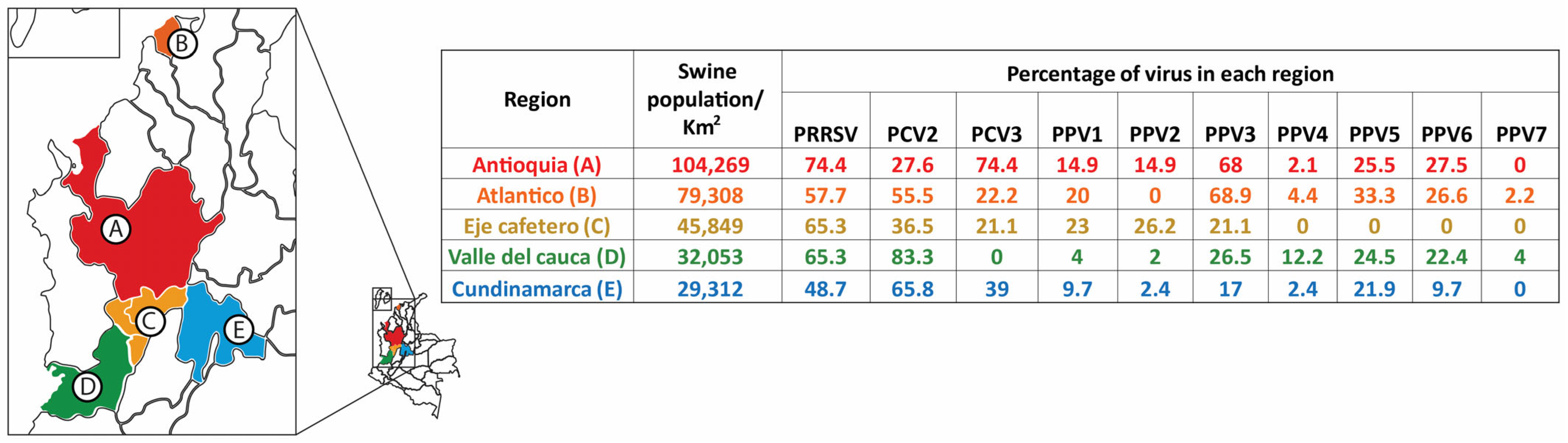
3.3. Virus Detection at the Herd Level and Infection at the Individual Level
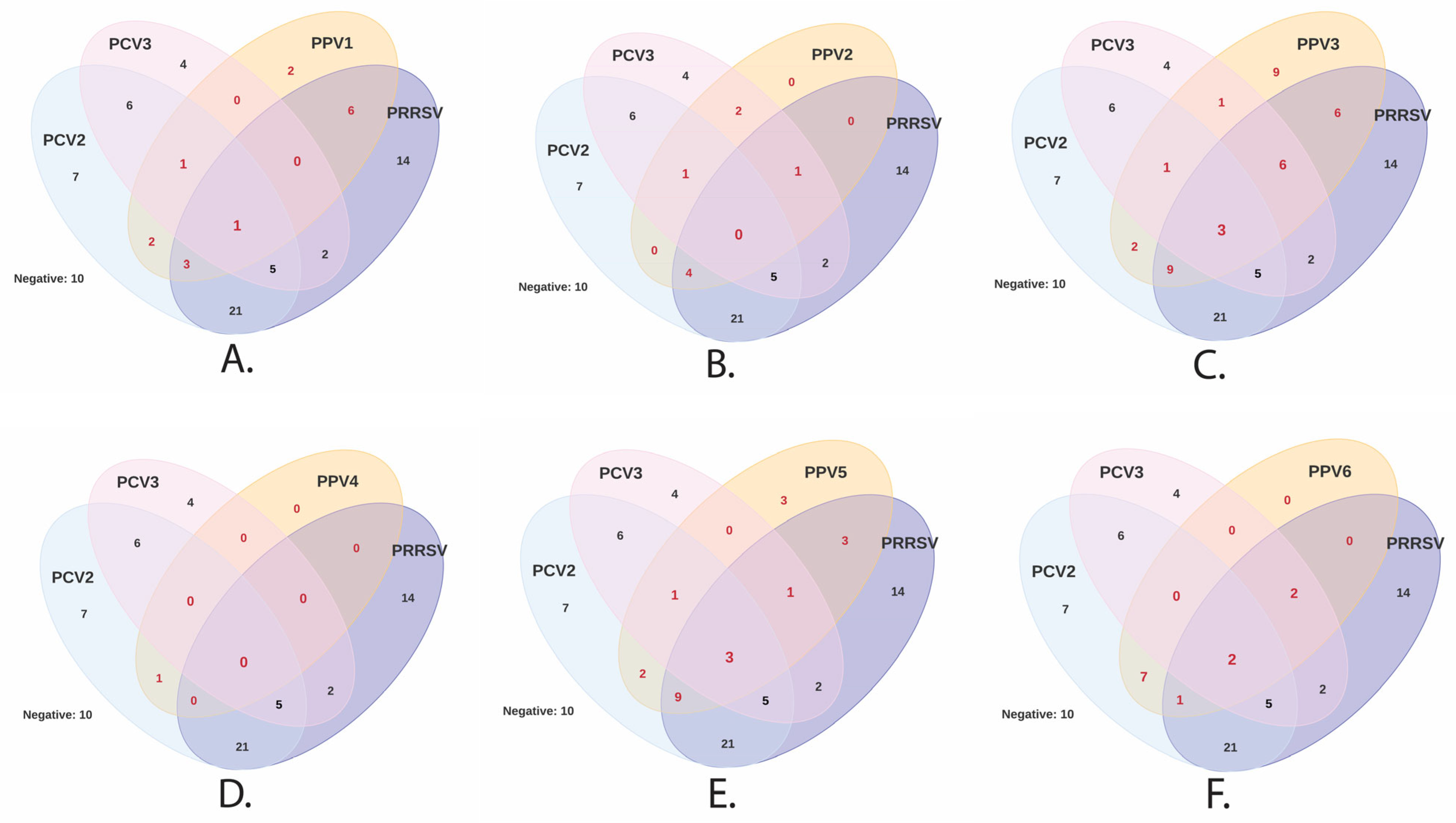
3.4. Association between PPVs (PPV1 through PPV7) and PCV2, PCV3, PCV4, or PRRSV
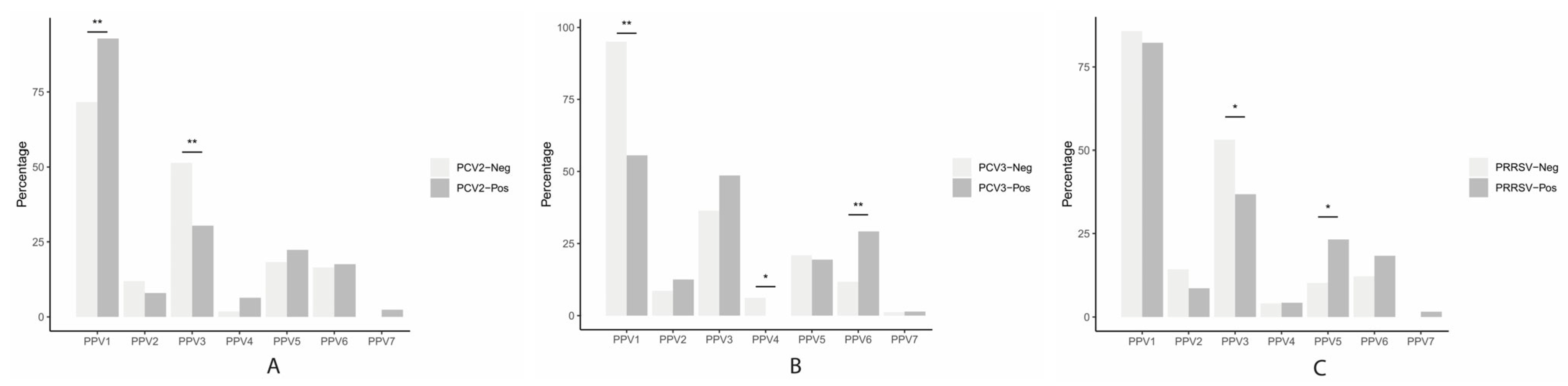
3.5. Association between PPVs, PCV2, PCV3, and PRRSV Status in Gilts and Performance Parameters in Sows
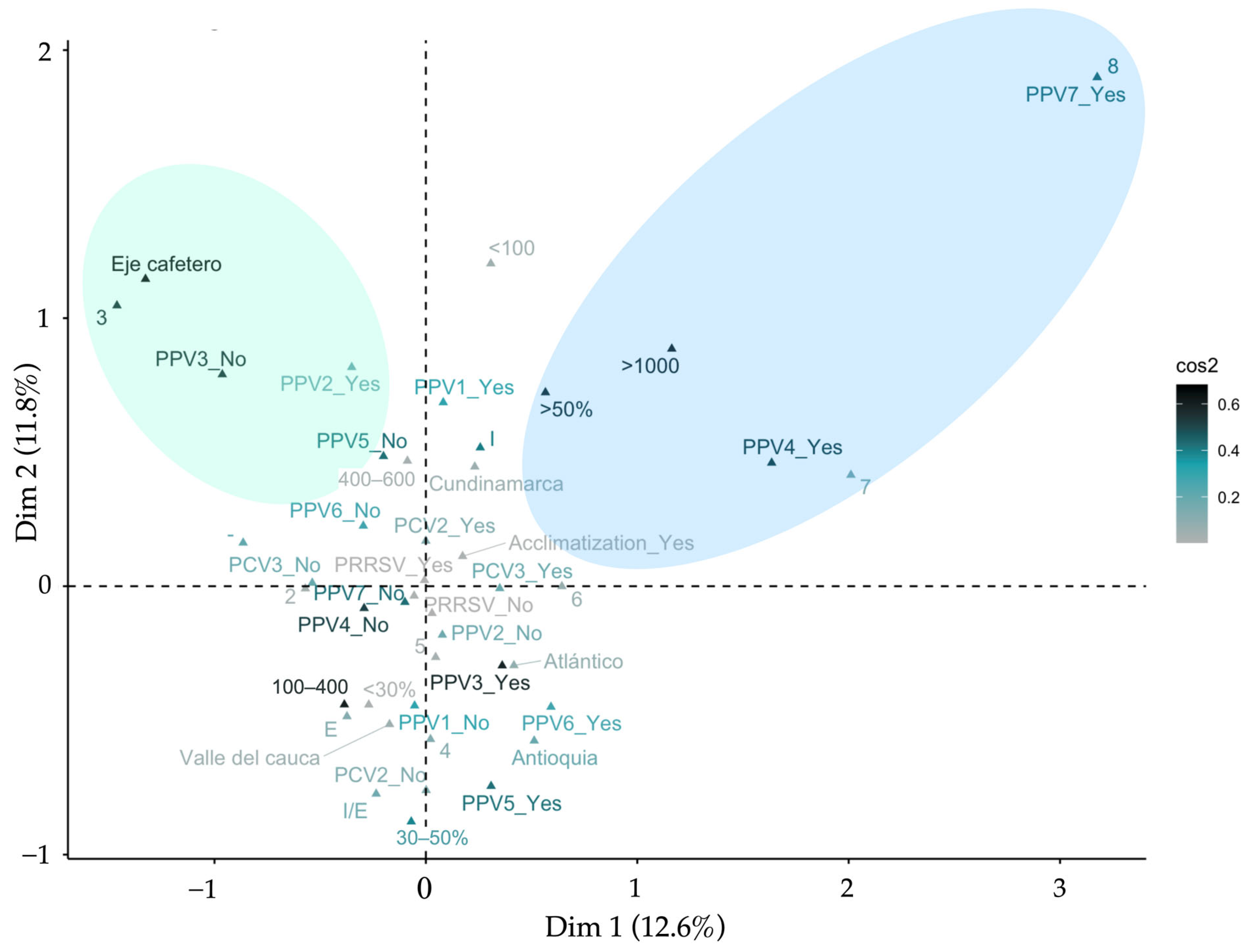
3.6. Multiple Correspondence Analysis—MCA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. Ictv Report Consortium ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.C.; Tomlinson, J.E.; Lopez-Astacio, R.A.; Parrish, C.R.; Van de Walle, G.R. Small but mighty: Old and new parvoviruses of veterinary significance. Virol. J. 2021, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Hijikata, M.; Abe, K.; Win, K.M.; Shimizu, Y.K.; Keicho, N.; Yoshikura, H. Identification of new parvovirus DNA sequence in swine sera from Myanmar. JPN J. Infect. Dis. 2001, 54, 244–245. [Google Scholar]
- Palinski, R.M.; Mitra, N.; Hause, B.M. Discovery of a novel Parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes 2016, 52, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yan, G.; Chen, S.; Han, H.; Li, J.; Zhang, H.; Luo, S.; Liu, M.; Wu, Q.; Li, Q.; et al. Identification and genomic characterization of a novel porcine parvovirus in China. Front. Vet. Sci. 2022, 9, 1009103. [Google Scholar] [CrossRef] [PubMed]
- Van Leengoed, L.A.; Vos, J.; Gruys, E.; Rondhuis, P.; Brand, A. Porcine Parvovirus infection: Review and diagnosis in a sow herd with reproductive failure. Vet. Q. 1983, 5, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Mogollon, J.D.; Franco-Rodriguez, C.; Jaime, J. The novel porcine parvoviruses: Current state of knowledge and their possible implications in clinical syndromes in pigs. Viruses 2023, 15, 2398. [Google Scholar] [CrossRef] [PubMed]
- Cságola, A.; Lőrincz, M.; Cadar, D.; Tombácz, K.; Biksi, I.; Tuboly, T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012, 157, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection Patterns of Porcine Parvovirus (PPV) and Novel Porcine Parvoviruses 2 through 6 (PPV2-PPV6) in Polish Swine Farms. Viruses 2019, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Saekhow, P.; Mawatari, T.; Ikeda, H. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol. Immunol. 2014, 58, 382–387. [Google Scholar] [CrossRef]
- Streck, A.F.; Homeier, T.; Foerster, T.; Fischer, S.; Truyen, U. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch. Virol. 2013, 158, 1173–1180. [Google Scholar] [CrossRef]
- Lagan Tregaskis, P.; Staines, A.; Gordon, A.; Sheridan, P.; McMenamy, M.; Duffy, C.; Collins, P.J.; Mooney, M.H.; Lemon, K. Co-infection status of novel parvovirus’s (PPV2 to 4) with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound. Emerg. Dis. 2021, 68, 1979–1994. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P.G. Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet. Microbiol. 2014, 173, 9–16. [Google Scholar] [CrossRef]
- Qin, S.; Ruan, W.; Yue, H.; Tang, C.; Zhou, K.; Zhang, B. Viral communities associated with porcine respiratory disease complex in intensive commercial farms in Sichuan province, China. Sci. Rep. 2018, 8, 13341. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Camacho, L.A.; Vargas-Ruiz, A.; Marin-Flamand, E.; Ramírez-Álvarez, H.; Brown, C. A retrospective study of DNA prevalence of porcine parvoviruses in Mexico and its relationship with porcine circovirus associated disease. Microbiol. Immunol. 2020, 64, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Novosel, D.; Cadar, D.; Tuboly, T.; Jungic, A.; Stadejek, T.; Ait-Ali, T.; Cságola, A. Investigating porcine parvoviruses genogroup 2 infection using in situ polymerase chain reaction. BMC Vet. Res. 2018, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Tse, H.; Fu, C.T.Y.; Au, W.-K.; Chen, X.-C.; Tsoi, H.-W.; Tsang, T.H.F.; Chan, J.S.Y.; Tsang, D.N.C.; et al. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008, 89, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, Y.; Liu, J.; Tang, Q.; Liu, C. Prevalence of porcine hokovirus and its co-infection with porcine circovirus 2 in China. Arch. Virol. 2013, 158, 1987–1991. [Google Scholar] [CrossRef]
- Adlhoch, C.; Kaiser, M.; Ellerbrok, H.; Pauli, G. High prevalence of porcine Hokovirus in German wild boar populations. Virol. J. 2010, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Cságola, A.; Kiss, T.; Tuboly, T. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol. Phylogenet. Evol. 2013, 66, 243–253. [Google Scholar] [CrossRef]
- Cheung, A.K.; Wu, G.; Wang, D.; Bayles, D.O.; Lager, K.M.; Vincent, A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010, 155, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhai, S.-L.; Cheung, A.K.; Zhang, H.-B.; Long, J.-X.; Yuan, S.-S. Detection of a novel porcine parvovirus, PPV4, in Chinese swine herds. Virol. J. 2010, 7, 333. [Google Scholar] [CrossRef] [PubMed]
- Gava, D.; Souza, C.K.; Schaefer, R.; Vincent, A.L.; Cantão, M.E.; Coldebella, A.; Ciacci-Zanella, J.R. A TaqMan-based real-time PCR for detection and quantification of porcine parvovirus 4. J. Virol. Methods 2015, 219, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; Giménez-Lirola, L.G.; Jiang, Y.-H.; Halbur, P.G.; Opriessnig, T. Characterization of a novel porcine parvovirus tentatively designated PPV5. PLoS ONE 2013, 8, e65312. [Google Scholar] [CrossRef]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol. J. 2014, 11, 203. [Google Scholar] [CrossRef] [PubMed]
- Cibulski, S.; Alves de Lima, D.; Fernandes Dos Santos, H.; Teixeira, T.F.; Tochetto, C.; Mayer, F.Q.; Roehe, P.M. A plate of viruses: Viral metagenomics of supermarket chicken, pork and beef from Brazil. Virology 2021, 552, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-C.; Kim, J.-H.; Kim, J.-Y.; Park, G.-S.; Jeong, C.-G.; Kim, W.-I. Prevalence of porcine parvovirus 1 through 7 (PPV1–PPV7) and co-factor association with PCV2 and PRRSV in Korea. BMC Vet. Res. 2022, 18, 133. [Google Scholar] [CrossRef]
- Li, J.; Xiao, Y.; Qiu, M.; Li, X.; Li, S.; Lin, H.; Li, X.; Zhu, J.; Chen, N. A Systematic Investigation Unveils High Coinfection Status of Porcine Parvovirus Types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 2021, 9, e0129421. [Google Scholar] [CrossRef] [PubMed]
- Ouh, I.-O.; Park, S.; Lee, J.-Y.; Song, J.Y.; Cho, I.-S.; Kim, H.-R.; Park, C.-K. First detection and genetic characterization of porcine parvovirus 7 from Korean domestic pig farms. J. Vet. Sci. 2018, 19, 855–857. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cao, L.; Sun, W.; Xin, J.; Zheng, M.; Tian, M.; Lu, H.; Jin, N. Sequence and phylogenetic analysis of novel porcine parvovirus 7 isolates from pigs in Guangxi, China. PLoS ONE 2019, 14, e0219560. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Stadejek, T. The detection and genetic diversity of novel porcine parvovirus 7 (PPV7) on Polish pig farms. Res. Vet. Sci. 2018, 120, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Rendon-Marin, S.; Ruiz-Saenz, J.; Mogollón, D.; Jaime, J. The first report of porcine parvovirus 7 (PPV7) in Colombia demonstrates the presence of variants associated with modifications at the level of the VP2-capsid protein. PLoS ONE 2021, 16, e0258311. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Podgórska, K.; Stadejek, T. Do porcine parvoviruses 1 through 7 (PPV1-PPV7) have an impact on porcine circovirus type 2 (PCV2) viremia in pigs? Vet. Microbiol. 2020, 242, 108613. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; Piscopo, N.; Pagnini, U.; Esposito, L.; Montagnaro, S. Detection of selected pathogens in reproductive tissues of wild boars in the Campania region, southern Italy. Acta Vet. Scand. 2024, 66, 9. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Díaz, A.; Mogollón, J.D.; Jaime, J. Longitudinal comparison of the humoral immune response and viral load of Porcine Circovirus Type 2 in pigs with different vaccination schemes under field conditions. [version 2; peer review: 2 approved, 1 approved with reservations]. F1000Research 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Batista, L.; Pijoan, C.; Dee, S.; Olin, M.; Molitor, T.; Joo, H.S.; Xiao, Z.; Murtaugh, M. Virological and immunological responses to porcine reproductive and respiratory syndrome virus in a large population of gilts. Can. J. Vet. Res. 2004, 68, 267–273. [Google Scholar] [PubMed]
- Corzo, C.A.; Mondaca, E.; Wayne, S.; Torremorell, M.; Dee, S.; Davies, P.; Morrison, R.B. Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermúdez, D.S.; Vargas-Pinto, M.A.; Mogollón, J.D.; Jaime, J. Field infection of a gilt and its litter demonstrates vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (PCV3). BMC Vet. Res. 2021, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Campos, F.S.; Bonil, L.; Mogollon, D.; Jaime, J. First detection of porcine circovirus type 3 in Colombia and the complete genome sequence demonstrates the circulation of PCV3a1 and PCV3a2. Vet. Med. Sci. 2019, 5, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, M.; Nunes, T.; Heuer, C.; Marshall, J.; Sanchez, J. Tools for the Analysis of Epidemiological Data. In Package EpiR: CRAN. 2017. Available online: http://cran.nexr.com/web/packages/epiR/epiR.pdf (accessed on 25 February 2024).
- Xiao, C.-T.; Giménez-Lirola, L.G.; Halbur, P.G.; Opriessnig, T. Increasing porcine PARV4 prevalence with pig age in the U.S. pig population. Vet. Microbiol. 2012, 160, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Biernacka, K.; Fan, J.; Gerber, P.F.; Stadejek, T.; Opriessnig, T. Circulation of Porcine Parvovirus Types 1 through 6 in Serum Samples Obtained from Six Commercial Polish Pig Farms. Transbound. Emerg. Dis. 2017, 64, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Zhou, H.; Tong, L.; Chen, Y.; Sun, Y.; Wang, H.; Zhang, G. First identification of porcine parvovirus 7 in China. Arch. Virol. 2018, 163, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Tramontana, S.; Bionaz, M.; Sharma, A.; Graugnard, D.E.; Cutler, E.A.; Ajmone-Marsan, P.; Hurley, W.L.; Loor, J.J. Internal controls for quantitative polymerase chain reaction of swine mammary glands during pregnancy and lactation. J. Dairy Sci. 2008, 91, 3057–3066. [Google Scholar] [CrossRef] [PubMed]
- Drigo, M.; Franzo, G.; Belfanti, I.; Martini, M.; Mondin, A.; Ceglie, L. Validation and comparison of different end point and real time RT-PCR assays for detection and genotyping of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2014, 201, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Kleiboeker, S.B.; Schommer, S.K.; Lee, S.-M.; Watkins, S.; Chittick, W.; Polson, D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Investig. 2005, 17, 165–170. [Google Scholar] [CrossRef]
- Song, C.; Zhu, C.; Zhang, C.; Cui, S. Detection of porcine parvovirus using a taqman-based real-time pcr with primers and probe designed for the NS1 gene. Virol. J. 2010, 7, 353. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Prickett, J.R.; Madson, D.M.; Shen, H.-G.; Juhan, N.M.; Pogranichniy, R.R.; Meng, X.-J.; Halbur, P.G. Porcine circovirus type 2 (PCV2)-infection and re-inoculation with homologous or heterologous strains: Virological, serological, pathological and clinical effects in growing pigs. Vet. Res. 2010, 41, 31. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.P.M.; Loiko, M.R.; Andrade, J.d.S.; Tochetto, C.; Cibulski, S.P.; Lima, D.A.; Weber, M.N.; Roehe, P.M.; Mayer, F.Q. Complete genome characterization of porcine circovirus 3 recovered from wild boars in Southern Brazil. Transbound. Emerg. Dis. 2021, 68, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.-B.; Zhao, Y.; Cui, J.-T.; Zheng, H.-H.; Xu, T.; Hou, C.-Y.; Wang, Z.-Y.; Li, X.-S.; Zheng, L.-L.; Chen, H.-Y. Molecular detection and phylogenetic analysis of Porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 2021, 68, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Kleinbaum, D.G.; Klein, M. Logistic Regression: A Self-Learning Text (Statistics for Biology and Health), 3rd ed.; Springer: New York, NY, USA, 2010; p. 719. ISBN 978-1-4419-1741-6. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, CRAN. R package version 1.0.7.; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Faustini, G.; Tucciarone, C.M.; Franzo, G.; Donneschi, A.; Boniotti, M.B.; Alborali, G.L.; Drigo, M. Molecular Survey on Porcine Parvoviruses (PPV1-7) and Their Association with Major Pathogens in Reproductive Failure Outbreaks in Northern Italy. Viruses 2024, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, S.; Barua, S.; Tripathi, B.N.; Rouse, B.T. Virological and immunological outcomes of coinfections. Clin. Microbiol. Rev. 2018, 31, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Dupont, K.; Nielsen, E.O.; Baekbo, P.; Larsen, L.E. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet. Microbiol. 2008, 128, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Kekarainen, T.; Montoya, M.; Dominguez, J.; Mateu, E.; Segalés, J. Porcine circovirus type 2 (PCV2) viral components immunomodulate recall antigen responses. Vet. Immunol. Immunopathol. 2008, 124, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Chase, C.; Lunney, J.K. Immune System. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 264–291. ISBN 9781119350859. [Google Scholar]
- Bisimwa, P.N.; Wasso, D.S.; Bantuzeko, F.; Aksanti, C.B.; Tonui, R.; Birindwa, A.B.; Bisimwa, E.B. First investigation on the presence of porcine parvovirus type 3 in domestic pig farms without reproductive failure in the Democratic Republic of Congo. Vet. Anim. Sci. 2021, 13, 100187. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Perez, A.; Sreevatsan, S.; Davies, P.; Culhane, M.; Torremorell, M. Association between Influenza A Virus Infection and Pigs Subpopulations in Endemically Infected Breeding Herds. PLoS ONE 2015, 10, e0129213. [Google Scholar] [CrossRef] [PubMed]
- Ouh, I.-O.; Lee, J.-Y.; Choi, H.; Moon, S.Y.; Song, J.Y.; Hyun, B.-H.; Kwak, D.; Lee, Y.-H.; Park, C.-K. Prevalence of Porcine Parvoviruses 1 to 6 and Porcine Circovirus 3 Infections and of Their Co-infections with Porcine Circovirus 2 in the Republic of Korea. Preprints 2023, 2023051112. [Google Scholar] [CrossRef]
- Valdes-Donoso, P.; Alvarez, J.; Jarvis, L.S.; Morrison, R.B.; Perez, A.M. Production losses from an endemic animal disease: Porcine reproductive and respiratory syndrome (PRRS) in selected midwest US sow farms. Front. Vet. Sci. 2018, 5, 102. [Google Scholar] [CrossRef]
- Madson, D.M.; Opriessnig, T. Effect of porcine circovirus type 2 (PCV2) infection on reproduction: Disease, vertical transmission, diagnostics and vaccination. Anim. Health Res. Rev. 2011, 12, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, K.J.; Askaa, J. Fetal infection with porcine parvovirus in herds with reproductive failure. Acta Vet. Scand. 1981, 22, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef] [PubMed]
- Allan, G.M.; Kennedy, S.; McNeilly, F.; Foster, J.C.; Ellis, J.A.; Krakowka, S.J.; Meehan, B.M.; Adair, B.M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 1999, 121, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Krakowka, S.; Ellis, J.A.; Meehan, B.; Kennedy, S.; McNeilly, F.; Allan, G. Viral wasting syndrome of swine: Experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet. Pathol. 2000, 37, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Eterpi, M.; McDonnell, G.; Thomas, V. Virucidal Activity of Disinfectants against Parvoviruses and Reference Viruses. Applied Biosafety 2010, 15, 165–171. [Google Scholar] [CrossRef]
- Streck, A.F.; Truyen, U. Porcine Parvovirus. Curr. Issues Mol. Biol. 2020, 37, 33–46. [Google Scholar] [CrossRef]
- Zeeuw, E.J.L.; Leinecker, N.; Herwig, V.; Selbitz, H.J.; Truyen, U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J. Gen. Virol. 2007, 88, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Koketsu, Y.; Tani, S.; Iida, R. Factors for improving reproductive performance of sows and herd productivity in commercial breeding herds. Porc. Health Manag. 2017, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Jara, M.; Rasmussen, D.A.; Corzo, C.A.; Machado, G. Porcine reproductive and respiratory syndrome virus dissemination across pig production systems in the United States. Transbound. Emerg. Dis. 2021, 68, 667–683. [Google Scholar] [CrossRef] [PubMed]
- Mastin, A.; Alarcon, P.; Pfeiffer, D.; Wood, J.; Williamson, S.; Brown, I.; COSI Consortium; Wieland, B. Prevalence and risk factors for swine influenza virus infection in the English pig population. PLoS Curr. Influenza 2011, 3, RRN1209. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. An Ancient Lineage of Highly Divergent Parvoviruses Infects both Vertebrate and Invertebrate Hosts. Viruses 2019, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Ndze, V.N.; Cadar, D.; Cságola, A.; Kisfali, P.; Kovács, E.; Farkas, S.; Ngu, A.F.; Esona, M.D.; Dán, Á.; Tuboly, T.; et al. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect. Genet. Evol. 2013, 17, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Parsyan, A.; Szmaragd, C.; Allain, J.-P.; Candotti, D. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J. Gen. Virol. 2007, 88, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Nelsen, A.; Lin, C.-M.; Hause, B.M. Porcine parvovirus 2 is predominantly associated with macrophages in porcine respiratory disease complex. Front. Vet. Sci. 2021, 8, 726884. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, S.; Moffett, D.; McNeilly, F.; Meehan, B.; Ellis, J.; Krakowka, S.; Allan, G.M. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 2000, 122, 9–24. [Google Scholar] [CrossRef] [PubMed]
| Variable | Variable Categories | Frequency |
|---|---|---|
| Herd size | Small (100 to 300 breeding females) | 16 (40%) |
| Medium (301 to 1000 breeding females) | 12 (30%) | |
| Large (≥1000 breeding females) | 5 (12.5%) | |
| No response * | 7 (17.5%) | |
| Acclimatization practice | Yes | 9 (22.5%) |
| No | 25 (62.5%) | |
| No response * | 6 (15%) | |
| Replacement rate | ≥50% | 9 (22.5%) |
| 30–50 | 16 (40%) | |
| ≤30 | 2 (5%) | |
| No response * | 13 (32.5%) | |
| Source of the gilts | External (from other farms) | 8 (20%) |
| Internal (from the same farm) | 19 (47.5%) | |
| Internal and external | 7 (17.5%) | |
| No response * | 6 (15%) | |
| Farrowing rate (mean 86.6%) | Herds with a mean <86.6 | 11 (27.5) |
| Herds with a mean >86.6 | 18 (45%) | |
| No response * | 11 (27.5%) | |
| Percentage of stillbirths (mean 5.4%) | Herds with a mean >5.4 | 15 (37.5%) |
| Herds with a mean <5.4 | 16 (40%) | |
| No response * | 9 (22.5%) | |
| Percentage of mummies (mean 5.1%) | Herds with a mean >5.1 | 10 (25%) |
| Herds with a mean <5.1 | 21 (52.5%) | |
| No response * | 9 (22.5%) | |
| Number of pigs weaned per sow (mean 11.2) | Herds with a mean >11.2 | 17 (42.5%) |
| Herds with a mean <11.2 | 13 (32.5%) | |
| No response * | 10 (25%) | |
| Number of pigs weaned per year (mean 27) | Herds with a mean >27 | 15 (37.5%) |
| Herds with a mean <27 | 9 (22.5%) | |
| No response * | 16 (40%) |
| PPVs | PCV2 n * (%) | PCV3 n (%) | PRRSV n (%) |
|---|---|---|---|
| PPV1 | 18 (7.7) | 10 (4.3) | 20 (8.5) |
| PPV2 | 10 (4.3) | 9 (3.9) | 15 (6.4) |
| PPV3 | 38 (16.2) | 35 (15) | 64 (27.3) |
| PPV4 | 8 (3.4) | 0 (0.0) | 7 (3) |
| PPV5 | 28 (12) | 14 (6) | 36 (15.4) |
| PPV6 | 22 (9.4) | 21 (9) | 37 (15.8) |
| PPV7 | 3 (1.3) | 1 (0.4) | 3 (1.3) |
| Variable | Regression Coefficient | p Value | OR * | 95% CI |
|---|---|---|---|---|
| Intercept | 0.4813 | 0.4487 | ||
| Positive PCV3 | −3.1804 | 0.0071 | 0.04 | 0.002–0.30 |
| Positive PPV4 | 2.2666 | 0.0993 | 9.64 | 0.84–266.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Bermudez, D.S.; Diaz, A.; Polo, G.; Mogollon, J.D.; Jaime, J. Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance. Vet. Sci. 2024, 11, 185. https://doi.org/10.3390/vetsci11050185
Vargas-Bermudez DS, Diaz A, Polo G, Mogollon JD, Jaime J. Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance. Veterinary Sciences. 2024; 11(5):185. https://doi.org/10.3390/vetsci11050185
Chicago/Turabian StyleVargas-Bermudez, Diana S., Andres Diaz, Gina Polo, Jose Dario Mogollon, and Jairo Jaime. 2024. "Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance" Veterinary Sciences 11, no. 5: 185. https://doi.org/10.3390/vetsci11050185
APA StyleVargas-Bermudez, D. S., Diaz, A., Polo, G., Mogollon, J. D., & Jaime, J. (2024). Infection and Coinfection of Porcine-Selected Viruses (PPV1 to PPV8, PCV2 to PCV4, and PRRSV) in Gilts and Their Associations with Reproductive Performance. Veterinary Sciences, 11(5), 185. https://doi.org/10.3390/vetsci11050185






