Detection and Phylogenetic Analysis of Caprine Arthritis Encephalitis Virus Using TaqMan-based qPCR in Eastern China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Primers and Probes
2.3. RNA Extraction and Reverse Transcription
2.4. The TaqMan RT-qPCR Method
2.5. Standard Curve Generation
2.6. Specificity and Sensitivity Analysis of TaqMan RT-qPCR Assay
2.7. Repeatability Determination of TaqMan RT-qPCR Assay
2.8. Detecting of Clinical Samples
2.9. Construction of Phylogenetic Tree
3. Results
3.1. Designing Specific Primers and Probes
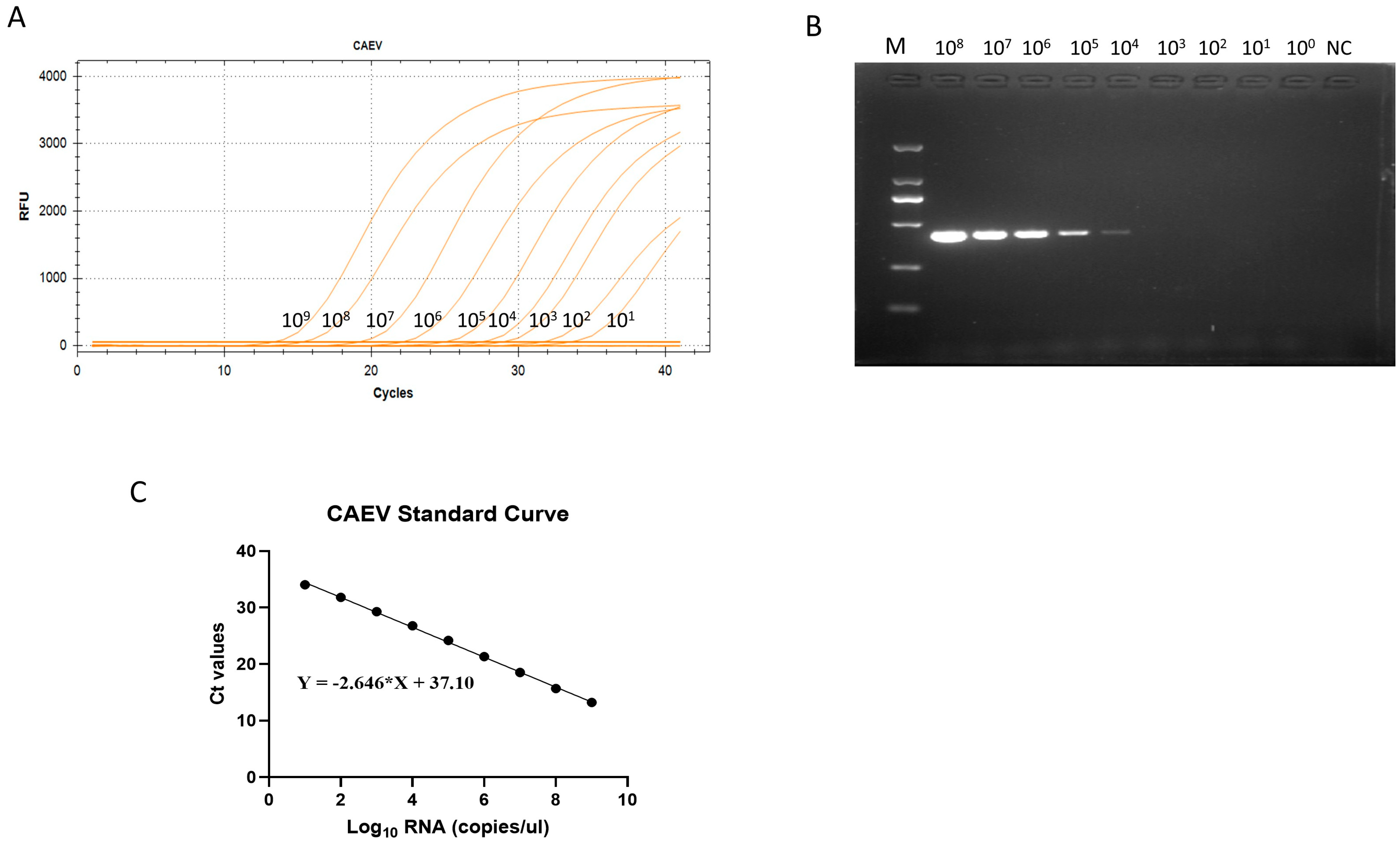
3.2. Preparation of the Standard Curve of Caprine Arthritis Encephalitis Virus
3.3. The Specificity of the RT-qPCR Assay
3.4. The Sensitivity of the RT-qPCR Assay
3.5. Intra-Assay and Inter-Assay Reproducibility of the RT-qPCR Assay
3.6. Detection of Samples from Farms in Eastern China
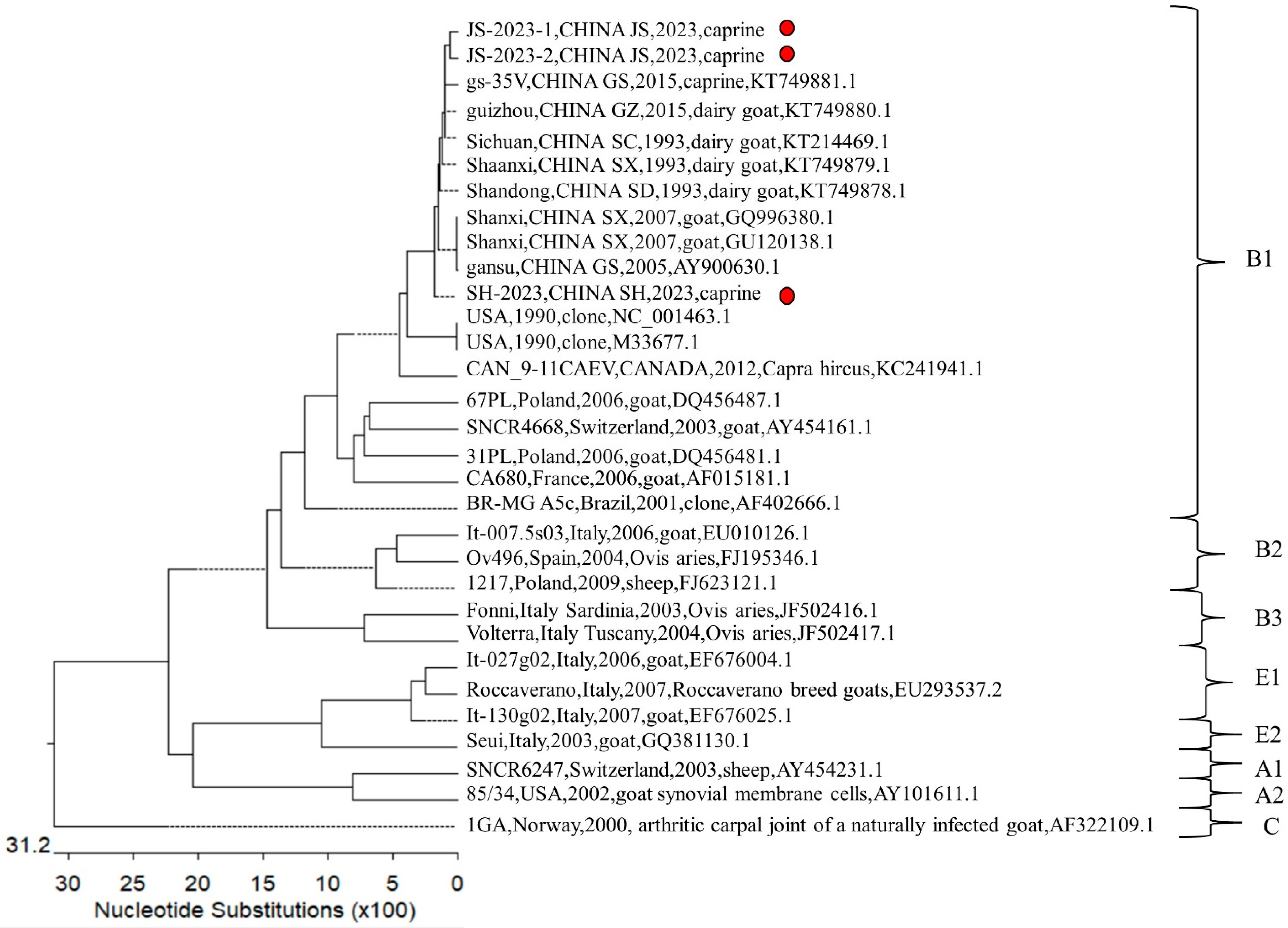
3.7. Phylogenetic Analyses of Caprine Arthritis Encephalitis Virus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Opendra Narayan, L.C.C. Lentiviral Diseases of Sheep and Goats: Chronic Pneumonia Leukoencephalomyelitis and Arthritis. Rev. Infect. Dis. 1985, 7, 89–98. [Google Scholar] [CrossRef]
- Liu, D.; Qiu, Z.; Pu, F.; Lin, J.; Li, J. Serologic diagnosis of Maedi visna. Chin. Vet. Sci. 1990, 9–12. [Google Scholar]
- Gong, C.; Zhou, T.; Hu, Z.; Lin, J.; Yue, M. Serologic diagnosis of Maedi visna in Xinjiang (preliminary report). Chin. J. Vet. Med. 1984, 12, 11–14. [Google Scholar]
- Cheevers, W.P.; Roberson, S.; Klevjer-Anderson, P.; Crawford, T.B. Characterization of Caprine Arthritis~Encephalitis Virus. Arch. Virol. 1981, 67, 111. [Google Scholar] [CrossRef] [PubMed]
- Olech, M.; Murawski, M.; Kuźmak, J. Molecular analysis of small-ruminant lentiviruses in Polish flocks reveals the existence of a novel subtype in sheep. Arch. Virol. 2019, 164, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yamamoto, T.; Shimada, T.; Shirafuji, H.; Kameyama, K.-I.; Sentsui, H.; Murakami, K. Development of Enzyme-Linked Immunosorbent Assay for Detection of Antibody against Caprine Arthritis Encephalitis Virus Using Recombinant Protein of the Precursor of the Major Core Protein, p55gag. J. Vet. Diagn. Investig. 2010, 22, 341–482. [Google Scholar] [CrossRef] [PubMed]
- Arcangeli, C.; Lucarelli, D.; Torricelli, M.; Sebastiani, C.; Ciullo, M.; Pellegrini, C.; Felici, A.; Costarelli, S.; Giammarioli, M.; Feliziani, F.; et al. First Survey of SNPs in TMEM154, TLR9, MYD88 and CCR5 Genes in Sheep Reared in Italy and Their Association with Resistance to SRLVs Infection. Viruses 2021, 13, 1290. [Google Scholar] [CrossRef]
- Braz, G.F.; Heinemann, M.B.; Reis, J.K.P.; Teixeira, B.M.; Cruz, J.C.M.; Rajão, D.S.; Oliveira, F.G.; Alves, F.; Castro, R.S.; Leite, R.C.; et al. Genetic and antigenic characterization of Brazilian SRLV strains: Natural small ruminant interspecies transmission from mixed herds. Infect. Genet. Evol. 2022, 103, 105322. [Google Scholar] [CrossRef]
- L’Homme, Y.; Ouardani, M.; Lévesque, V.; Bertoni, G.; Simard, C.; Pisoni, G. Molecular characterization and phylogenetic analysis of small ruminant lentiviruses isolated from Canadian sheep and goats. Virol. J. 2011, 8, 271. [Google Scholar] [CrossRef]
- Pisoni, G.; Quasso, A.; Moroni, P. Phylogenetic analysis of small-ruminant lentivirus subtype B1 in mixed flocks: Evidence for natural transmission from goats to sheep. Virology 2005, 339, 147–152. [Google Scholar] [CrossRef]
- Crawford, T.B.; Adams, D.S.; Cheevers, W.P.; Cork, L.C. Chronic Arthritis in Goats Caused by a Retrovirus. Science 1980, 207, 997. [Google Scholar] [CrossRef]
- Zhang, G. Studies on caprine arthritis-encephalitis—I. Isolation and preliminary characterization of CAEV89-GB1026 strain. Chin. J. Prev. Vet. Med. 1991, 2, 3–6. [Google Scholar]
- Zhao, Z. Molecular Mechanisms of the Interaction between Host Restrictive Factors and RNA Viruses. Ph.D. Thesis, Jinlin University, Changchun, China, 2021. [Google Scholar]
- Michiels, R.; Van Mael, E.; Quinet, C.; Adjadj, N.; Cay, A.; De Regge, N. Comparative Analysis of Different Serological and Molecular Tests for the Detection of Small Ruminant Lentiviruses (SRLVs) in Belgian Sheep and Goats. Viruses 2018, 10, 696. [Google Scholar] [CrossRef]
- Shah, C.; Böni, J.; Huder, J.B.; Vogt, H.-R.; Mühlherr, J.; Zanoni, R.; Miserez, R.; Lutz, H.; Schüpbach, J. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: Evidence for regular sheep-to-goat transmission and worldwide propagation through livestock trade. Virology 2004, 319, 12–26. [Google Scholar] [CrossRef]
- Pérez, M.; Biescas, E.; de Andrés, X.; Leginagoikoa, I.; Salazar, E.; Berriatua, E.; Reina, R.; Bolea, R.; de Andrés, D.; Juste, R.A.; et al. Visna/maedi virus serology in sheep: Survey, risk factors and implementation of a successful control programme in Aragón (Spain). Vet. J. 2010, 186, 221–225. [Google Scholar] [CrossRef]
- Michiels, R.; Van Mael, E.; Quinet, C.; Welby, S.; Cay, A.B.; De Regge, N. Seroprevalence and risk factors related to small ruminant lentivirus infections in Belgian sheep and goats. Prev. Vet. Med. 2018, 151, 13–20. [Google Scholar] [CrossRef]
- Brajon, G.; Mandas, D.; Liciardi, M.; Taccori, F.; Meloni, M.; Corrias, F.; Montaldo, C.; Coghe, F.; Casciari, C.; Giammarioli, M.; et al. Development and Field Testing of a Real-Time PCR Assay for Caprine Arthritis-Encephalitis-Virus (CAEV). Open Virol. J. 2012, 6, 82–90. [Google Scholar] [CrossRef]
- Le Jan, C.; Bellaton, C.; Greenland, T.; Mornex, J.-F. Mammary transmission of caprine arthritis encephalitis virus a 3D model for in vitro study. Reprod. Nutr. Dev. 2005, 45, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Blacklaws, B.A.; Berriatua, E.; Torsteinsdottir, S.; Watt, N.J.; de Andres, D.; Klein, D.; Harkiss, G.D. Transmission of small ruminant lentiviruses. Vet. Microbiol. 2004, 101, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y. Study on Molecular Detection Techniques of Caprine Arthritis Encephalitis Virus. Master’s Thesis, Tianjin University, Tianjin, China, 2011. [Google Scholar]
- Adams, D.S.; Crawford, T.B.; Klevjer-Anderson, P. A pathogenetic study of the early connective tissue lesions of viral caprine arthritis-encephalitis. Am. J. Pathol. 1980, 99, 257–278. [Google Scholar] [PubMed]
- Toplu, N.; Oğuzoğlu, T.Ç. Caprine arthritis encephalitis virus-induced apoptosis associated with brain lesions in naturally infected kids. J. Comp. Pathol. 2023, 206, 36–43. [Google Scholar] [CrossRef]
- Santry, L.A.; de Jong, J.; Gold, A.C.; Walsh, S.R.; Menzies, P.I.; Wootton, S.K. Genetic characterization of small ruminant lentiviruses circulating in naturally infected sheep and goats in Ontario, Canada. Virus Res. 2013, 175, 30–44. [Google Scholar] [CrossRef]
- Heckert, R.A.; McNab, W.B.; Richardson, S.M.; Briscoe, M.R. Evaluation of an enzyme-linked immunosorbent assay for the detection of antibodies to caprine arthritis-encephalitis virus in goat serum. Can. J. Vet. Res. 1992, 56, 237. [Google Scholar]
- Marinho, R.C.; Martins, G.R.; Souza, K.C.; Sousa, A.L.M.; Silva, S.T.C.; Nobre, J.A.; Teixeira, M.F.S. Duplex nested-PCR for detection of small ruminant lentiviruses. Braz. J. Microbiol. 2018, 49, 83–92. [Google Scholar] [CrossRef]
- De Regge, N.; Cay, B. Development, validation and evaluation of added diagnostic value of a q(RT)-PCR for the detection of genotype A strains of small ruminant lentiviruses. J. Virol. Methods 2013, 194, 250–257. [Google Scholar] [CrossRef]
- DeMartini, J.C.; Halsey, W.; Boshoff, C.; York, D.; Howell, M.D. Comparison of a maedi-visna virus CA-TM fusion protein ELISA with other assays for detecting sheep infected with North American ovine lentivirus strains. Vet. Immunol. Immunopathol. 1999, 71, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.S.; Leite, R.C.; Resende, M.; Gouveia, A.M.G. Gouveia. A Labelled Avidin-Biotin ELISA to Detect Antibodies to Caprine Arthritis-encephalitis Virus in Goats’ Sera. Vet. Res. Commun. 1999, 23, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Varea, R.; Monleon, E.; Pacheco, C.; Luja’n, L.; Bolea, R.; Vargas, M.A.; Van Eynde, G.; Saman, E.; Dickson, L.; Harkiss, G.; et al. Early detection of maedi-visna (ovine progressive pneumonia) virus seroconversion in field sheep samples. J. Vet. Diagn. Investig. 2001, 13, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Reina, R.; Berriatua, E.; Luján, L.; Juste, R.; Sánchez, A.; de Andrés, D.; Amorena, B. Prevention strategies against small ruminant lentiviruses: An update. Vet. J. 2009, 182, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. Epidemics and prevention of caprine arthritis-encephalitis in China. Grass-Feed. Livest. 1995, 1, 54–56. [Google Scholar]
- Li, X. TaqMan PCR for Detection of Caprine Arthritis Encephalitis Virus & Eukaryotic Expression of Recombinant SU. Master’s Thesis, Tianjin University, Tianjin, China, 2012. [Google Scholar]
- Li, Y.; Zhou, F.; Li, X.; Wang, J.; Zhao, X.; Huang, J. Development of TaqMan-based qPCR method for detection of caprine arthritis–encephalitis virus (CAEV) infection. Arch. Virol. 2013, 158, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y. Establishment and Preliminary Application for Quantitative Real-Time RT-PCR Method of Small Ruminant Lentiviruses. Master’s Thesis, Shihezi University, Shihezi, China, 2018. [Google Scholar]
- Yu, S.; Lin, Y.; Chuan, W.; Hao, D.; Tianyi, Y.; Chen, Z.; Song, X.H. Serological investigation of caprine arthritis encephalitis and ovine progressive pneumoniina in 11 provinces of China. Chin. Vet. Sci. 2018, 48, 34–38. [Google Scholar] [CrossRef]
- Huang, J.; Sun, Y.; Liu, Y.; Xiao, H.; Zhuang, S. Development of a loop-mediated isothermal amplification method for rapid detection of caprine arthritis-encephalitis virus proviral DNA. Arch. Virol. 2012, 157, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Eltahir, Y.M.; Dovas, C.I.; Papanastassopoulou, M.; Koumbati, M.; Giadinis, N.; Verghese-Nikolakaki, S.; Koptopoulos, G. Development of a semi-nested PCR using degenerate primers for the generic detection of small ruminant lentivirus proviral DNA. J. Virol. Methods 2006, 135, 240–246. [Google Scholar] [CrossRef] [PubMed]
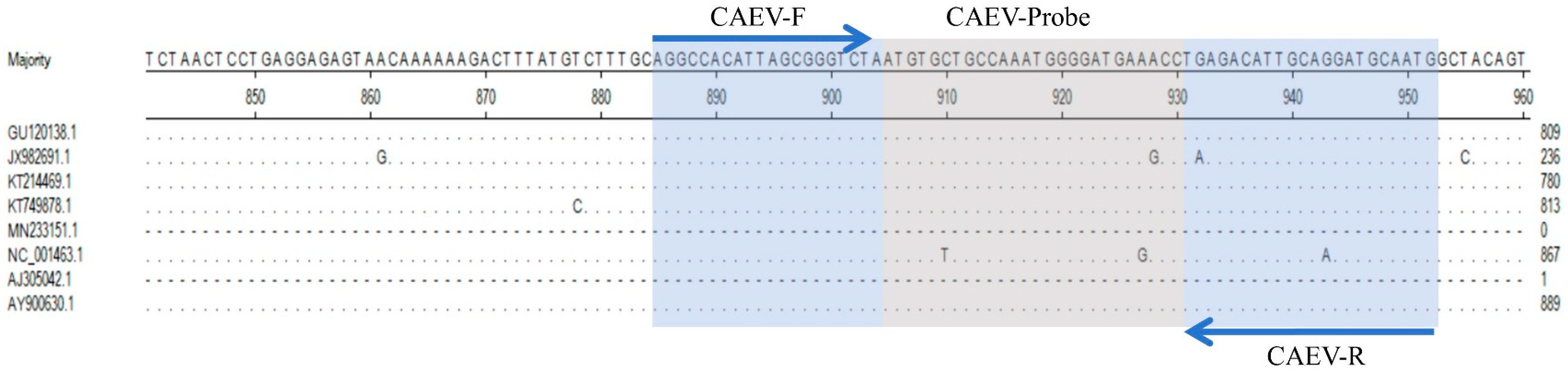

| Virus | Location | Primer/Probe | Sequences (5′-3′) | Length (bp) |
|---|---|---|---|---|
| CAEV | gag | Primer-CAEV-F | ATGTCTTTGCAGGCCACATT | 68 |
| Primer-CAEV-R | TGCAATGTCTCAGGTTTCATCC | |||
| Probe-CAEV | ROX-CCATTTGGCAGCACATTAGACCCGC-BHQ−1 | |||
| CAEV | gag | CAEV-F | AACTGGAAAGCAGTAGAC | 571 |
| CAEV-R | TACACTAGCTTGTTGCAC |
| Virus Standards | Copy Number | Intra-Assay | Inter-Assay | ||
|---|---|---|---|---|---|
| CT Value (Mean ± SD) | CV (%) | CT Value (Mean ± SD) | CV (%) | ||
| CAEV | 109 | 13.16 ± 0.06 | 0.3636 | 14.86 ± 0.19 | 0.6621 |
| 108 | 15.55 ± 0.14 | 0.7356 | 18.15 ± 0.07 | 0.2131 | |
| 107 | 18.44 ± 0.18 | 0.6902 | 21.87 ± 0.54 | 1.3212 | |
| 106 | 21.30 ± 0.05 | 0.1232 | 25.13 ± 0.20 | 0.4617 | |
| 105 | 24.23 ± 0.06 | 0.1856 | 28.29 ± 0.05 | 0.1212 | |
| 104 | 26.56 ± 0.23 | 0.6158 | 31.93 ± 0.49 | 0.8793 | |
| 103 | 29.38 ± 0.25 | 0.6095 | 34.34 ± 0.26 | 0.7571 | |
| 102 | 31.70 ± 0.42 | 0.9550 | 36.18 ± 0.47 | 1.2991 | |
| 101 | 34.34 ± 0.11 | 0.2615 | 38.56 ± 0.28 | 0.7261 | |
| 100 | NaN | NaN | NaN | NaN | |
| NC | NaN | NaN | NaN | NaN | |
| Location | Farm | Farm Size | Type of Sample | Sample Collection | Clinical Symptoms | NO. of Samples | NO. of Positive Samples |
|---|---|---|---|---|---|---|---|
| Jiangsu | JS-1 | 500–1000 | Goat Blood | Jugular | No | 34 | 2 (JS-2023-1, JS-2023-2) |
| JS-2 | >2000 | Goat Blood | No | 77 | 0 | ||
| JS-3 | >2000 | Goat Blood | No | 35 | 0 | ||
| Shanghai | SH-1 | >2000 | Sheep Blood | Jugular | No | 352 | 3 (SH-2023) |
| Sheep Brain | Dissection | Yes | 4 | 0 | |||
| SH-2 | 500–1000 | Sheep Blood | Jugular | No | 150 | 0 | |
| SH-3 | 500–1000 | Sheep Blood | Jugular | No | 128 | 1 | |
| Total | 780 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Zhang, H.; Zhang, Y.; Zhang, X.; Guan, Z.; Zhang, J.; Qiu, Y.; Li, B.; Liu, K.; Li, Z.; et al. Detection and Phylogenetic Analysis of Caprine Arthritis Encephalitis Virus Using TaqMan-based qPCR in Eastern China. Vet. Sci. 2024, 11, 138. https://doi.org/10.3390/vetsci11030138
Tian Y, Zhang H, Zhang Y, Zhang X, Guan Z, Zhang J, Qiu Y, Li B, Liu K, Li Z, et al. Detection and Phylogenetic Analysis of Caprine Arthritis Encephalitis Virus Using TaqMan-based qPCR in Eastern China. Veterinary Sciences. 2024; 11(3):138. https://doi.org/10.3390/vetsci11030138
Chicago/Turabian StyleTian, Yutong, Hailong Zhang, Yan Zhang, Xinya Zhang, Zhilei Guan, Junjie Zhang, Yafeng Qiu, Beibei Li, Ke Liu, Zongjie Li, and et al. 2024. "Detection and Phylogenetic Analysis of Caprine Arthritis Encephalitis Virus Using TaqMan-based qPCR in Eastern China" Veterinary Sciences 11, no. 3: 138. https://doi.org/10.3390/vetsci11030138
APA StyleTian, Y., Zhang, H., Zhang, Y., Zhang, X., Guan, Z., Zhang, J., Qiu, Y., Li, B., Liu, K., Li, Z., Shao, D., Li, P., Ma, Z., & Wei, J. (2024). Detection and Phylogenetic Analysis of Caprine Arthritis Encephalitis Virus Using TaqMan-based qPCR in Eastern China. Veterinary Sciences, 11(3), 138. https://doi.org/10.3390/vetsci11030138







