Molecular Epidemiology of Foot-and-Mouth Disease Viruses in the Emirate of Abu Dhabi, United Arab Emirates
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Outbreaks Investigations
2.2. Clinical Samples
2.3. Detection of FMDV via Real-Time Quantitative PCR (RT-qPCR) and Serotyping
2.4. Sanger Sequencing and Phylogenetic Analysis
3. Results
3.1. FMD-Positive Samples
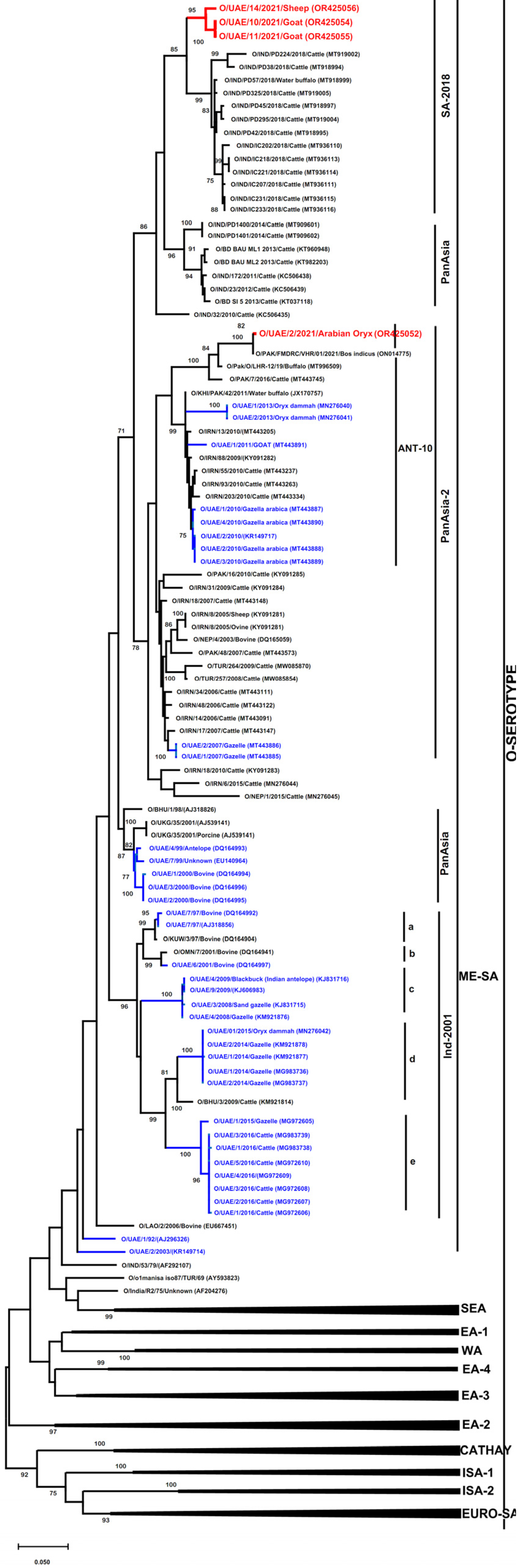
3.2. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sutmoller, P.; Olascoaga, R.C. Unapparent foot and mouth disease infection (sub-clinical infections and carriers): Implications for control. Rev. Sci. Tech. Off. Int. Épizoot. 2002, 21, 519–524. [Google Scholar] [CrossRef] [PubMed]
- WOAH. 2023. Available online: https://www.woah.org/en/disease/foot-and-mouth-disease/ (accessed on 3 March 2023).
- Grubman, M.J.; Baxt, B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004, 17, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Alkheraije, K.A. The prevalence of foot-and-mouth disease in Asia. Front. Vet. Sci. 2023, 10, 1201578. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Pacheco, J.M.; Rodriguez, L.L.; Arzt, J. Early events in the pathogenesis of foot-and-mouth disease in pigs; identification of oropharyngeal tonsils as sites of primary and sustained viral replication. PLoS ONE 2014, 9, e106859. [Google Scholar] [CrossRef] [PubMed]
- Barnett, P.; Cox, S. The role of small ruminants in the epidemiology and transmission of foot-and-mouth disease. Vet. J. 1999, 158, 6–13. [Google Scholar] [CrossRef]
- Jemberu, W.T.; Molla, W.; Fentie, T. A randomized controlled field trial assessing foot and mouth disease vaccine effectiveness in Gondar Zuria district, Northwest Ethiopia. Prev. Vet. Med. 2020, 183, 105136. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, A.; Sellers, R. Foot-and-mouth disease. Dis. Sheep 2000, 3, 254–258. [Google Scholar]
- OIE. Foot and Mouth Disease, Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Bees); World Organization for Animal Health: Paris, France, 2012; Volume 1, pp. 145–173. [Google Scholar]
- Brocchi, E.; Bergmann, I.; Dekker, A.; Paton, D.; Sammin, D.; Greiner, M.; Grazioli, S.; De Simone, F.; Yadin, H.; Haas, B. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine 2006, 24, 6966–6979. [Google Scholar] [CrossRef]
- Barnett, P.; Geale, D.; Clarke, G.; Davis, J.; Kasari, T. A review of OIE country status recovery using vaccinate-to-live versus vaccinate-to-die foot-and-mouth disease response policies I: Benefits of higher potency vaccines and associated NSP DIVA test systems in post-outbreak surveillance. Transbound. Emerg. Dis. 2015, 62, 367–387. [Google Scholar] [CrossRef]
- Jamal, S.M.; Belsham, G.J. Foot-and-mouth disease: Past, present and future. Vet. Res. 2013, 44, 116. [Google Scholar] [CrossRef]
- FAO; World Organisation for Animal Health. Ressources documentaires de l’OMSA. In Proceedings of the 2023 5th GF-TADs Middle East Roadmap and 2nd Epidemiology and Laboratory Networks Meeting for Foot-and-Mouth Disease, Virtual, 6–9 December 2021. [Google Scholar] [CrossRef]
- Madin, B. An evaluation of Foot-and-Mouth Disease outbreak reporting in mainland South-East Asia from 2000 to 2010. Prev. Vet. Med. 2011, 102, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Paton, D.J.; Gubbins, S.; King, D.P. Understanding the transmission of foot-and-mouth disease virus at different scales. Curr. Opin. Virol. 2018, 28, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Ramanoon, S.Z.; Robertson, I.D.; Edwards, J.; Hassan, L.; Md Isa, K. Outbreaks of foot-and-mouth disease in Peninsular Malaysia from 2001 to 2007. Trop. Anim. Health Prod. 2013, 45, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Young, J.R.; Nampanya, S.; Khounsy, S.; Singanallur, N.B.; Vosloo, W.; Abila, R.; Hamilton, S.A.; Bush, R.D.; Windsor, P.A. Risk factors for emergence of exotic foot-and-mouth disease O/ME-SA/Ind-2001d on smallholder farms in the Greater Mekong Subregion. Prev. Vet. Med. 2018, 159, 115–122. [Google Scholar] [CrossRef]
- Ringa, N.; Bauch, C.T. Dynamics and control of foot-and-mouth disease in endemic countries: A pair approximation model. J. Theor. Biol. 2014, 357, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Roche, S.; Garner, M.; Sanson, R.; Cook, C.; Birch, C.; Backer, J.; Dube, C.; Patyk, K.; Stevenson, M.; Yu, Z. Evaluating vaccination strategies to control foot-and-mouth disease: A model comparison study. Epidemiol. Infect. 2015, 143, 1256–1275. [Google Scholar] [CrossRef]
- Knowles, N.J.; Wadsworth, J.; Bachanek-Bankowska, K.; King, D. VP1 sequencing protocol for foot and mouth disease virus molecular epidemiology. Rev. Sci. Tech. 2016, 35, 741–755. [Google Scholar] [CrossRef]
- Brito, B.; Rodriguez, L.; Hammond, J.; Pinto, J.; Perez, A. Review of the global distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound. Emerg. Dis. 2017, 64, 316–332. [Google Scholar] [CrossRef]
- Di Nardo, A.; Knowles, N.; Paton, D. Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia. Rev. Sci. Tech. OIE 2011, 30, 63. [Google Scholar] [CrossRef]
- Sumption, K.; Domenech, J.; Ferrari, G. Progressive control of FMD on a global scale. Vet. Rec. 2012, 170, 637–639. [Google Scholar] [CrossRef]
- Knowles, N.; Nazem Shirazi, M.; Wadsworth, J.; Swabey, K.; Stirling, J.; Statham, R.; Li, Y.; Hutchings, G.; Ferris, N.; Parlak, Ü. Recent spread of a new strain (A-Iran-05) of foot-and-mouth disease virus type A in the Middle East. Transbound. Emerg. Dis. 2009, 56, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Bachanek-Bankowska, K.; Di Nardo, A.; Wadsworth, J.; Henry, E.K.; Parlak, Ü.; Timina, A.; Mischenko, A.; Qasim, I.A.; Abdollahi, D.; Sultana, M. Foot-and-mouth disease in the middle east caused by an A/ASIA/G-VII virus lineage, 2015–2016. Emerg. Infect. Dis. 2018, 24, 1073. [Google Scholar] [CrossRef] [PubMed]
- Bachanek-Bankowska, K.; Di Nardo, A.; Wadsworth, J.; Mioulet, V.; Pezzoni, G.; Grazioli, S.; Brocchi, E.; Kafle, S.C.; Hettiarachchi, R.; Kumarawadu, P.L. Reconstructing the evolutionary history of pandemic foot-and-mouth disease viruses: The impact of recombination within the emerging O/ME-SA/Ind-2001 lineage. Sci. Rep. 2018, 8, 14693. [Google Scholar] [CrossRef]
- FAO. Foot-and-Mouth Disease: Quarterly Report. Rome. January–March 2023. 2023. Available online: https://www.fao.org/documents/card/en/c/cc6065en (accessed on 5 March 2023).
- WAHIS. 2023. Available online: https://wahis.woah.org/#/dashboards/country-or-disease-dashboard (accessed on 3 March 2023).
- Lignereux, L.; Chaber, A.L.; Saegerman, C.; Heath, L.; Knowles, N.J.; Wadsworth, J.; Mioulet, V.; King, D.P. Foot-and-mouth disease outbreaks in captive scimitar-horned oryx (Oryx dammah). Transbound. Emerg. Dis. 2020, 67, 1716–1724. [Google Scholar] [CrossRef]
- FAO. The Progressive Control Pathway for Foot-and-Mouth Disease Control (PCP-FMD): Principles, Stage Descriptions and Standards; FAO: Rome, Italy, 2018. [Google Scholar]
- Callahan, J.D.; Brown, F.; Osorio, F.A.; Sur, J.H.; Kramer, E.; Long, G.W.; Lubroth, J.; Ellis, S.J.; Shoulars, K.S.; Gaffney, K.L. Use of a portable real-time reverse transcriptasepolymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 2002, 220, 1636–1642. [Google Scholar] [CrossRef]
- Shaw, A.E.; Reid, S.M.; Ebert, K.; Hutchings, G.H.; Ferris, N.P.; King, D.P. Implementation of a one-step real-time RT-PCR protocol for diagnosis of foot-and-mouth disease. J. Virol. Methods 2007, 143, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.R.; Wood, B.A.; Henry, E.; King, D.P.; Mioulet, V. Elimination of Non-cytopathic Bovine Viral Diarrhea Virus From the LFBK-αvβ6 Cell Line. Front. Vet. Sci. 2021, 8, 715120. [Google Scholar] [CrossRef]
- LaRocco, M.; Krug, P.W.; Kramer, E.; Ahmed, Z.; Pacheco, J.M.; Duque, H.; Baxt, B.; Rodriguez, L.L. A continuous bovine kidney cell line constitutively expressing bovine αvβ6 integrin has increased susceptibility to foot-and-mouth disease virus. J. Clin. Microbiol. 2013, 51, 1714–1720. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Sumption, K. The progressive control pathway for FMD (PCP-FMD): A tool for developing sustainable long term national and regional FMD control. In Proceedings of the FAO/OIE Global Conference on Foot Mouth Disease Control, Bangkok, Thailand, 27–29 June 2012. [Google Scholar]
- Mielke, S.R.; Garabed, R. Environmental persistence of foot-and-mouth disease virus applied to endemic regions. Transbound. Emerg. Dis. 2020, 67, 543–554. [Google Scholar] [CrossRef]
- Ulziibat, G.; Raizman, E.; Lkhagvasuren, A.; Bartels, C.J.; Oyun-Erdene, O.; Khishgee, B.; Browning, C.; King, D.P.; Ludi, A.B.; Lyons, N.A. Comparison of vaccination schedules for foot-and-mouth disease among cattle and sheep in Mongolia. Front. Vet. Sci. 2023, 10, 990043. [Google Scholar] [CrossRef] [PubMed]
- Buckle, K.; Bueno, R.; McFadden, A.; van Andel, M.; Spence, R.; Hamill, C.; Roe, W.; Vallee, E.; Castillo-Alcala, F.; Abila, R. Detection of foot-and-mouth disease virus in the absence of clinical disease in cattle and buffalo in South East Asia. Front. Vet. Sci. 2021, 8, 691308. [Google Scholar] [CrossRef] [PubMed]
- Colenutt, C.; Brown, E.; Paton, D.J.; Mahapatra, M.; Parida, S.; Nelson, N.; Maud, J.; Motta, P.; Sumption, K.; Adhikari, B. Environmental sampling for the detection of foot-and-mouth disease virus and peste des petits ruminants virus in a live goat market, Nepal. Transbound. Emerg. Dis. 2022, 69, 3041–3046. [Google Scholar] [CrossRef]
- Ostrowski, S.; Anajariyah, S. Middle East Arabian Oryx Disease Survey; National Wildlife Research Center: Taif, Saudi Arabia, 2002. [Google Scholar]
- Ulziibat, G.; Maygmarsuren, O.; Khishgee, B.; Basan, G.; Sandag, B.; Ruuragc, S.; Limon, G.; Wilsden, G.; Browning, C.; King, D.P. Immunogenicity of imported foot-and-mouth vaccines in different species in Mongolia. Vaccine 2020, 38, 1708–1714. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, T.; el-Gadir, F. Studies on foot and mouth disease in the eastern region of Abu Dhabi, United Arab Emirates. Rev. Sci. Tech. Int. Off. Epizoot. 1993, 12, 831–837. [Google Scholar] [CrossRef]
- FAO. World Reference Laboratory for FMD Genotyping Report. Country Reports: 2022 | World Reference Laboratory for Foot-and-Mouth Disease. Available online: https://wrlfmd.org (accessed on 13 January 2023).
- Raouf, Y.A.; Wadsworth, J.; Bin-Tarif, A.; Gray, A.R.; Habiela, M.; Almutalb, A.A.; Yousif, H.; Ragab, M.; Alfouz, W.; Ahmed, N.H. Genotyping of foot-and-mouth disease viruses collected in Sudan between 2009 and 2018. Transbound. Emerg. Dis. 2022, 69, e1393–e1406. [Google Scholar] [CrossRef]
- Maree, F.F.; Kasanga, C.J.; Scott, K.A.; Opperman, P.A.; Melanie, C.; Sangula, A.K.; Raphael, S.; Yona, S.; Wambura, P.N.; King, D.P. Challenges and prospects for the control of foot-and-mouth disease: An African perspective. Vet. Med. Res. Rep. 2014, 5, 119–138. [Google Scholar]


| Location of Cases | Notification Date | Livestock Population on the Farm (species) | FMD Affected Animals | Age of FMD Cases | Deaths | Morbidity Rate (%) | Mortality Rate (%) | Case Fatality Rate (%) | FMD Vaccination | |
|---|---|---|---|---|---|---|---|---|---|---|
| Farm A | Abu Dhabi | 10 April 2021 | 132 (sheep 24, Goat 93, Arabian oryx 15) | 4 Arabian oryx | 3 years | 1 Arabian oryx | 3 | 0.76 | 25 | Yes (one dose) 1 |
| Farm B | Abu Dhabi | 20 April 2021 | 44 (sheep 27, goats 17) | 3 sheep | <3 months | 0 | 6.8 | 0 | 0 | No |
| Farm C | Abu Dhabi | 21 April 2021 | 407 (sheep 202, goats 205) | 4 sheep | 1 year | 1 sheep | 1 | 0.24 | 25 | Yes (7 February 2021) |
| Farm D | Al Ain | 18 November 2021 | 529 2 (sheep 81, goats 448) | 94 (80 goats, 14 sheep) | 1 year | 9 (1 sheep, 8 goats) | 0.18 | 1.70 | 9.60 | No |
| Farm E | Al Ain | 19 December 2021 | 150 (sheep 100, goats 50) | 20 sheep | 1.5 years | 5 sheep | 0.11 | 0.03 | 25 | No |
| Market F | Al Ain | 1 December 2021 | 309 (cattle pens only) | 11 cattle | 4 years | 1 cattle | 3.56 | 0.3 | 9 | No |
| Farm Name | Date of Sample Collection | Sample Type | ADAFSA Label | WRLFMD Ref. No. | Animal Species Infected |
|---|---|---|---|---|---|
| A | April 2021 | Mouth swab | ADAFSA-2 | UAE/2/2021 | Arabian oryx |
| B | April 2021 | Mouth swab | ADAFSA-9 ADAFSA-10 | UAE/3/2021 UAE/4/2021 | Sheep |
| C | April 2021 | Mouth swab | ADAFSA-11 ADAFSA-12 ADAFSA-13 | UAE/7/2021 UAE/5/2021 UAE/6/2021 | Sheep |
| D | November 2021 | Mouth swab | ADAFSA-4 ADAFSA-5 | UAE/10/2021 UAE/11/2021 | Goat |
| E | December 2021 | Mouth swab | ADAFSA-14 | UAE/14/2021 | Sheep |
| F | December 2021 | Mouth swab | ADAFSA-6 ADAFSA-7 ADAFSA-8 | UAE/8/2021 UAE/12/2021 UAE/13/2021 | Cattle |
| O/UAE/2/2021/Arabian oryx | O/UAE/10/2021/Goat | O/UAE/11/2021/Goat | O/UAE/14/2021/Sheep | |
|---|---|---|---|---|
| O/UAE/2/2021/Arabian oryx | 88% | 88% | 88% | |
| O/UAE/10/2021/Goat | 88% | 100% | 98% | |
| O/UAE/11/2021/Goat | 88% | 100% | 98% | |
| O/UAE/14/2021/Sheep | 88% | 98% | 98% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eltahir, Y.M.; Ishag, H.Z.A.; Wadsworth, J.; Hicks, H.M.; Knowles, N.J.; Mioulet, V.; King, D.P.; Mohamed, M.S.; Bensalah, O.K.; Yusof, M.F.; et al. Molecular Epidemiology of Foot-and-Mouth Disease Viruses in the Emirate of Abu Dhabi, United Arab Emirates. Vet. Sci. 2024, 11, 32. https://doi.org/10.3390/vetsci11010032
Eltahir YM, Ishag HZA, Wadsworth J, Hicks HM, Knowles NJ, Mioulet V, King DP, Mohamed MS, Bensalah OK, Yusof MF, et al. Molecular Epidemiology of Foot-and-Mouth Disease Viruses in the Emirate of Abu Dhabi, United Arab Emirates. Veterinary Sciences. 2024; 11(1):32. https://doi.org/10.3390/vetsci11010032
Chicago/Turabian StyleEltahir, Yassir M., Hassan Zackaria Ali Ishag, Jemma Wadsworth, Hayley M. Hicks, Nick J. Knowles, Valérie Mioulet, Donald P. King, Meera Saeed Mohamed, Oum Keltoum Bensalah, Mohd Farouk Yusof, and et al. 2024. "Molecular Epidemiology of Foot-and-Mouth Disease Viruses in the Emirate of Abu Dhabi, United Arab Emirates" Veterinary Sciences 11, no. 1: 32. https://doi.org/10.3390/vetsci11010032
APA StyleEltahir, Y. M., Ishag, H. Z. A., Wadsworth, J., Hicks, H. M., Knowles, N. J., Mioulet, V., King, D. P., Mohamed, M. S., Bensalah, O. K., Yusof, M. F., Gasim, E. F. M., Hammadi, Z. M. A., Shah, A. A. M., Abdelmagid, Y. A., Gahlan, M. A. m. E., Kassim, M. F., Kayaf, K., Zahran, A., & Nuaimat, M. M. A. (2024). Molecular Epidemiology of Foot-and-Mouth Disease Viruses in the Emirate of Abu Dhabi, United Arab Emirates. Veterinary Sciences, 11(1), 32. https://doi.org/10.3390/vetsci11010032








