Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification of Bovine IgG1 and IgG2
2.2. HRP Labeling of the Bovine IgG
2.3. Design and Synthesis of boFcγRIII Peptides
2.4. Conjugation of the boFcγRIII Peptides
2.5. Peptide IgG-Binding Assay
2.6. Bovine IgG-Blocking Assay
2.7. Truncation and Mutation of the Fc-Binding Peptide
2.8. Model Building of boFcγRIII
3. Results
3.1. Bovine IgG1 Binding to the boFcγRIII Peptides
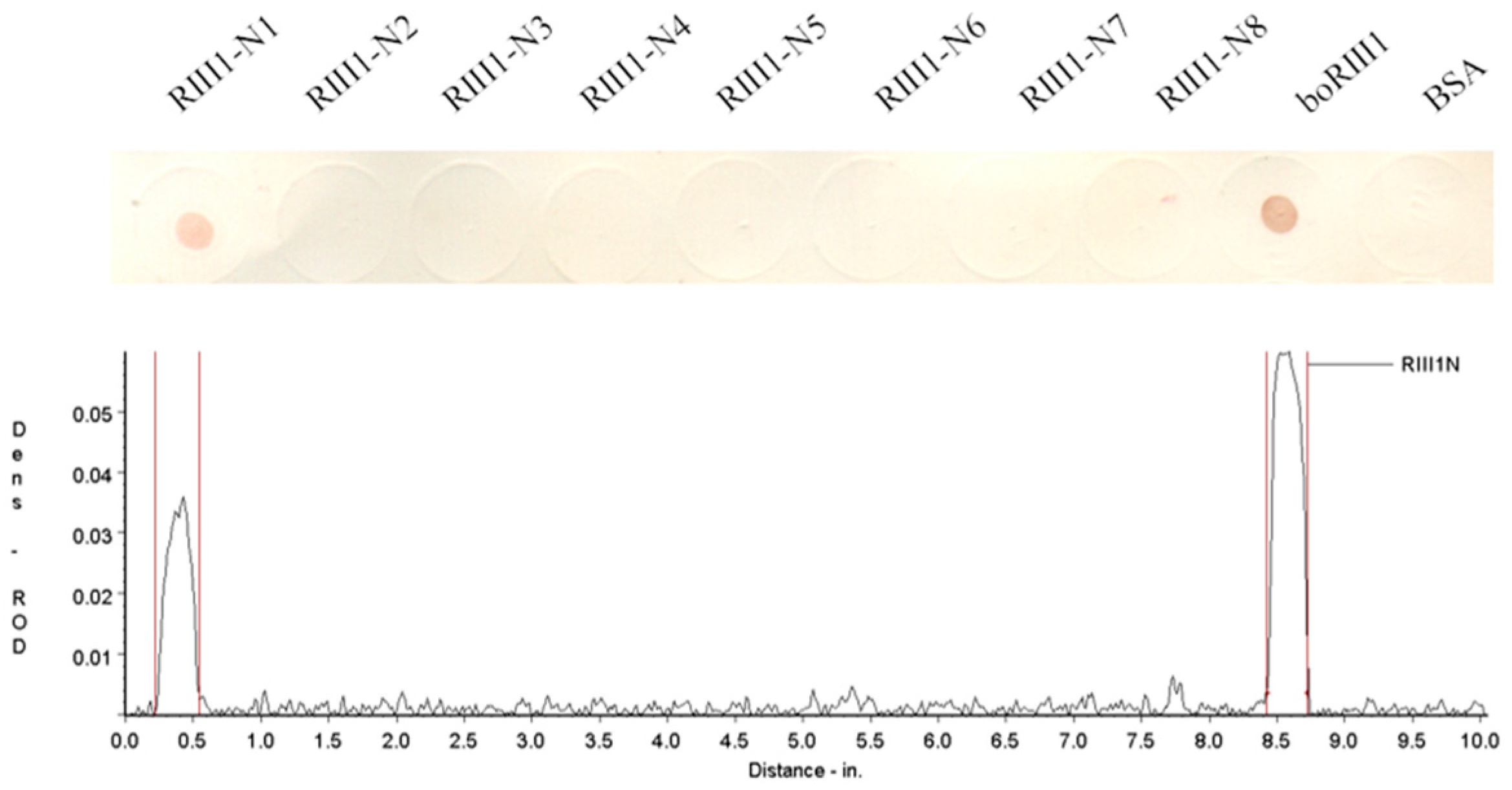
3.2. Determination of the Fc-Binding Site for Bovine IgG1
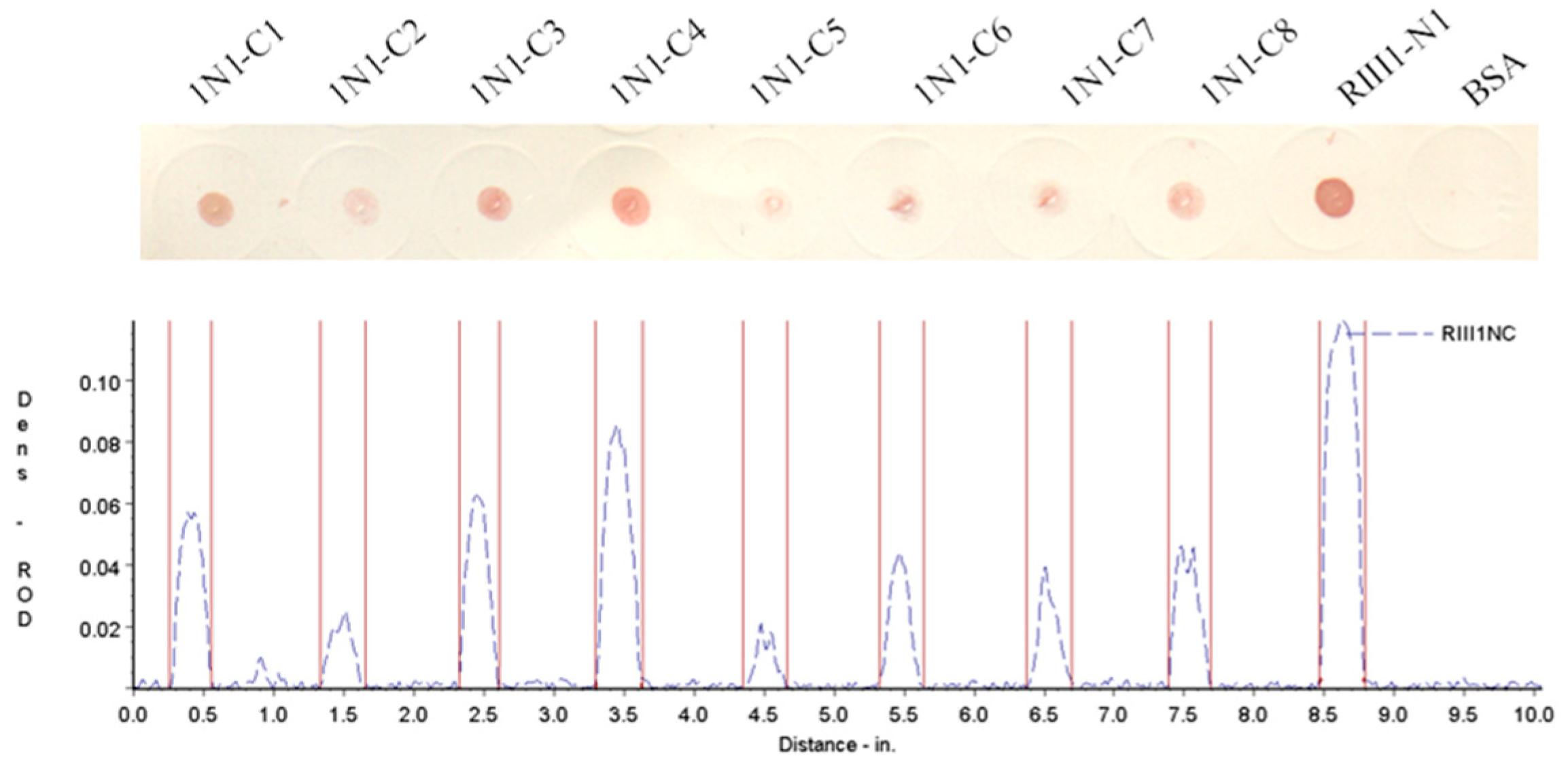
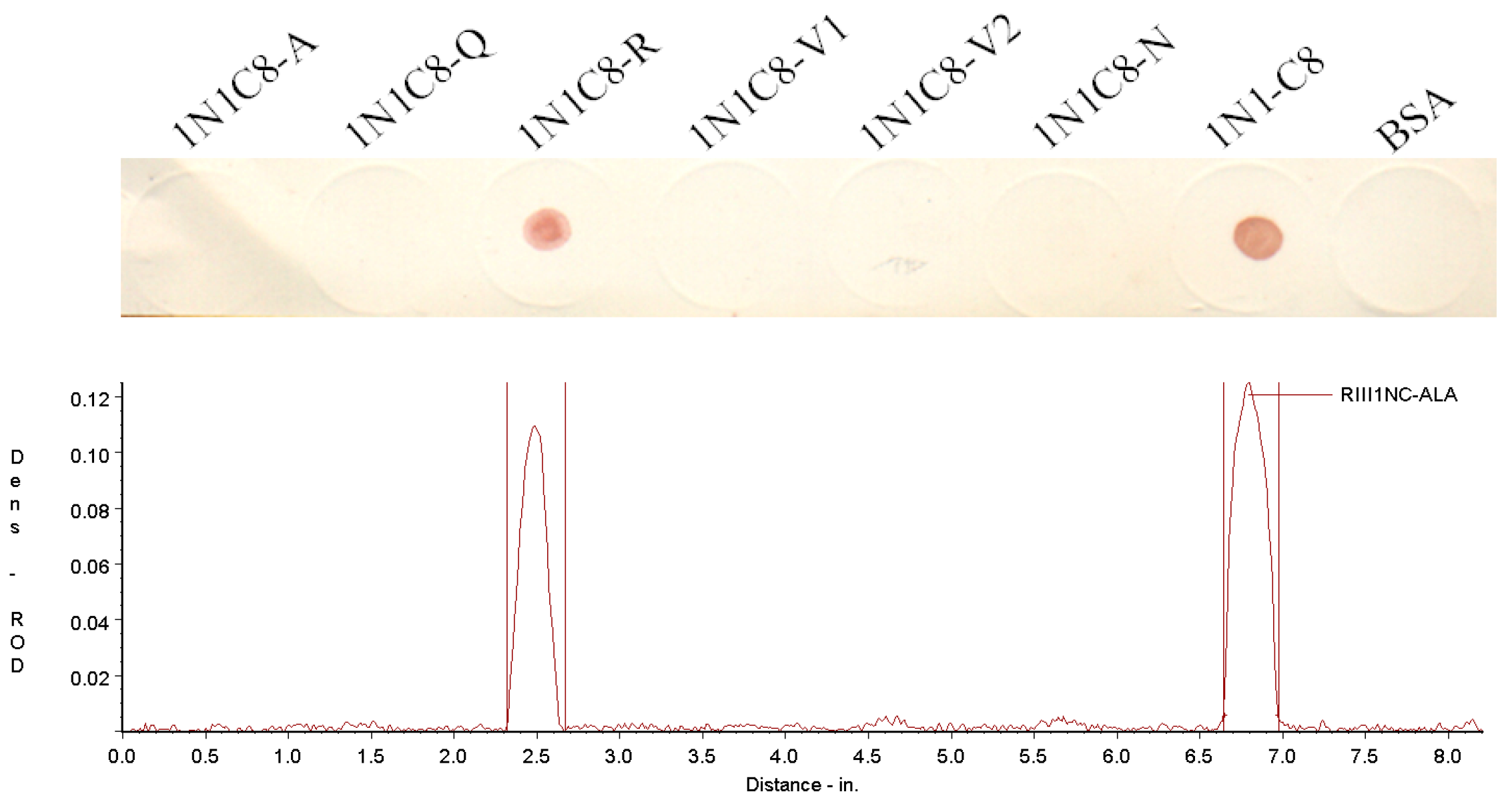
3.3. Crucial Residues for Bovine IgG1 in the Fc-Binding Site
3.4. Structural Characteristic of the Fc-Binding Site on boFcγRIII
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bournazos, S. IgG Fc receptors: Evolutionary considerations. In Fc Mediated Activity of Antibodies; Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2019; Volume 423, pp. 1–11. [Google Scholar]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Keeler, S.P.; Fox, J.M. Requirement of Fc-Fc gamma receptor interaction for antibody-based protection against emerging virus infections. Viruses 2021, 13, 1037. [Google Scholar] [CrossRef]
- Zhu, H.L.; Zhao, X.W.; Wang, X.Z.; Qi, Y.X.; Huang, D.W.; Cheng, G.L.; Zhao, H.L.; Yang, Y.X. Changes in expression of antimicrobial peptides and Fc receptors in the small intestines of neonatal calves during the passive immunity period. J. Dairy Sci. 2020, 103, 9515–9524. [Google Scholar] [CrossRef]
- Holtrop, T.; Budding, K.; Brandsma, A.M.; Leusen, J.H.W. Targeting the high affinity receptor, FcγRI, in autoimmune disease, neuropathy, and cancer. Immunother. Adv. 2022, 2, ltac011. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Bjorkman, P.J. Fc receptors and their interactions with immunoglobulins. Annu. Rev. Cell Dev. Biol. 1996, 12, 181–220. [Google Scholar] [CrossRef] [PubMed]
- Flesch, B.K.; Neppert, J. Functions of the Fc receptors for immunoglobulin G. J. Clin. Lab. Anal. 2000, 14, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Sondermann, P.; Huber, R.; Oosthuizen, V.; Jacob, U. The 3.2-Å crystal structure of the human IgG1 Fc fragment-FcγRIII complex. Nature 2000, 406, 267–273. [Google Scholar] [CrossRef]
- Zhang, Y.; Boesen, C.C.; Radaev, S.; Brooks, A.G.; Fridman, W.H.; Sautes-Fridman, C.; Sun, P.D. Crystal structure of the extracellular domain of a human FcγRIII. Immunity 2000, 13, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Radaev, S.; Motyka, S.; Fridman, W.H.; Sautes-Fridman, C.; Sun, P.D. The structure of a human type III Fcγ receptor in complex with Fc. J. Biol. Chem. 2001, 276, 16469–16477. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Keremane, S.R.; Vielmetter, J.; Bjorkman, P.J. Structural characterization of GASDALIE Fc bound to the activating Fc receptor FcγRIIIa. J. Struct. Biol. 2016, 194, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Barb, A.W. A single amino acid distorts the Fcγ receptor IIIb/CD16b structure upon binding immunoglobulin G1 and reduces affinity relative to CD16a. J. Biol. Chem. 2018, 293, 19899–19908. [Google Scholar] [CrossRef]
- Tolbert, W.D.; Gohain, N.; Kremer, P.G.; Hederman, A.P.; Nguyen, D.N.; Van, V.; Sherburn, R.; Lewis, G.K.; Finzi, A.; Pollara, J.; et al. Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques. Front. Immunol. 2022, 13, 960411. [Google Scholar] [CrossRef] [PubMed]
- Uray, K.; Medgyesi, D.; Hilbert, A.; Sármay, G.; Gergely, J.; Hudecz, F. Synthesis and receptor binding of IgG1 peptides derived from the IgG Fc region. J. Mol. Recognit. 2004, 17, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Cendron, A.C.; Wines, B.D.; Brownlee, R.T.; Ramsland, P.A.; Pietersz, G.A.; Hogarth, P.M. An FcγRIIa-binding peptide that mimics the interaction between FcγRIIa and IgG. Mol. Immunol. 2008, 45, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, S.; Spadola, L.; Buchanan, A.G.; Jermutus, L.; Lund, J. Identification of cyclic peptides able to mimic the functional epitope of IgG1-Fc for human FcγRI. FASEB J. 2009, 23, 575–585. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, G.; Chen, C.; Li, X.; Li, Q. Bovine FcγRIII with a single extracellular domain. Res. Vet. Sci. 2000, 68, 115–118. [Google Scholar] [CrossRef]
- Liu, Y.; Qiao, S.; Wang, A.; Chang, J.; Chen, Y.; Yang, S.; Deng, R.; Zhang, G. Cloning and characterization of ovine immunoglobulin G Fc receptor III (FcγRIII). Vet. Immunol. Immunopathol. 2011, 139, 282–288. [Google Scholar] [CrossRef]
- Collins, R.A.; Gelder, K.I.; Howard, C.J. Nucleotide sequence of cattle FcGRIII: Its identification in γδ T cells. Immunogenetics 1997, 45, 440–443. [Google Scholar] [CrossRef]
- Zhang, G.; Young, J.R.; Tregaskes, C.R.; Howard, C.J. Cattle FcγRII: Molecular cloning and ligand specificity. Immunogenetics 1994, 39, 423–427. [Google Scholar] [CrossRef]
- Cao, S.Z. Veterinary Microbiology and Immunology Techniques; Beijing Agricultural University Press: Beijing, China, 1992; pp. 335–337. [Google Scholar]
- Chan, W.C.; White, P.D. Fmoc Solid Phase Peptides Synthesis: A Practical Approach; Oxford University Press: Oxford, UK, 1999; pp. 9–76. [Google Scholar]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3-A versatile homology modelling toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, M.L.; Tolvanen, M.; Carpen, O. Membrane-proximal Ig-like domain of FcγRIII (CD16) contains residues critical for ligand binding. J. Immunol. 1994, 152, 4466–4474. [Google Scholar] [CrossRef] [PubMed]
- Tamm, A.; Kister, A.; Nolte, K.U.; Gessner, J.E.; Schmidt, R.E. The IgG binding site of human FcγRIIIB receptor involves CC’ and FG loops of the membrane-proximal domain. J. Biol. Chem. 1996, 271, 3659–3666. [Google Scholar] [CrossRef] [PubMed]
- Radaev, S.; Sun, P. Recognition of immunoglobulins by Fcγ receptors. Mol. Immunol. 2001, 38, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Woof, J.M.; Burton, D.R. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat. Rev. Immunol. 2004, 4, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Coënon, L.; Villalba, M. From CD16a biology to antibody-dependent cell-mediated cytotoxicity improvement. Front. Immunol. 2022, 13, 913215. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.R.; Rodriguez Benavente, M.C.; Lorenz, W.W.; Mace, E.M.; Barb, A.W. Fcγ receptor IIIa/CD16a processing correlates with the expression of glycan-related genes in human natural killer cells. J. Biol. Chem. 2021, 296, 100183. [Google Scholar] [CrossRef]
- Aguilar, O.A.; Gonzalez-Hinojosa, M.D.R.; Arakawa-Hoyt, J.S.; Millan, A.J.; Gotthardt, D.; Nabekura, T.; Lanier, L.L. The CD16 and CD32b Fc-gamma receptors regulate antibody-mediated responses in mouse natural killer cells. J. Leukoc. Biol. 2023, 113, 27–40. [Google Scholar] [CrossRef]
- Mackin, S.R.; Desai, P.; Whitener, B.M.; Karl, C.E.; Liu, M.; Baric, R.S.; Edwards, D.K.; Chicz, T.M.; McNamara, R.P.; Alter, G.; et al. Fc-γR-dependent antibody effector functions are required for vaccine-mediated protection against antigen-shifted variants of SARS-CoV-2. Nat. Microbiol. 2023, 8, 569–580. [Google Scholar] [CrossRef]
- Rojas-Ortega, D.A.; Rojas-Hernández, S.; Sánchez-Mendoza, M.E.; Gómez-López, M.; Sánchez-Camacho, J.V.; Rosales-Cruz, E.; Yépez, M.M.C. Role of FcγRIII in the nasal cavity of BALB/c mice in the primary amebic meningoencephalitis protection model. Parasitol. Res. 2023, 122, 1087–1105. [Google Scholar] [CrossRef]
- Xi, J.; Zhang, G.P.; Qiao, S.L.; Guo, J.Q.; Wang, X.N.; Yang, Y.Y.; Zhang, L.N.; Miao, X.W.; Zhao, D.; Zhi, Y.B.; et al. Increased survival and reduced renal injury in MRL/lpr mice treated with a human Fcγ receptor II (CD32) peptide. Immunology 2012, 136, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Hao, J.; Li, N.; Xing, G.; Hu, M.; Zhang, G. Integrated system for purification and assembly of PCV Cap nano vaccine based on targeting peptide ligand. Int. J. Nanomed. 2020, 15, 8507–8517. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Deng, Z.F.; Lu, Y.; Fang, W.H.; He, F. A modular and self-adjuvanted multivalent vaccine platform based on porcine circovirus virus-like nanoparticles. J. Nanobiotechnol. 2022, 20, 493. [Google Scholar] [CrossRef] [PubMed]
- Kirstgen, M.; Müller, S.F.; Lowjaga, K.A.A.T.; Goldmann, N.; Lehmann, F.; Alakurtti, S.; Yli-Kauhaluoma, J.; Baringhaus, K.H.; Krieg, R.; Glebe, D.; et al. Identification of novel HBV/HDV entry inhibitors by pharmacophore- and QSAR-guided virtual screening. Viruses 2021, 13, 1489. [Google Scholar]



| Name | Sequence | Length (aa) | Mass (Da) | Isoelectric Point | IgG1 Binding b | Predicted Position c |
|---|---|---|---|---|---|---|
| boRIII1 | 97(C) a VAQRVVNVGKPIRLK111 | 16 | 1780.14 | 11.66 | + | A–B loop |
| boRIII2 | 112CHSWKKTPVAKV123 | 12 | 1383.62 | 10.58 | − | B–C loop |
| boRIII3 | 124(C)QYFRNGRGKKYS135 | 13 | 1606.77 | 10.58 | − | C–C’ loop |
| boRIII4 | 136(C)HGNSDFHIPE145 | 11 | 1255.28 | 5.07 | − | D–E loop |
| boRIII5 | 146(C)AKLEHSGSYF155 | 11 | 1241.33 | 7.15 | − | E–F loop |
| boRIII6 | 156CRGIIGSKNESSESVQ171 | 16 | 1693.77 | 6.45 | − | F–G loop |
| Name | Sequence | Length (aa) | Mass (Da) | Isoelectric Point | IgG1 Binding b |
|---|---|---|---|---|---|
| RIII1-N1 | 98(C) a AQRVVNVGKPIRLK111 | 15 | 1681.01 | 11.66 | + |
| RIII1-N2 | 99(C)QRVVNVGKPIRLK111 | 14 | 1609.94 | 11.66 | − |
| RIII1-N3 | 100(C)RVVNVGKPIRLK111 | 13 | 1481.81 | 11.66 | − |
| RIII1-N4 | 101(C)VVNVGKPIRLK111 | 12 | 1325.63 | 10.89 | − |
| RIII1-N5 | 102(C)VNVGKPIRLK111 | 11 | 1226.50 | 10.89 | − |
| RIII1-N6 | 103(C)NVGKPIRLK111 | 10 | 1127.37 | 10.89 | − |
| RIII1-N7 | 104(C)VGKPIRLK111 | 9 | 1013.27 | 10.89 | − |
| RIII1-N8 | 105(C)GKPIRLK111 | 8 | 914.14 | 10.89 | − |
| Name | Sequence | Length (aa) | Mass (Da) | Isoelectric Point | IgG1 Binding b |
|---|---|---|---|---|---|
| 1N1-C1 | 98(C) a AQRVVNVGKPIRL110 | 14 | 1552.84 | 11.59 | + |
| 1N1-C2 | 98(C)AQRVVNVGKPIR109 | 13 | 1439.69 | 11.59 | + |
| 1N1-C3 | 98(C)AQRVVNVGKPI108 | 12 | 1283.51 | 10.64 | + |
| 1N1-C4 | 98(C)AQRVVNVGKP107 | 11 | 1170.36 | 10.64 | + |
| 1N1-C5 | 98(C)AQRVVNVGK106 | 10 | 1073.25 | 10.64 | + |
| 1N1-C6 | 98(C)AQRVVNVG105 | 9 | 945.08 | 9.55 | + |
| 1N1-C7 | 98(C)AQRVVNV104 | 8 | 888.03 | 9.55 | + |
| 1N1-C8 | 98(C)AQRVVN103 | 7 | 788.90 | 9.55 | + |
| Name | Sequence | Length (aa) | Mass (Da) | Isoelectric Point | IgG1 Binding b |
|---|---|---|---|---|---|
| 1N1C8-A | 98(C) a GQRVVN103 | 7 | 774.88 | 9.55 | − |
| 1N1C8-Q | 98(C)AARVVN103 | 7 | 731.84 | 9.55 | − |
| 1N1C8-R | 98(C)AQAVVN103 | 7 | 703.79 | 5.12 | + |
| 1N1C8-V1 | 98(C)AQRAVN103 | 7 | 760.84 | 9.55 | − |
| 1N1C8-V2 | 98(C)AQRVAN103 | 7 | 760.84 | 9.55 | − |
| 1N1C8-N | 98(C)AQRVVA103 | 7 | 745.87 | 9.55 | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Guo, J.; Li, G.; Wang, X.; Yang, J.; Li, Q.; Zhang, G. Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides. Vet. Sci. 2024, 11, 24. https://doi.org/10.3390/vetsci11010024
Wang R, Guo J, Li G, Wang X, Yang J, Li Q, Zhang G. Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides. Veterinary Sciences. 2024; 11(1):24. https://doi.org/10.3390/vetsci11010024
Chicago/Turabian StyleWang, Ruining, Junqing Guo, Ge Li, Xun Wang, Jifei Yang, Qingmei Li, and Gaiping Zhang. 2024. "Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides" Veterinary Sciences 11, no. 1: 24. https://doi.org/10.3390/vetsci11010024
APA StyleWang, R., Guo, J., Li, G., Wang, X., Yang, J., Li, Q., & Zhang, G. (2024). Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides. Veterinary Sciences, 11(1), 24. https://doi.org/10.3390/vetsci11010024






