RT-LAMP as Diagnostic Tool for Influenza—A Virus Detection in Swine
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Primer and gBlock DNA Design
2.2. RT-qPCR
2.3. Fluorescent RT-LAMP
2.4. Laboratory Virus Culture and Extraction
2.5. Clinical Sample Collection and Processing
2.6. Statistical Analysis
3. Results
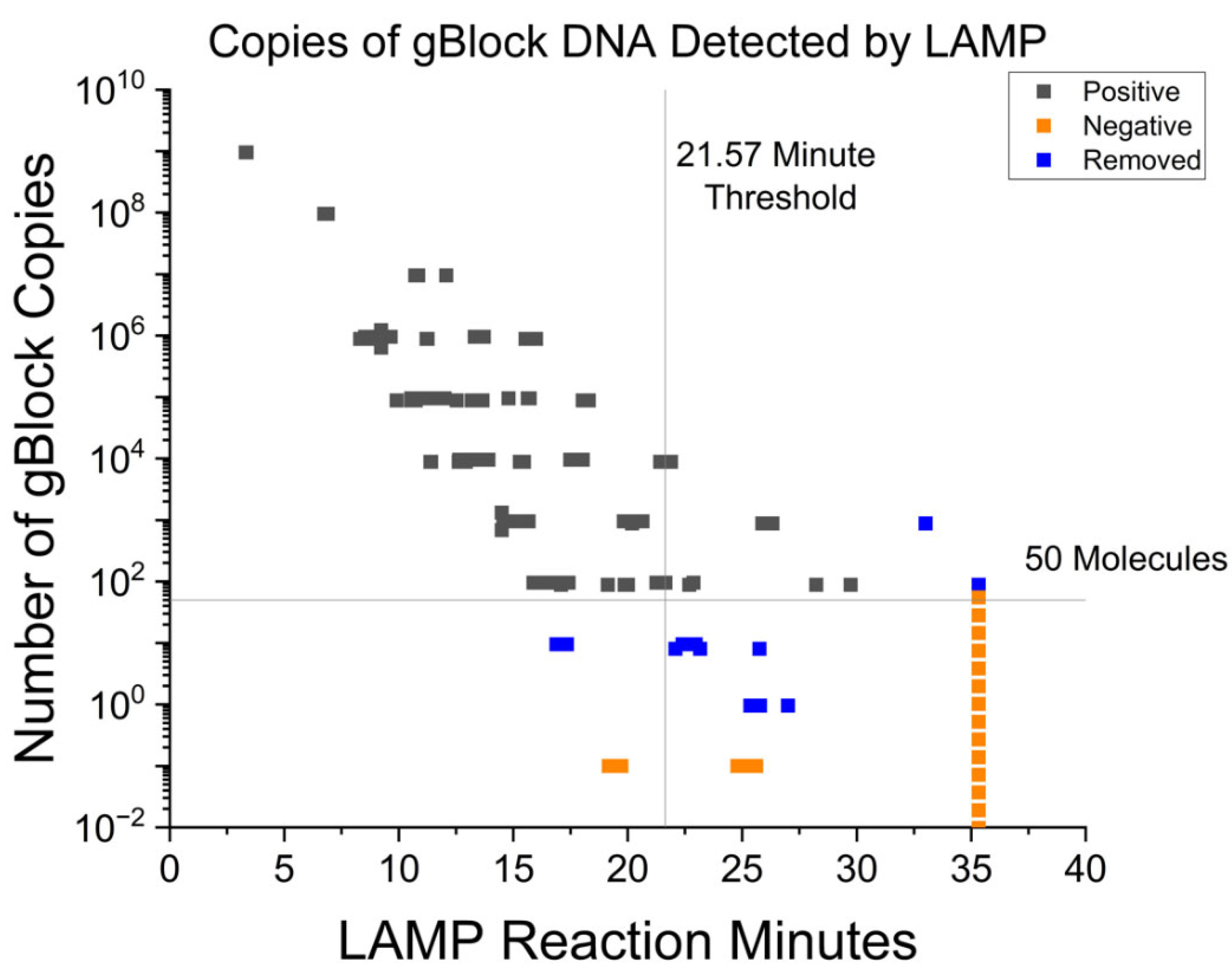
3.1. Fluorescent RT-LAMP Limit of Detection in Syntethic DNA
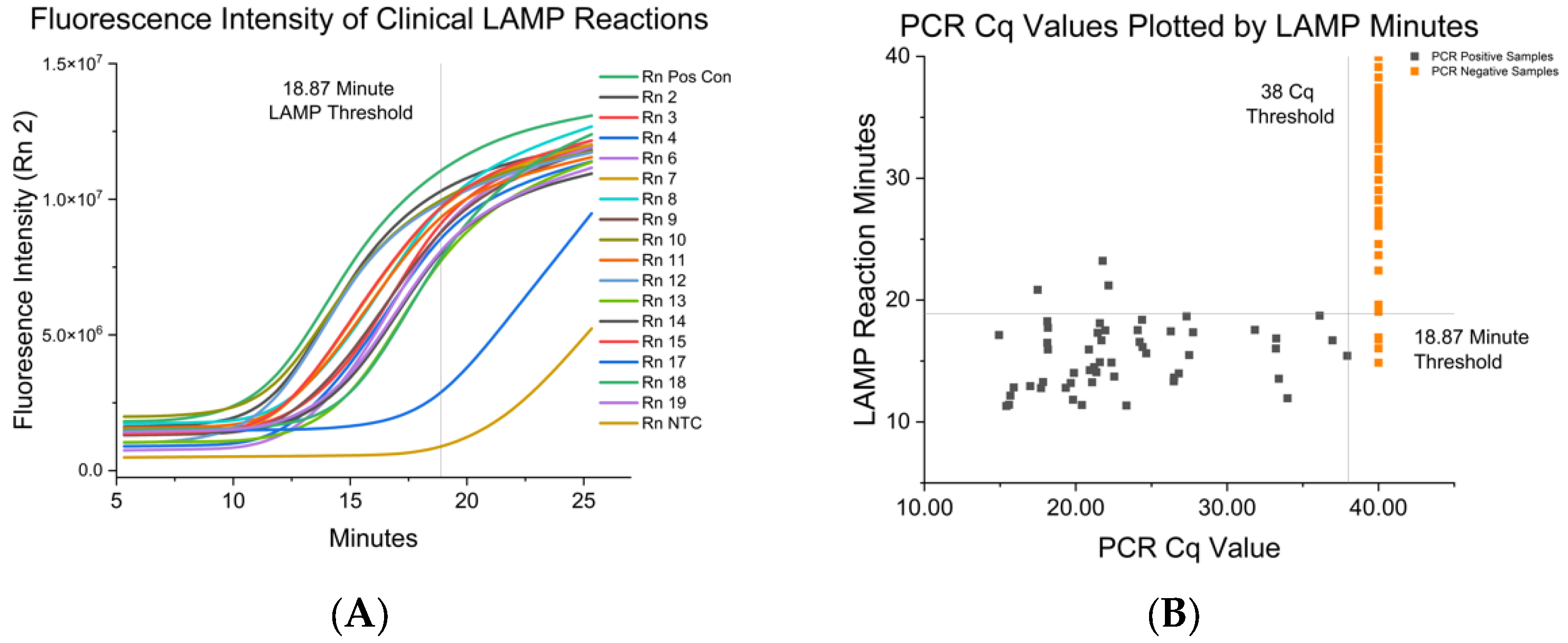
3.2. Fluorescent RT-LAMP Limit of Detection in IAV either Grown in Cell Culutre or from Clinical Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CDC. Disease Burden of Flu. Available online: https://www.cdc.gov/flu/about/burden/index.html (accessed on 9 November 2022).
- Kessler, S.; Harder, T.C.; Schwemmle, M.; Ciminski, K. Influenza A Viruses and Zoonotic Events—Are We Creating Our Own Reservoirs? Viruses 2021, 13, 2250. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R.; Murcia, P.R.; Holmes, E.C. Influenza Virus Reservoirs and Intermediate Hosts: Dogs, Horses, and New Possibilities for Influenza Virus Exposure of Humans. J. Virol. 2015, 89, 2990. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.S.; Webby, R.; Gilchrist, M.J.R.; Gray, G.C. Avian Influenza among Waterfowl Hunters and Wildlife Professionals. Emerg. Infect. Dis. 2006, 12, 1284. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Reeves, A.B.; Lagassé, B.J.; Patil, V.; Hubbard, L.E.; Kolpin, D.W.; McCleskey, R.B.; Repert, D.A.; Stallknecht, D.E.; Poulson, R.L. Evidence for Interannual Persistence of Infectious Influenza A Viruses in Alaska Wetlands. Sci. Total Environ. 2022, 803, 150078. [Google Scholar] [CrossRef] [PubMed]
- Brooke, C.B. Population Diversity and Collective Interactions during Influenza Virus Infection. J. Virol. 2017, 91, e01164-17. [Google Scholar] [CrossRef] [PubMed]
- Barbezange, C.; Jones, L.; Blanc, H.; Isakov, O.; Celniker, G.; Enouf, V.; Shomron, N.; Vignuzzi, M.; van der Werf, S. Seasonal Genetic Drift of Human Influenza A Virus Quasispecies Revealed by Deep Sequencing. Front. Microbiol. 2018, 9, 2596. [Google Scholar] [CrossRef]
- Abdelwhab, E.M.; Abdel-Moneim, A.S. Orthomyxoviruses. In Recent Advances in Animal Virology; Springer: Singapore, 1996; pp. 351–378. [Google Scholar] [CrossRef]
- Couceiro, J.N.S.S.; Paulson, J.C.; Baum, L.G. Influenza Virus Strains Selectively Recognize Sialyloligosaccharides on Human Respiratory Epithelium; the Role of the Host Cell in Selection of Hemagglutinin Receptor Specificity. Virus Res. 1993, 29, 155–165. [Google Scholar] [CrossRef]
- Ma, W.; Kahn, R.E.; Richt, J.A. The Pig as a Mixing Vessel for Influenza Viruses: Human and Veterinary Implications. J. Mol. Genet. Med. 2009, 3, 158. [Google Scholar] [CrossRef]
- Rajao, D.S.; Vincent, A.L.; Perez, D.R. Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 2019, 5, 347. [Google Scholar] [CrossRef]
- Supply Shortages Impacting COVID-19 and Non-COVID Testing. Available online: https://asm.org/Articles/2020/September/Clinical-Microbiology-Supply-Shortage-Collecti-1 (accessed on 4 January 2022).
- Hagen, A. Laboratory Supply Shortages Are Impacting COVID-19 and Non-COVID Diagnostic Testing. Available online: https://asm.org/Articles/2020/September/Laboratory-Supply-Shortages-Are-Impacting-COVID-19 (accessed on 28 June 2022).
- Cantera, J.L.; White, H.; Diaz, M.H.; Beall, S.G.; Winchell, J.M.; Lillis, L.; Kalnoky, M.; Gallarda, J.; Boyle, D.S. Assessment of Eight Nucleic Acid Amplification Technologies for Potential Use to Detect Infectious Agents in Low-Resource Settings. PLoS ONE 2019, 14, e0215756. [Google Scholar] [CrossRef]
- McNerney, R. Diagnostics for Developing Countries. Diagnostics 2015, 5, 200–209. [Google Scholar] [CrossRef] [PubMed]
- The Use of a Commercial Loop-Mediated Isothermal Amplification Assay (TB-Lamp) for the Detection of Tuberculosis: Expert Group Meeting Report. Available online: https://www.who.int/publications/i/item/WHO-HTM-TB-2013.05 (accessed on 29 December 2022).
- Eiken Chemical. Molecular Test “Lamp”—Products. Available online: https://www.eiken.co.jp/en/products/lamp/ (accessed on 29 December 2022).
- Sadeghi, Y.; Kananizadeh, P.; Moghadam, S.O.; Alizadeh, A.; Pourmand, M.R.; Mohammadi, N.; Afshar, D.; Ranjbar, R. The Sensitivity and Specificity of Loop-Mediated Isothermal Amplification and PCR Methods in Detection of Foodborne Microorganisms: A Systematic Review and Meta-Analysis. Iran. J. Public Health 2021, 50, 2172. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Notomi, T.; Mori, Y.; Notomi, T. Loop-Mediated Isothermal Amplifi Cation (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; Stewart de Ramirez, S.A.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, 12, 7075. [Google Scholar] [CrossRef]
- Park, J.Y.; Park, S.; Park, Y.R.; Kang, D.Y.; Kim, E.M.; Jeon, H.S.; Kim, J.J.; Kim, W.I.; Lee, K.T.; Kim, S.H.; et al. Reverse-Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for the Visual Detection of European and North American Porcine Reproductive and Respiratory Syndrome Viruses. J. Virol. Methods 2016, 237, 10–13. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Nam, J.; Mihn, D.C.C.; Sung, H.W.; Lim, C.S.; Kim, J. Development of a Multiplex Isothermal Amplification Molecular Diagnosis Method for On-Site Diagnosis of Influenza. PLoS ONE 2020, 15, e0238615. [Google Scholar] [CrossRef]
- Bakre, A.A.; Jones, L.P.; Bennett, H.K.; Bobbitt, D.E.; Tripp, R.A. Detection of Swine Influenza Virus in Nasal Specimens by Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP). J. Virol. Methods 2021, 288, 114015. [Google Scholar] [CrossRef]
- Stellrecht, K.A. History of Matrix Genes Mutations within PCR Target Regions among Circulating Influenza H3N2 Clades over Ten-plus-Years. J. Clin. Virol. 2018, 107, 11–18. [Google Scholar] [CrossRef]
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J.; et al. Influenza Research Database: An Integrated Bioinformatics Resource for Influenza Virus Research. Nucleic Acids Res. 2017, 45, D466–D474. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Hardinge, P.; Kiddle, G.; Tisi, L.; Murray, J.A.H. Optimised LAMP Allows Single Copy Detection of 35Sp and NOSt in Transgenic Maize Using Bioluminescent Assay in Real Time (BART). Sci. Rep. 2018, 8, 17590. [Google Scholar] [CrossRef] [PubMed]
- Breslauer, K.J.; Frank, R.; Blocker, H.; Marky, L.A. Predicting DNA Duplex Stability from the Base Sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A.; Tanner, N. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv 2020. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- WHO. Information for the Molecular Detection of Influenza Viruses. Available online: http://www.who.int/influenza/gisrs_laboratory/collaborating_centres/list/en/index.html (accessed on 10 April 2022).
- Klein, S.; Müller, T.G.; Khalid, D.; Sonntag-Buck, V.; Heuser, A.M.; Glass, B.; Meurer, M.; Morales, I.; Schillak, A.; Freistaedter, A.; et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-QPCR and RT-LAMP. Viruses 2020, 12, 863. [Google Scholar] [CrossRef]
- NEB. WarmStart Colorimetric LAMP 2X Master Mix Typical LAMP Protocol (M1800). Available online: https://www.neb.com/protocols/2016/08/15/warmstart-colorimetric-lamp-2x-master-mix-typical-lamp-protocol-m1800 (accessed on 21 February 2023).
- Balish, A.L.; Kat, J.M.; Klimov, A.I. Influenza: Propagation, Quantification, and Storage. Curr. Protoc. Microbiol. 2013, 29, 15G.1.1–15G.1.24. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty per Cent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Spackman, E.; Pedersen, J.C.; McKinley, E.T.; Gelb, J. Optimal Specimen Collection and Transport Methods for the Detection of Avian Influenza Virus and Newcastle Disease Virus. BMC Vet. Res. 2013, 9, 35. [Google Scholar] [CrossRef]
- Nolting, J.M.; Szablewski, C.M.; Edwards, J.L.; Nelson, S.W.; Bowman, A.S. Nasal Wipes for Influenza A Virus Detection and Isolation from Swine. J. Vis. Exp. 2015, 2015, 53313. [Google Scholar] [CrossRef]
- Zou, K.H.; O’Malley, A.J.; Mauri, L. Receiver-Operating Characteristic Analysis for Evaluating Diagnostic Tests and Predictive Models. Circulation 2007, 115, 654–657. [Google Scholar] [CrossRef]
- Prioritizing Diseases for Research and Development in Emergency Contexts. Available online: https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts (accessed on 22 November 2022).
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Dutta, J.; Cortes-Fernández, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovão, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 Influenza Pandemic in Swine in Mexico. eLife 2016, 5, e16777. [Google Scholar] [CrossRef] [PubMed]
- Goodell, C.K.; Prickett, J.; Kittawornrat, A.; Zhou, F.; Rauh, R.; Nelson, W.; O’connell, C.; Burrell, A.; Wang, C.; Yoon, K.-J.; et al. Probability of Detecting Influenza A Virus Subtypes H1N1 and H3N2 in Individual Pig Nasal Swabs and Pen-Based Oral Fluid Specimens over Time. Vet. Microbiol. 2013, 166, 450–460. [Google Scholar] [CrossRef] [PubMed]
- USDA APHIS|Influenza A Virus in Swine Surveillance Information. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_animal_disease_information/sa_swine_health/ct_siv_surveillance/ (accessed on 27 March 2022).
- Zeller, M.A.; Anderson, T.K.; Walia, R.W.; Vincent, A.L.; Gauger, P.C. ISU FLUture: A Veterinary Diagnostic Laboratory Web-Based Platform to Monitor the Temporal Genetic Patterns of Influenza A Virus in Swine. BMC Bioinform. 2018, 19, 397. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, A.; Ali, W.R.; Asif, M.; Ahmed, N.; Khan, W.S.; Mansoor, S.; Bajwa, S.Z.; Amin, I. Development of a LAMP Assay Using a Portable Device for the Real-Time Detection of Cotton Leaf Curl Disease in Field Conditions. Biol. Methods Protoc. 2021, 6, bpab010. [Google Scholar] [CrossRef]
- Khodaparast, M.; Sharley, D.; Best, N.; Marshall, S.; Beddoe, T. In-Field LAMP Assay for Rapid Detection of Human Faecal Contamination in Environmental Water. Environ. Sci. 2022, 8, 2641–2651. [Google Scholar] [CrossRef]
- Gärtner, K.; Meleke, H.; Kamdolozi, M.; Chaima, D.; Samikwa, L.; Paynter, M.; Nyirenda Nyang’Wa, M.; Cloutman-Green, E.; Nastouli, E.; Klein, N.; et al. A Fast Extraction-Free Isothermal LAMP Assay for Detection of SARS-CoV-2 with Potential Use in Resource-Limited Settings. Virol. J. 2022, 19, 77. [Google Scholar] [CrossRef]
- Jankelow, A.M.; Lee, H.; Wang, W.; Hoang, T.H.; Bacon, A.; Sun, F.; Chae, S.; Kindratenko, V.; Koprowski, K.; Stavins, R.A.; et al. Smartphone Clip-on Instrument and Microfluidic Processor for Rapid Sample-to-Answer Detection of Zika Virus in Whole Blood Using Spatial RT-LAMP. Analyst 2022, 147, 3838–3853. [Google Scholar] [CrossRef]
| Primer | Sequence (5’–3’) | Concentration in 10 × Mix (µM) | Matrix SequencePosition |
|---|---|---|---|
| F3 | CAA GGA GGT GTC ACT AAG C | 2 | 361–374 |
| B3 | CAT CTG CCT AGT CTG ATT AGC | 2 | 641–661 |
| FIP | GTG AGA CCG ATG CTG TGA ATC AGG AAC AGT GAC CAC AGA AG | 16 | 496–511 |
| BIP | ACC ACC AAT CCA CTA ATC AGG CGC CAT CTG TTC CAT AGC C | 16 | 527–545 |
| LoopF | TCT GTT CAC AAG TGG CAC A | 4 | 461–485 |
| LoopB | ACA GAA TGG TGC TGG CTA G | 4 | 555–573 |
| Subtype | Strain and Accession Numbers | H1/H3 | N1/N2 |
|---|---|---|---|
| H1N1 | A/California/07/2009 641809.71 | H1N1pdm09 | Pandemic |
| H1N1 | A/swine/Minnesota/A02245728/2020 | Gamma | Classic |
| H1N1 | A/swine/Iowa/A02479151/2020 | Delta1a | Classic |
| H1N1 | A/swine/Texas/A02245632/2020 | Beta | Classic |
| H1N2 | A/swine/Colorado/A02245414/2020 | Delta1b | 2002A |
| H1N2 | A/swine/Indiana/A02478520/2019 | Delta2 | 1998B |
| H1N2 | A/swine/Iowa/A02478576/2019 | Alpha | 2002B |
| H3N2 | A/swine/Nebraska/A02524799/2020 | 2010.1 | 2002B |
| H3N2 | A/swine/Iowa/A02524878/2020 | IV-A | 2002B |
| H3N2 | A/swine/NewYork/A01104005/2011 | IV-A | 2002A |
| IAV-S Strain | RT-LAMP Mean and SD in Minutes to Positive | Paired RT-PCR Cq |
|---|---|---|
| H1N1 IA | 12.85 ± 0.67 | 21.02 |
| H1N1 MN | 14.39 ± 0.62 | 19.50 |
| H1N1 TX | 12.66 ± 0.22 | 18.39 |
| H1N2 CO | 14.83 ± 0.71 | 19.92 |
| H1N2 IA | 12.83 ± 0.30 | 21.04 |
| H1N2 IN | 10.97 ± 0.12 | 20.90 |
| H3N2 IA | 11.85 ± 0.79 | 23.13 |
| H3N2 NE | 13.45 ± 0.88 | 19.99 |
| H3N2 NY | 10.24 ± 1.98 | 18.70 |
| Test Cutoff Time | Sensitivity% | Specificity % | 95% CI | Asym. Probability |
|---|---|---|---|---|
| <18.87 min | 94.34 | 94.85 | 95.35–99.62 | 2.97 × 10−22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Storms, S.M.; Shisler, J.; Nguyen, T.H.; Zuckermann, F.A.; Lowe, J.F. RT-LAMP as Diagnostic Tool for Influenza—A Virus Detection in Swine. Vet. Sci. 2023, 10, 220. https://doi.org/10.3390/vetsci10030220
Storms SM, Shisler J, Nguyen TH, Zuckermann FA, Lowe JF. RT-LAMP as Diagnostic Tool for Influenza—A Virus Detection in Swine. Veterinary Sciences. 2023; 10(3):220. https://doi.org/10.3390/vetsci10030220
Chicago/Turabian StyleStorms, Suzanna M., Joanna Shisler, Thanh H. Nguyen, Federico A. Zuckermann, and James F. Lowe. 2023. "RT-LAMP as Diagnostic Tool for Influenza—A Virus Detection in Swine" Veterinary Sciences 10, no. 3: 220. https://doi.org/10.3390/vetsci10030220
APA StyleStorms, S. M., Shisler, J., Nguyen, T. H., Zuckermann, F. A., & Lowe, J. F. (2023). RT-LAMP as Diagnostic Tool for Influenza—A Virus Detection in Swine. Veterinary Sciences, 10(3), 220. https://doi.org/10.3390/vetsci10030220






