A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Vaccine Strains and Clinical Samples
2.2. Design of Primers and Probes
2.3. Nucleic Acid Extraction
2.4. Construction of the Standard Plasmids
2.5. Optimization of the Multiplex qRT-PCR
2.6. Construction of Standard Curves
2.7. Specificity Analysis
2.8. Sensitivity Analysis
2.9. Repeatability Analysis
2.10. Detection of the Clinical Samples
3. Results
3.1. Construction of Standard Plasmids
3.2. Determination of the Optimal Reaction Conditions
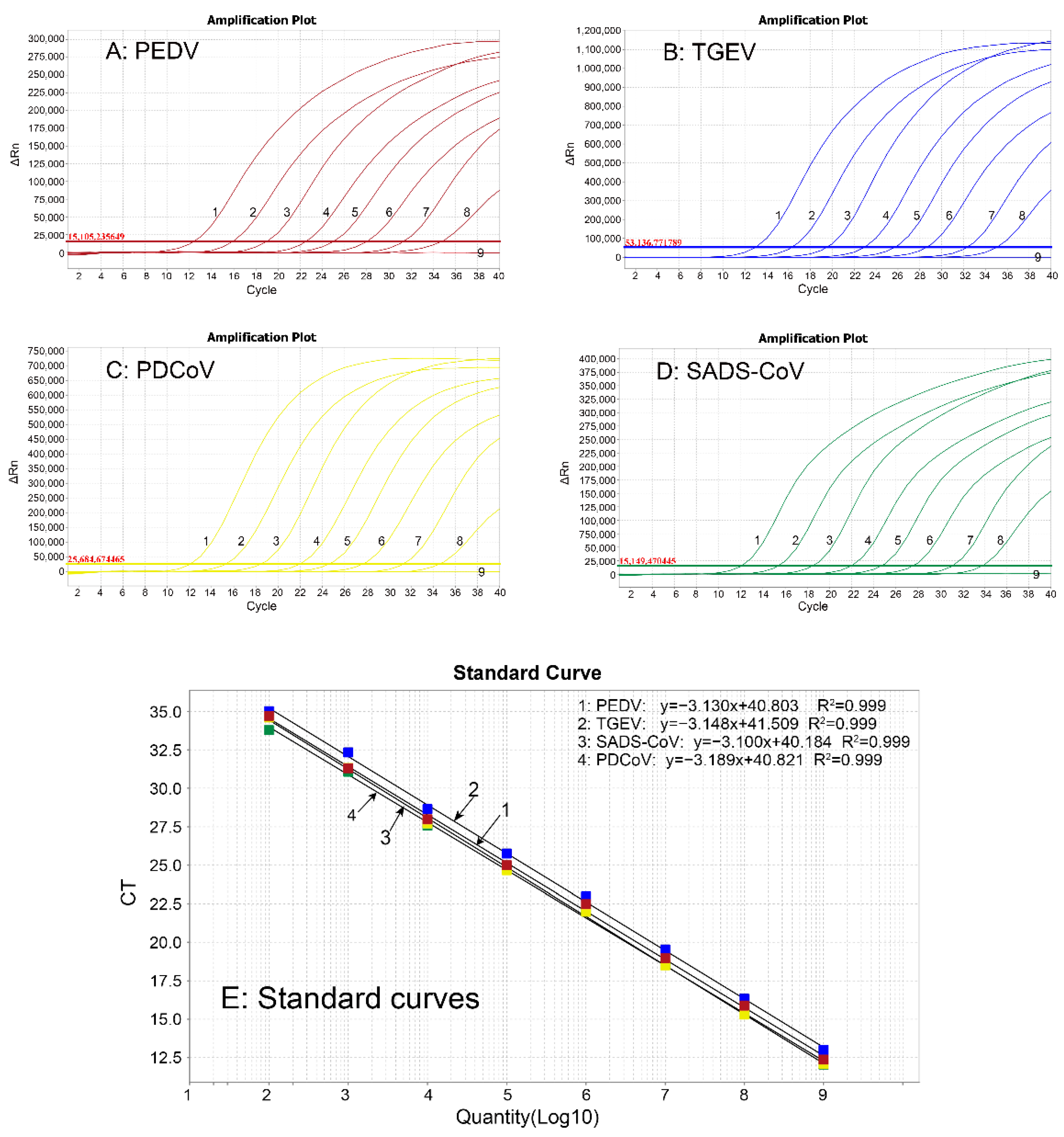
3.3. Generation of Standard Curves
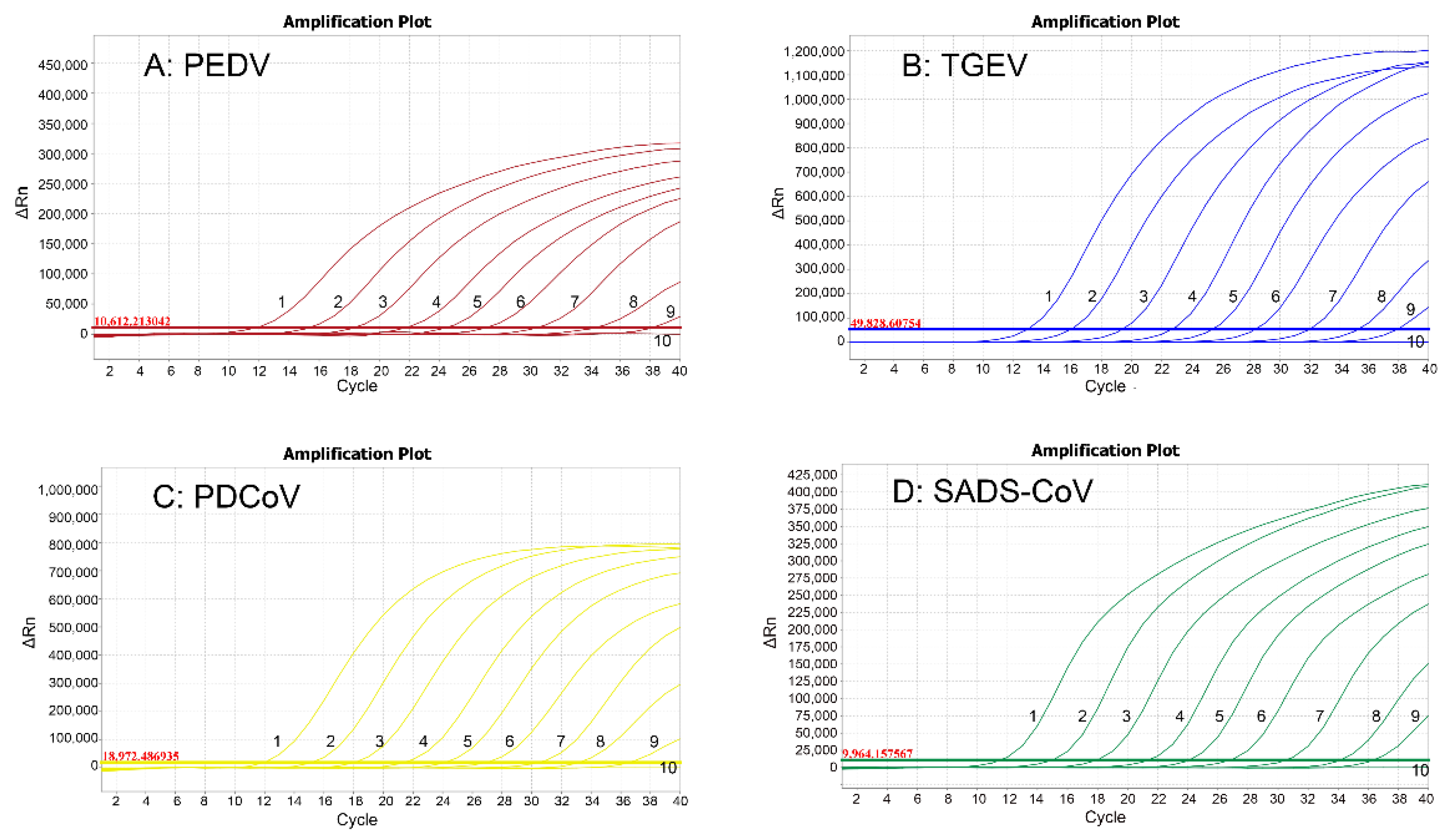
3.4. Specificity Analysis
3.5. Sensitivity Analysis
3.6. Repeatability Analysis
3.7. Detection Results of the Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, H.Y. Porcine enteric coronaviruses: An updated overview of the pathogenesis, prevalence, and diagnosis. Vet. Res. Commun. 2021, 45, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhou, G.Z.; Zhang, Y.; Peng, L.H.; Zou, L.P.; Yang, Y.S. Coronaviruses and gastrointestinal diseases. Mil. Med. Res. 2020, 7, 49. [Google Scholar] [CrossRef]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef]
- Sun, R.Q.; Cai, R.J.; Chen, Y.Q.; Liang, P.S.; Chen, D.K.; Song, C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013, 25, 649–654. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef]
- Lin, C.M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Porcine coronaviruses: Overview of the state of the art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Yan, Z.; Li, M.; Wang, Y.; Su, M.; Sun, D. Isolation and characterization of a porcine transmissible gastroenteritis coronavirus in northeast China. Front. Vet. Sci. 2021, 8, 611721. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, H.; Geng, C.; Yang, K.; Liu, W.; Liu, Z.; Yuan, F.; Gao, T.; Wang, S.; Wen, P.; et al. Epidemic and evolutionary characteristics of swine enteric viruses in South-Central China from 2018 to 2021. Viruses 2022, 14, 1420. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A New Bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef]
- Yang, Y.L.; Yu, J.Q.; Huang, Y.W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 2020, 285, 198024. [Google Scholar] [CrossRef]
- Xing, J.; Xu, Z.Y.; Gao, H.; Xu, S.J.; Liu, J.; Zhu, D.H.; Guo, Y.F.; Yang, B.S.; Chen, X.N.; Zheng, Z.Z.; et al. Re-emergence of severe acute diarrhea syndrome coronavirus (SADS-CoV) in Guangxi, China, 2021. J. Infect. 2022, 85, e130–e133. [Google Scholar]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar]
- Wang, L.; Byrum, B.; Zhang, Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014, 20, 1594–1595. [Google Scholar] [CrossRef]
- Duan, C. An updated review of porcine deltacoronavirus in terms of prevalence, pathogenicity, pathogenesis and antiviral strategy. Front. Vet. Sci. 2021, 8, 811187. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Kenney, S.P.; Jung, K.; Wang, Q.; Saif, L.J. Deltacoronavirus evolution and transmission: Current scenario and evolutionary perspectives. Front. Vet. Sci. 2020, 7, 626785. [Google Scholar] [CrossRef] [PubMed]
- More-Bayona, J.A.; Ramirez-Velasquez, M.; Hause, B.; Nelson, E.; Rivera-Geronimo, H. First isolation and whole genome characterization of porcine deltacoronavirus from pigs in Peru. Transbound. Emerg. Dis. 2022, 69, e1561–e1573. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Liu, X.; Sun, Y.; Zeng, W.; Li, Y.; Zhao, F.; Wu, K.; Fan, S.; Zhao, M.; Chen, J.; et al. Swine enteric coronavirus: Diverse pathogen-host interactions. Int. J. Mol. Sci. 2022, 23, 3953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, B.; Tao, J.; Cheng, J.; Liu, H. The complex co-infections of multiple porcine diarrhea viruses in local area based on the Luminex xTAG multiplex detection method. Front. Vet. Sci. 2021, 8, 602866. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.; Choi, C.; Kim, B.; Chae, C. Detection and differentiation of porcine epidemic diarrhoea virus and transmissible gastroenteritis virus in clinical samples by multiplex RT-PCR. Vet. Rec. 2000, 146, 637–640. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, B.J.; Huang, X.G.; Peng, M.Y.; Zhang, X.M.; He, D.N.; Pang, R.; Zhou, B.; Chen, P.Y. A multiplex RT-PCR assay for rapid and differential diagnosis of four porcine diarrhea associated viruses in field samples from pig farms in East China from 2010 to 2012. J. Virol. Methods 2013, 194, 107–112. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, F.; Li, Q.; Wu, M.; Lei, L.; Pan, Z. A multiplex RT-PCR assay for rapid and simultaneous detection of four RNA viruses in swine. J. Virol. Methods 2019, 269, 38–42. [Google Scholar] [CrossRef]
- Hu, H.; Jung, K.; Wang, Q.; Saif, L.J.; Vlasova, A.N. Development of a one-step RT-PCR assay for detection of pancoronaviruses (α-, β-, γ-, and δ-coronaviruses) using newly designed degenerate primers for porcine and avian fecal samples. J. Virol. Methods 2018, 256, 116–122. [Google Scholar] [CrossRef]
- Liu, G.; Jiang, Y.; Opriessnig, T.; Gu, K.; Zhang, H.; Yang, Z. Detection and differentiation of five diarrhea related pig viruses utilizing a multiplex PCR assay. J. Virol. Methods 2019, 263, 32–37. [Google Scholar] [CrossRef]
- Nan, P.; Wen, D.; Opriessnig, T.; Zhang, Q.; Yu, X.; Jiang, Y. Novel universal primer-pentaplex PCR assay based on chimeric primers for simultaneous detection of five common pig viruses associated with diarrhea. Mol. Cell Probes 2021, 58, 101747. [Google Scholar] [CrossRef] [PubMed]
- Si, G.; Niu, J.; Zhou, X.; Xie, Y.; Chen, Z.; Li, G.; Chen, R.; He, D. Use of dual priming oligonucleotide system-based multiplex RT-PCR assay to detect five diarrhea viruses in pig herds in South China. AMB Express 2021, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Fu, Y.; Li, B.; Chen, J.; Wang, J.; Yin, B.; Sha, W.; Liu, G. Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound. Emerg. Dis. 2020, 67, 678–685. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, I.J.; Pyo, H.M.; Tark, D.S.; Song, J.Y.; Hyun, B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods 2007, 146, 172–177. [Google Scholar] [CrossRef]
- Masuda, T.; Tsuchiaka, S.; Ashiba, T.; Yamasato, H.; Fukunari, K.; Omatsu, T.; Furuya, T.; Shirai, J.; Mizutani, T.; Nagai, M. Development of one-step real-time reverse transcriptase-PCR-based assays for the rapid and simultaneous detection of four viruses causing porcine diarrhea. Jpn. J. Vet. Res. 2016, 64, 5–14. [Google Scholar]
- Jia, S.; Feng, B.; Wang, Z.; Ma, Y.; Gao, X.; Jiang, Y.; Cui, W.; Qiao, X.; Tang, L.; Li, Y.; et al. Dual priming oligonucleotide (DPO)-based real-time RT-PCR assay for accurate differentiation of four major viruses causing porcine viral diarrhea. Mol. Cell Probes 2019, 47, 101435. [Google Scholar] [CrossRef]
- Huang, X.; Chen, J.; Yao, G.; Guo, Q.; Wang, J.; Liu, G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019, 103, 4943–4952. [Google Scholar] [CrossRef]
- Pan, Z.; Lu, J.; Wang, N.; He, W.T.; Zhang, L.; Zhao, W.; Su, S. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef]
- Zhu, J.H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y.L.; Huang, Y.W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J. Development and clinical applications of a 5-Plex real-time RT-PCR for swine enteric coronaviruses. Viruses 2022, 14, 1536. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, K.; Zhao, J.; Yin, Y.; Chen, Y.; Si, H.; Qu, S.; Long, F.; Lu, W. Development of a one-step multiplex qRT-PCR assay for the detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. BMC Vet. Res. 2022, 18, 43. [Google Scholar] [CrossRef]
- Su, M.; Li, C.; Qi, S.; Yang, D.; Jiang, N.; Yin, B.; Guo, D.; Kong, F.; Yuan, D.; Feng, L.; et al. A molecular epidemiological investigation of PEDV in China: Characterization of co-infection and genetic diversity of S1-based genes. Transbound. Emerg. Dis. 2020, 67, 1129–1140. [Google Scholar] [CrossRef]
- Guo, R.; Fan, B.; Chang, X.; Zhou, J.; Zhao, Y.; Shi, D.; Yu, Z.; He, K.; Li, B. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology 2020, 545, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Saeng-Chuto, K.; Madapong, A.; Kaeoket, K.; Piñeyro, P.E.; Tantituvanont, A.; Nilubol, D. Coinfection of porcine deltacoronavirus and porcine epidemic diarrhea virus increases disease severity, cell trophism and earlier upregulation of IFN-α and IL12. Sci. Rep. 2021, 11, 3040. [Google Scholar] [CrossRef]
- Zhang, H.; Han, F.; Shu, X.; Li, Q.; Ding, Q.; Hao, C.; Yan, X.; Xu, M.; Hu, H. Co-infection of porcine epidemic diarrhoea virus and porcine deltacoronavirus enhances the disease severity in piglets. Transbound. Emerg. Dis. 2022, 69, 1715–1726. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, Y.; Lan, T.; Wu, R.; Chen, J.; Wu, Z.; Xie, Q.; Zhang, X.; Ma, J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound. Emerg. Dis. 2019, 66, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, Q.N.; Su, J.N.; Chen, G.H.; Wu, Z.X.; Luo, Y.; Wu, R.T.; Sun, Y.; Lan, T.; Ma, J.Y. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound. Emerg. Dis. 2019, 66, 2180–2183. [Google Scholar] [CrossRef] [PubMed]

| Primer/Probe | Sequence (5′→3′) | Product/bp |
|---|---|---|
| PEDV(N)-U | CTGGAATGAGCAAATTCGCTG | 140 |
| PEDV(N)-D | CAACCCAGAAAACACCCTCAG | |
| PEDV(N)-P | JOE-AGCGAATTGAACAACCTTCCAATTGGCA-BHQ1 | |
| TGEV(M)-U | GCAATTCTTTGCGTTAGTGCAT | 102 |
| TGEV(M)-D | AGCGTACAAATTCCCTGAAAGC | |
| TGEV(M)-P | Texas Red-CTTCCTCTCGAAGGTGTGCCAACTGG-BHQ2 | |
| PDCoV(M)-U | ATCGACCACATGGCTCCAA | 72 |
| PDCoV(M)-D | CAGCTCTTGCCCATGTAGCTT | |
| PDCoV(M)-P | FAM-CACACCAGTCGTTAAGCATGGCAAGCT-BHQ1 | |
| SADS-CoV(N)-U | TACTGGTCCTCACGCAGATG | 120 |
| SADS-CoV(N)-D | ACGATTGCGAACACCAAGAC | |
| SADS-CoV(N)-P | Cy5-CAACAGCGACCCAATGCACACCCT-BHQ3 |
| Reagent | Volume (µL) | Final Concentration (nM) |
|---|---|---|
| 2× One-Step RT-PCR Buffer III (TaKaRa) | 10 | / |
| Ex Taq HS (5 U/μL) (TaKaRa) | 0.4 | / |
| PrimeScript RT Enzyme Mix II (TaKaRa) | 0.4 | / |
| PEDV(N)-U (20 pmol/μL) | 0.2 | 200 |
| PEDV(N)-D (20 pmol/μL) | 0.2 | 200 |
| PEDV(N)-P (20 pmol/μL) | 0.3 | 300 |
| TGEV(M)-U (20 pmol/μL) | 0.3 | 300 |
| TGEV(M)-D (20 pmol/μL) | 0.3 | 300 |
| TGEV(M)-P (20 pmol/μL) | 0.3 | 300 |
| PDCoV(M)-U (20 pmol/μL) | 0.2 | 200 |
| PDCoV(M)-D (20 pmol/μL) | 0.2 | 200 |
| PDCoV(M)-P (20 pmol/μL) | 0.2 | 200 |
| SADS-CoV(N)-U (20 pmol/μL) | 0.3 | 300 |
| SADS-CoV(N)-D (20 pmol/μL) | 0.3 | 300 |
| SADS-CoV(N)-P (20 pmol/μL) | 0.3 | 300 |
| Nucleic acid template | 2.0 | / |
| RNase-free distilled H2O | Up to 20 | / |
| Plasmid | Concentration (Copies/μL) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|---|
| SD | CV (%) | SD | CV (%) | ||||
| p-PEDV | 1.21 × 109 | 11.96 | 0.13 | 1.09 | 11.88 | 0.04 | 0.34 |
| 1.21 × 107 | 18.36 | 0.14 | 0.76 | 18.30 | 0.18 | 0.98 | |
| 1.21 × 105 | 24.44 | 0.06 | 0.25 | 24.52 | 0.16 | 0.65 | |
| p-TGEV | 1.21 × 109 | 12.97 | 0.05 | 0.39 | 12.87 | 0.12 | 0.93 |
| 1.21 × 107 | 19.26 | 0.03 | 0.16 | 19.12 | 0.17 | 0.89 | |
| 1.21 × 105 | 25.12 | 0.19 | 0.76 | 25.10 | 0.09 | 0.36 | |
| p-PDCoV | 1.21 × 109 | 11.84 | 0.10 | 0.84 | 11.72 | 0.12 | 1.02 |
| 1.21 × 107 | 18.27 | 0.27 | 1.48 | 18.55 | 0.10 | 0.54 | |
| 1.21 × 105 | 24.41 | 0.14 | 0.57 | 24.67 | 0.15 | 0.61 | |
| p-SADS-CoV | 1.21 × 109 | 11.51 | 0.02 | 0.17 | 11.79 | 0.22 | 1.87 |
| 1.21 × 107 | 17.81 | 0.06 | 0.34 | 17.95 | 0.18 | 1.00 | |
| 1.21 × 105 | 24.51 | 0.35 | 1.43 | 24.75 | 0.11 | 0.44 | |
| Date | Number | Number of Positive Samples | |||||
|---|---|---|---|---|---|---|---|
| PEDV (%) | TGEV (%) | PDCoV (%) | SADS-CoV (%) | PEDV + TGEV (%) | PEDV + PDCoV (%) | ||
| October, 2020 | 200 | 26 (13.00%) | 3 (1.50%) | 30 (15.00%) | 0 | 0 | 15 (7.50%) |
| March, 2021 | 112 | 32 (28.57%) | 1 (0.89%) | 71 (63.39%) | 0 | 1 (0.89%) | 12 (10.71%) |
| October, 2021 | 37 | 6 (16.22%) | 0 | 0 | 0 | 0 | 0 |
| November, 2021 | 217 | 3 (1.38%) | 0 | 1 (0.46%) | 0 | 0 | 0 |
| January, 2022 | 314 | 90 (28.66%) | 0 | 64 (20.38%) | 0 | 0 | 4 (1.27%) |
| February, 2022 | 394 | 110 (27.92%) | 0 | 74 (18.78%) | 0 | 0 | 1 (0.25%) |
| March, 2022 | 244 | 16 (6.56%) | 0 | 40 (16.39%) | 0 | 0 | 0 |
| April, 2022 | 378 | 95 (25.13%) | 1 (0.26%) | 26 (6.88%) | 0 | 0 | 2 (0.53%) |
| May, 2022 | 30 | 4 (13.33%) | 0 | 0 | 0 | 0 | 0 |
| June, 2022 | 496 | 58 (11.69%) | 0 | 2 (0.40%) | 0 | 0 | 0 |
| July, 2022 | 287 | 47 (16.38%) | 0 | 33 (11.50%) | 0 | 0 | 4 (1.39%) |
| August, 2022 | 76 | 14 (18.42%) | 2 (2.63%) | 11 (14.47%) | 0 | 0 | 0 |
| September, 2022 | 92 | 23 (25.00%) | 2 (2.17%) | 16 (17.39%) | 1 (1.09%) | 0 | 2 (2.17%) |
| October, 2022 | 359 | 67 (18.66%) | 6 (1.67%) | 58 (16.16%) | 4 (1.11%) | 1 (0.28%) | 6 (1.67%) |
| Total | 3236 | 591 (18.26%) | 15 (0.46%) | 426 (13.16%) | 5 (0.15%) | 2 (0.06%) | 46 (1.42%) |
| Method | Positive Samples | |||
|---|---|---|---|---|
| PEDV (%) | TGEV (%) | PDCoV (%) | SADS-CoV (%) | |
| The developed quadruplex qRT-PCR | 591/3236 (18.26%) | 15/3236 (0.46%) | 426/3236 (13.16%) | 5/3236 (0.15%) |
| The reference multiplex qRT-PCR | 572/3236 (17.67%) | 15/3236 (0.46%) | 417/3236 (12.89%) | 5/3236 (0.15%) |
| Agreements | 99.41% | 100% | 99.72% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Shi, K.; Long, F.; Zhao, K.; Feng, S.; Yin, Y.; Xiong, C.; Qu, S.; Lu, W.; Li, Z. A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses. Vet. Sci. 2022, 9, 634. https://doi.org/10.3390/vetsci9110634
Zhou H, Shi K, Long F, Zhao K, Feng S, Yin Y, Xiong C, Qu S, Lu W, Li Z. A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses. Veterinary Sciences. 2022; 9(11):634. https://doi.org/10.3390/vetsci9110634
Chicago/Turabian StyleZhou, Hongjin, Kaichuang Shi, Feng Long, Kang Zhao, Shuping Feng, Yanwen Yin, Chenyong Xiong, Sujie Qu, Wenjun Lu, and Zongqiang Li. 2022. "A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses" Veterinary Sciences 9, no. 11: 634. https://doi.org/10.3390/vetsci9110634
APA StyleZhou, H., Shi, K., Long, F., Zhao, K., Feng, S., Yin, Y., Xiong, C., Qu, S., Lu, W., & Li, Z. (2022). A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses. Veterinary Sciences, 9(11), 634. https://doi.org/10.3390/vetsci9110634








