Emergent Molecular Techniques Applied to the Detection of Porcine Viruses
Abstract
:Simple Summary
Abstract
1. Introduction
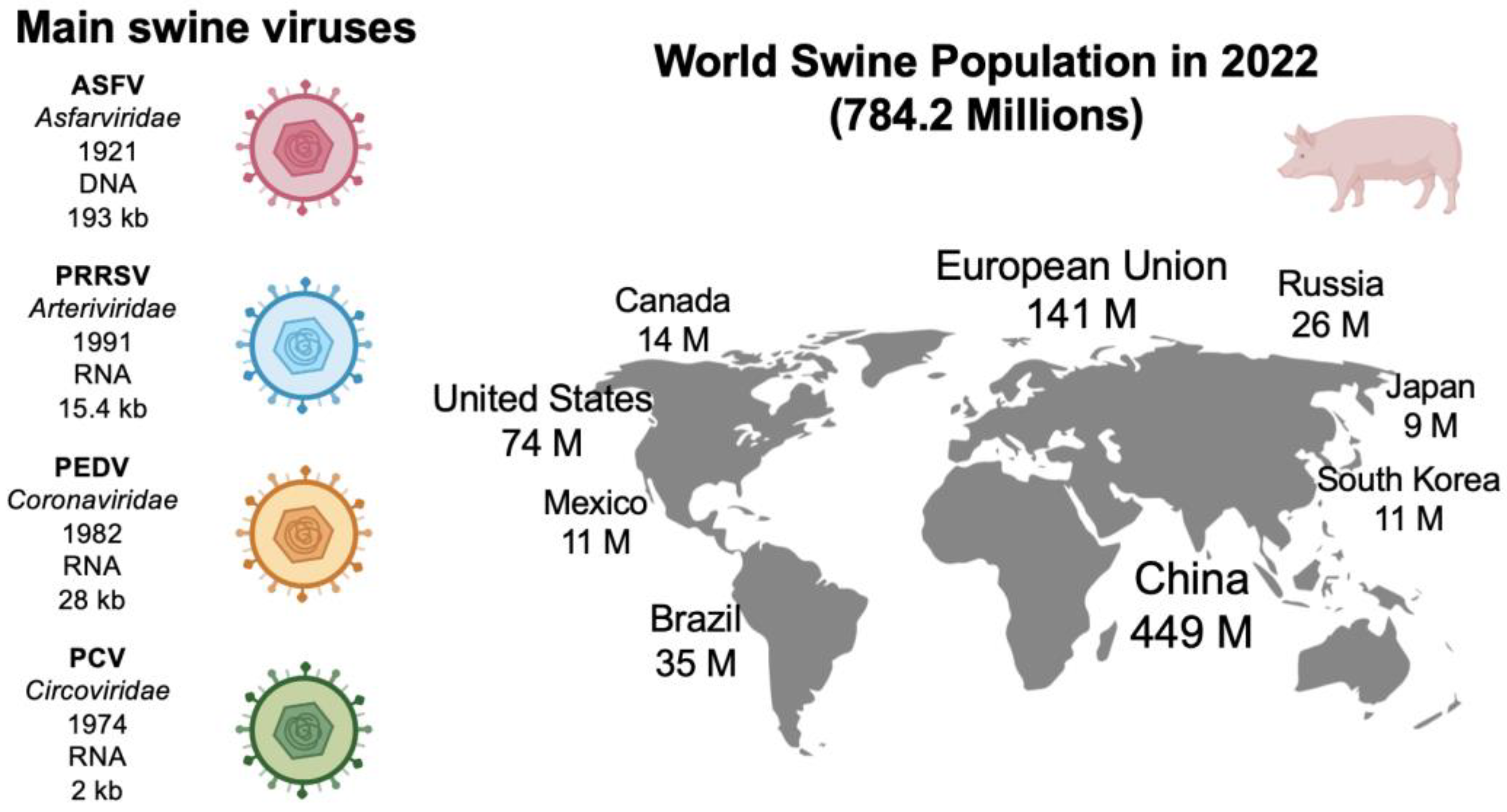
2. Main Swine Viral Diseases
3. Multiplex qPCR
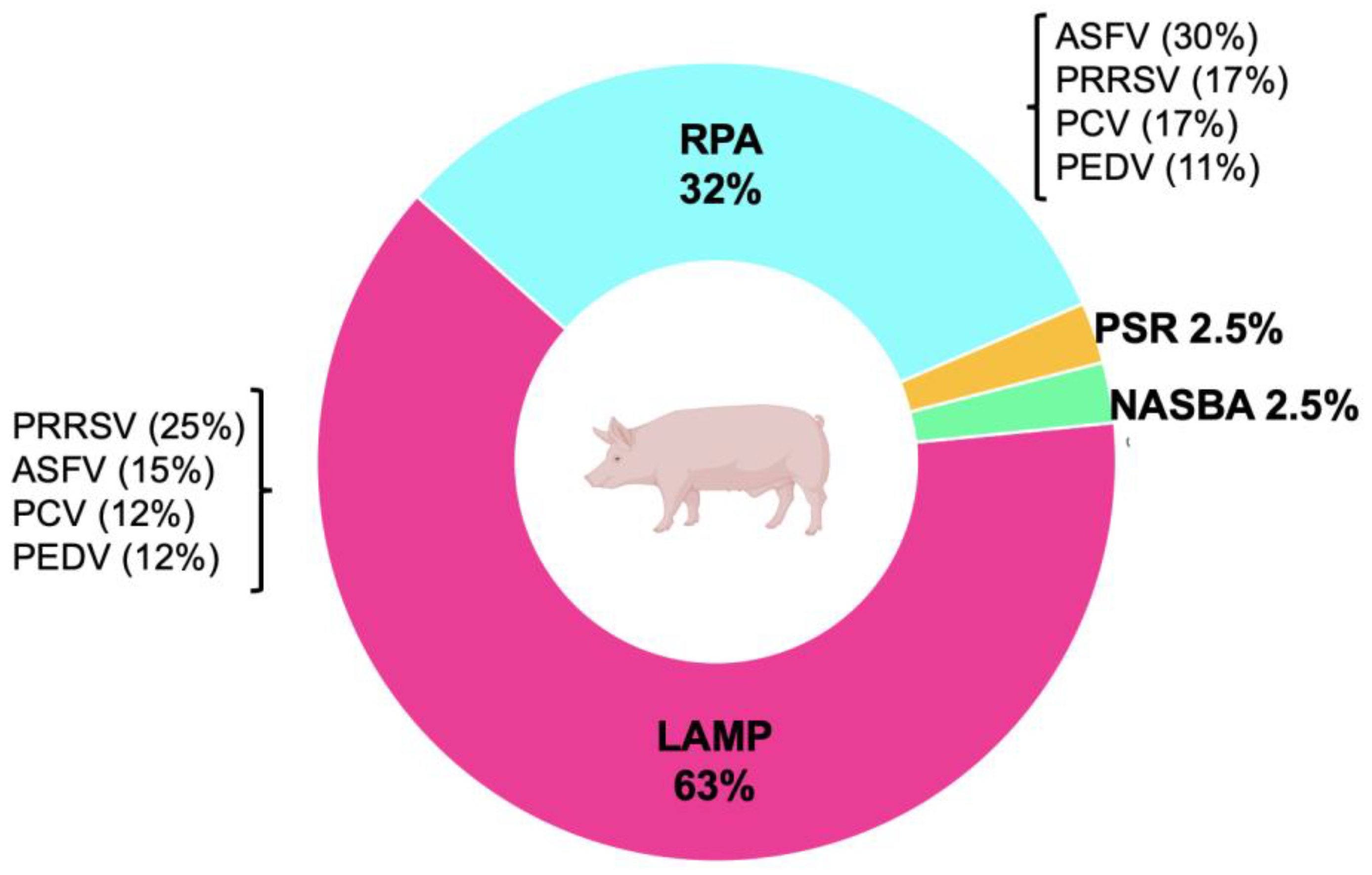
4. Isothermal Methods
| Isothermal Method | Enzymes | Conditions | Target | Detection Signal | LOD | Detection Time (min) | Reference |
| LAMP | Bst | 25–60 min at 60–65 °C | ASFV | Fluorescence (Carbon nanodots) | 15.21 copies/µL | 30 | [48] |
| ASFV | Fluorescence | 330 copies/µL | 25 | [62] | |||
| ASFV | Fluorescence (FAM) | 13 copies/µL | 40 | [49] | |||
| PRRSV | Fluorescence (Picogreen) | 80 fg/µL | 40 | [63] | |||
| PRRSV | Fluorescence and precipitate formation (Ethidium bromide, Picogreen and magnesium pyrophosphate precipitate) | 1 × 100 to 1 × 101 copies/reaction | 70 | [64] | |||
| PRRSV | Colorimetric (HBN) | 103 copies/reaction | 60 | [65] | |||
| PRRSV | Turbidity | 0.1 TCID50 | 62 | [66] | |||
| PRRSV | Fluorescence (SYBR) | 0.01 ng/µL | 60 | [67] | |||
| PRRSV | Fluorescence (SYBR) | 0.001 TCID50 | 50 | [68] | |||
| PRRSV | Electrophoresis | 5 copies/tube | 47 | [69] | |||
| PRRSV | Turbidity and fluorescence (SYBR) | 102 to 104 TCID50/mL | 50 to 60 | [70] | |||
| PRRSV | Colorimetric (HBN) | 0.1 to 1 TCID50/0.1 mL | 40 | [71] | |||
| PCV1 | Turbidity | 10 copies/µL | 62 | [72] | |||
| PCV3 | Fluorescence SYTO-9 | 1 × 101 copies/µL | 70 | [73] | |||
| PCV2- PEDV | Lateral flow dipstick (LFD) | 0.246 ng/µL for PCV2 and 0.1 ng/µL for PEDV | 90 | [50] | |||
| PCV3 | Fluorescence (FAM) | 50 copies/reaction | 17.34 ± 4.45 | [74] | |||
| PEDV | Fluorescence | 2 × 100 TCID50/mL to 2.8 × 101 TCID50/mL | 50 | [75] | |||
| PEDV | Fluorescence (SYBR) | 0.0001 ng/µL | 62 | [76] | |||
| IAV | Lateral flow dipstick (LFD) | 7.8 pg/µL | 30 | [77] | |||
| CSFV | Colorimetric (HBN) | 100 copy numbers | 60 | [78] | |||
| PDCoV | Fluorescence (FAM) | 25 copies/µL | <40 | [79] | |||
| PDCoV | Fluorescence (SYBR) | 1 × 101 copy numbers | 70 | [80] | |||
| PPV | Electrophoresis | 12 fg | 45 | [81] | |||
| Getah | Fluorescence (SYBR) | 2.61 copies/µL | 50 | [82] | |||
| PToV | Fluorescence (SYBR) | 1 × 101 copies/μL | 70 | [83] | |||
| SVDV | Fluorescence (SYBR) | 50 copies per assay | 30 to 60 | [84] | |||
| NASBA | AMV-RT, RNase H, and T7 polymerase | 120 min at 41–42 °C | CSFV | Fluorescence (ThT) | 2 copies/µL | 120 | [85] |
| JEV | Fluorescence (FAM) | 6 copies/reaction | 10 to 50 | [52] | |||
| CSFV | Colorimetric (ABTS) | 10 copies/mL | 180 | [53] | |||
| RPA | Recombinase, DNA polymerase, and SSB | 10–60 min at 37–42 °C | ASFV | Fluorescence (FAM) | 93.4 copies/reaction | 16 | [86] |
| ASFV | Lateral flow dipstick (LFD) | 150 copies/reaction | 10 | [56] | |||
| EMCV | Fluorescence (FAM) and lateral flow dipstick (LFD) | 1 × 102 copies for fluorescent RPA and 1 × 101 copies for LFD | 20 | [57] | |||
| PCV2 | Fluorescence (FAM) and lateral flow dipstick (LFD) | 102 copies/reaction | 20 | [87] | |||
| PCV2 | Electrophoresis | 102 copies | ~30 | [88] | |||
| PDCoV | Fluorescence (FAM) | 100 copies/reaction | 20 | [89] | |||
| PDCoV | Lateral flow dipstick (LFD) | 1 × 102 copies/µL | 10 | [90] | |||
| PEDV | Lateral flow dipstick (LFD) | 1 × 102 copies/µL | 30 | [91] | |||
| PEDV | Lateral flow dipstick (LFD) | 102 TCID50/mL | 25 | [92] | |||
| PRRSV | Lateral flow dipstick (LFD) | 5.6 × 10−1 TCID50 | 30 | [93] | |||
| SADS-CoV | Fluorescence (FAM) | 74 copies/µL | 30 | [58] | |||
| PSR | Bst | 61–65 min at 45–60 °C | ASFV | Fluorescence (SYBR) | 7.2 × 102 copies/µL | 45 | [60] |
| PEDV | Colorimetric (Phenol red and cresol red) | 1 fg/mL | 50 | [61] | |||
| PCV3 | Colorimetric (Phenol red and cresol red) | 1.13 × 102 copies/µL | 50 | [94] |
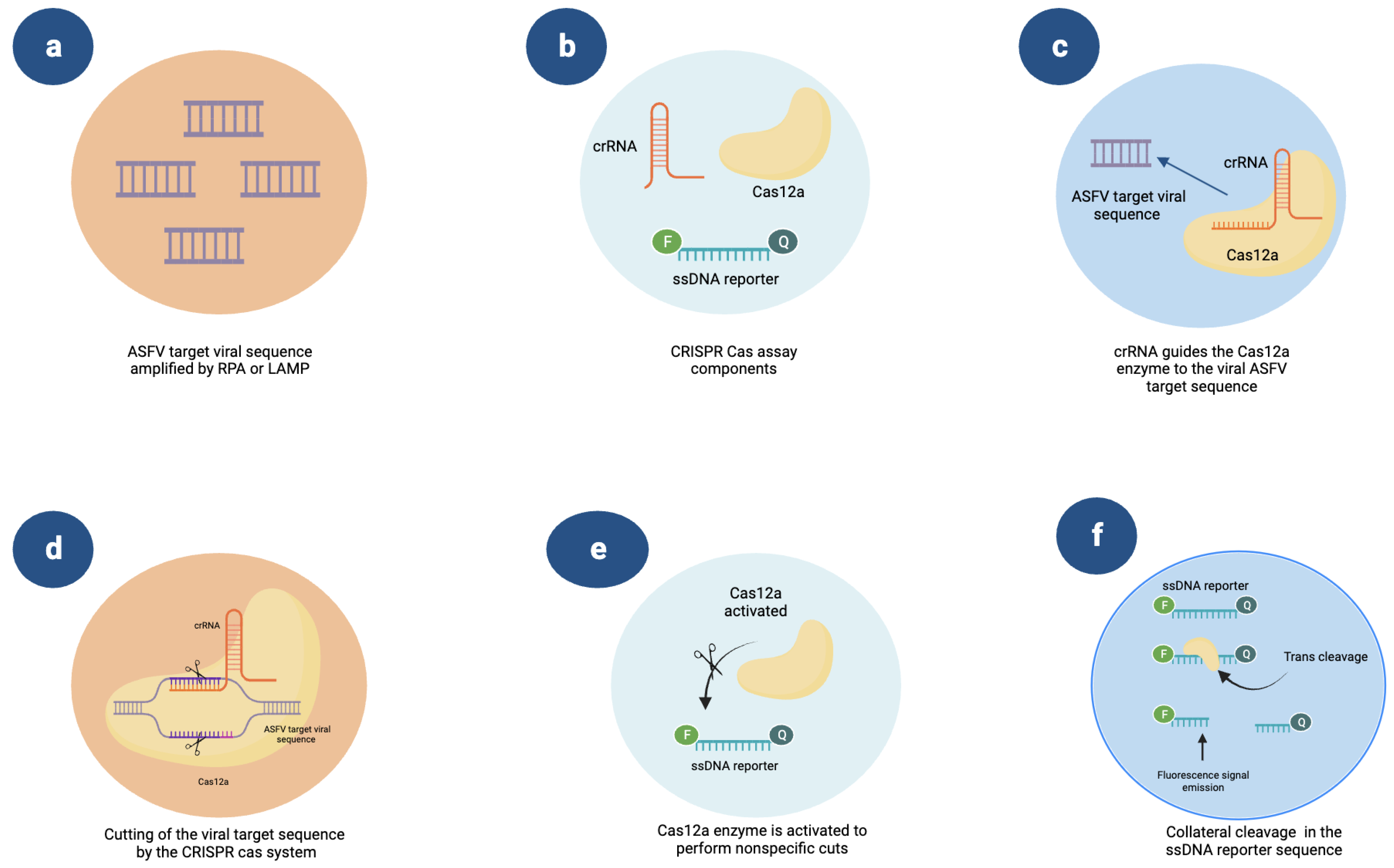
5. Novel Methods: CRISPR-Cas and Microfluidics Platforms
6. Current Challenges and Future Recommendations
- Price. The price of tests for analyzing animal health is between 50 and 70% cheaper than for human health. The average price of qPCR tests for the swine industry is around USD 22–35. If the technology is prohibitively expensive, it might not be appropriate for the animal industry, even if it meets all the technical requirements. Price is critical to maintaining ongoing diagnosis and monitoring.
- Turnaround time. Turnaround times for qPCR tests are 1–6 days. The processing time is a key parameter, since the capacity for action on the farm increases with the rapidity and accessibility of the test.
- Reliability. Tests must be sensitive and specific; for example, qPCR tests can detect 100 to 1 copies/mL. This range can be used as a reference for new technologies that seek to be implemented [125].
- Supply Chain. Suppliers of these technologies must be prepared to offer mass production, guarantee low prices, and maintain a continuous supply. The supply chains for the newest technologies are typically not prepared; as a result, the supply is complicated and expensive [126].
- Import costs. Local or regional suppliers should be identified to avoid excessive import and distribution costs [127].
- No cold chain. New emerging technologies should avoid refrigerated logistics since they increase the product’s costs and reduce the viability and lifetime [126].
- Easy to perform and interpret. The ideal technologies would be on-site systems that are fast and easy to process without requiring specialized technicians to perform the procedure and interpret the results [128].
- Available infrastructures. The necessary equipment and facilities must be available to perform the methodologies or implement the new technologies on farms or in local laboratories.
- Controls to avoid false results. New technologies require controls to avoid false positives and controls to detect when the result is a false negative.
- Detection of multiple pathogens. The diagnostic process includes sample collection, shipment, analysis, and the results report; thus, it is a significant effort and investment to detect only one pathogen. New technologies that seek to be implemented in the sector should be able to detect at least three pathogens in the same test to support issues of differential diagnosis, monitoring, and surveillance [129].
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, V.R.; Miller, R.S.; McKee, S.C.; Ernst, K.H.; Didero, N.M.; Maison, R.M.; Grady, M.J.; Shwiff, S.A. Risks of introduction and economic consequences associated with African swine fever, classical swine fever and foot-and-mouth disease: A review of the literature. Transbound. Emerg. Dis. 2021, 68, 1910–1965. [Google Scholar] [CrossRef] [PubMed]
- Number of Pigs. 2021. Available online: https://ourworldindata.org/grapher/pig-livestock-count-heads (accessed on 11 April 2023).
- Cornelison, A.S.; Karriker, L.A.; Williams, N.H.; Haberl, B.J.; Stalder, K.J.; Schulz, L.L.; Patience, J.F. Impact of health challenges on pig growth performance, carcass characteristics, and net returns under commercial conditions. Transl. Anim. Sci. 2018, 2, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Ghale-Noie, Z.N.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef] [PubMed]
- Montagnese, C.; Barattini, P.; Giusti, A.; Balka, G.; Bruno, U.; Bossis, I.; Gelasakis, A.; Bonasso, M.; Philmis, P.; Dénes, L.; et al. A Diagnostic Device for In-Situ Detection of Swine Viral Diseases: The SWINOSTICS Project. Sensors 2019, 19, 407. [Google Scholar] [CrossRef]
- Mota, D.; Guimarães, J.; Gandarilla, A.; Filho, J.; Brito, W.; Mariúba, L. Recombinase polymerase amplification in the molecular diagnosis of microbiological targets and its applications. Can. J. Microbiol. 2022, 68, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; ICTV Report Consortium. ICTV Virus Taxonomy Profile Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.; Goatley, L.C.; Guinat, C.; Netherton, C.L.; Gubbins, S.; Dixon, L.K.; Reis, A.L. Survival of African Swine Fever Virus in Excretions from Pigs Experimentally Infected with the Georgia 2007/1 Isolate. Transbound. Emerg. Dis. 2015, 64, 425–431. [Google Scholar] [CrossRef]
- Zimmerman, J.J. Diseases of Swine. 2019, p. 1108. Available online: https://www.wiley.com/en-us/Diseases+of+Swine%2C+11th+Edition-p-9781119350859 (accessed on 8 March 2023).
- Terrestrial Manual Online Access—WOAH—World Organisation for Animal Health. Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ (accessed on 8 March 2023).
- Terpstra, C.; Wensvoort, G.; Pol, J. Experimental reproduction of porcine epidemic abortion and respiratory syndrome (mystery swine disease) by infection with Lelystad vims: Koch’s postulates fulfilled. Vet. Q. 1991, 13, 131–136. [Google Scholar] [CrossRef]
- Bloemraad, M.; de Kluijver, E.P.; Petersen, A.; Burkhardt, G.E.; Wensvoort, G. Porcine reproductive and respiratory syndrome: Temperature and pH stability of Lelystad virus and its survival in tissue specimens from viraemic pigs. Vet. Microbiol. 1994, 42, 361–371. [Google Scholar] [CrossRef]
- Antas, M.; Woźniakowski, G. Current status of porcine epidemic diarrhoea (PED) in European pigs. J. Vet. Res. 2019, 63, 465–470. [Google Scholar] [CrossRef]
- Allan, G.M.; Ellis, J.A. Porcine Circoviruses: A Review. J. Vet. Diagn. Investig. 2000, 12, 3–14. [Google Scholar] [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2019, 67, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Mosolygó, T.; Laczi, K.; Spengler, G.; Burián, K. A Practical Approach for Quantitative Polymerase Chain Reaction, the Gold Standard in Microbiological Diagnosis. Sci 2022, 4, 4. [Google Scholar] [CrossRef]
- Filchakova, O.; Dossym, D.; Ilyas, A.; Kuanysheva, T.; Abdizhamil, A.; Bukasov, R. Review of COVID-19 testing and diagnostic methods. Talanta 2022, 244, 123409. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.S.; Gibbs, R.A.; Rainer, J.E.; Nguyen, P.N.; Thomas, C. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988, 16, 11141–11156. [Google Scholar] [CrossRef]
- Zhu, J.-H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y.-L.; Huang, Y.-W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J. Development and Clinical Applications of a 5-Plex Real-Time RT-PCR for Swine Enteric Coronaviruses. Viruses 2022, 14, 1536. [Google Scholar] [CrossRef]
- Liu, H.; Shi, K.; Sun, W.; Zhao, J.; Yin, Y.; Si, H.; Qu, S.; Lu, W. Development a multiplex RT-PCR assay for simultaneous detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. J. Virol. Methods 2021, 287, 114006. [Google Scholar] [CrossRef]
- Hu, L.; Lin, X.; Yang, Z.; Yao, X.; Li, G.; Peng, S.; Wang, Y. A multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, highly pathogenic porcine reproductive and respiratory syndrome virus, porcine reproductive and respiratory syndrome virus and pseudorabies in swines. Pol. J. Vet. Sci. 2015, 18, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; Agüero, M.; Fernández, J.; Romero, L.J.; Zamora, M.J.; Sánchez, C.; Belák, S.; Sánchez-Vizcaíno, J.M. A highly sensitive and specific gel-based multiplex RT-PCR assay for the simultaneous and differential diagnosis of African swine fever and Classical swine fever in clinical samples. Vet. Res. 2004, 35, 551–563. [Google Scholar] [CrossRef]
- Xu, X.-G.; Chen, G.-D.; Huang, Y.; Ding, L.; Li, Z.-C.; Chang, C.-D.; Wang, C.-Y.; Tong, D.-W.; Liu, H.-J. Development of multiplex PCR for simultaneous detection of six swine DNA and RNA viruses. J. Virol. Methods 2012, 183, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Giammarioli, M.; Pellegrini, C.; Casciari, C.; De Mia, G.M. Development of a novel hot-start multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, porcine circovirus type 2, porcine reproductive and respiratory syndrome virus and porcine parvovirus. Vet. Res. Commun. 2007, 32, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Jiao, Z.; Zhou, D.; Guo, R.; Duan, Z.; Tian, Y. Development of a multiplex PCR to detect and discriminate porcine circoviruses in clinical specimens. BMC Infect. Dis. 2019, 19, 778. [Google Scholar] [CrossRef]
- Sámi, L.; Ursu, K.; McKillen, J.; Kecskeméti, S.; Belák, S.; Kiss, I. Simultaneous detection of three porcine viruses by multiplex PCR. Acta Vet. Hung. 2007, 55, 267–276. [Google Scholar] [CrossRef]
- Ogawa, H.; Taira, O.; Hirai, T.; Takeuchi, H.; Nagao, A.; Ishikawa, Y.; Tuchiya, K.; Nunoya, T.; Ueda, S. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J. Virol. Methods 2009, 160, 210–214. [Google Scholar] [CrossRef]
- Kim, S.-C.; Jeong, C.-G.; Nazki, S.; Lee, S.-I.; Baek, Y.-C.; Jung, Y.-J.; Kim, W.-I. Evaluation of a multiplex PCR method for the detection of porcine parvovirus types 1 through 7 using various field samples. PLoS ONE 2021, 16, e0245699. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, W.; Wang, D.; Zhu, X.; Chen, Y.; Ouyang, K.; Wei, Z.; Liu, H.; Huang, W. Establishment of a Multiplex RT-PCR Method for the Detection of Five Known Genotypes of Porcine Astroviruses. Front. Vet. Sci. 2021, 8, 684279. [Google Scholar] [CrossRef]
- Zheng, L.-L.; Wang, Y.-B.; Li, M.-F.; Chen, H.-Y.; Guo, X.-P.; Geng, J.-W.; Wang, Z.-Y.; Wei, Z.-Y.; Cui, B.-A. Simultaneous detection of porcine parvovirus and porcine circovirus type 2 by duplex real-time PCR and amplicon melting curve analysis using SYBR Green. J. Virol. Methods 2013, 187, 15–19. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, K.; Liu, H.; Yin, Y.; Zhao, J.; Long, F.; Lu, W.; Si, H. Development of a multiplex qRT-PCR assay for detection of African swine fever virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus. J. Vet. Sci. 2021, 22, e87. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, K.; Zhao, J.; Yin, Y.; Chen, Y.; Si, H.; Qu, S.; Long, F.; Lu, W. Development of a one-step multiplex qRT–PCR assay for the detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. BMC Vet. Res. 2022, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shi, K.; Long, F.; Zhao, K.; Feng, S.; Yin, Y.; Xiong, C.; Qu, S.; Lu, W.; Li, Z. A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses. Vet. Sci. 2022, 9, 634. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Lu, J.; Wang, N.; He, W.-T.; Zhang, L.; Zhao, W.; Su, S. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Shi, K.; Zhou, Q.; Xiong, C.; Mo, S.; Zhou, H.; Long, F.; Wei, H.; Hu, L.; Mo, M. The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals 2022, 12, 1754. [Google Scholar] [CrossRef]
- Zou, J.; Liu, H.; Chen, J.; Zhang, J.; Li, X.; Long, Y.; Jiang, Y.; Li, W.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet. Sci. 2022, 10, 29. [Google Scholar] [CrossRef]
- Kim, H.-R.; Park, Y.-R.; Lim, D.-R.; Park, M.-J.; Park, J.-Y.; Kim, S.-H.; Lee, K.-K.; Lyoo, Y.S.; Park, C.-K. Multiplex real-time polymerase chain reaction for the differential detection of porcine circovirus 2 and 3. J. Virol. Methods 2017, 250, 11–16. [Google Scholar] [CrossRef]
- Li, X.; Qiao, M.; Sun, M.; Tian, K. A Duplex Real-Time PCR Assay for the Simultaneous Detection of Porcine Circovirus 2 and Circovirus 3. Virol. Sin. 2018, 33, 181–186. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Y.; Zheng, W.; Noll, L.; Porter, E.; Potter, M.; Cino, G.; Peddireddi, L.; Liu, X.; Anderson, G.; et al. A multiplex real-time PCR assay for the detection and differentiation of the newly emerged porcine circovirus type 3 and continuously evolving type 2 strains in the United States. J. Virol. Methods 2019, 269, 7–12. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- González-González, E.; Lara-Mayorga, I.M.; Rodríguez-Sánchez, I.P.; Zhang, Y.S.; Martínez-Chapa, S.O.; Santiago, G.T.-D.; Alvarez, M.M. Colorimetric loop-mediated isothermal amplification (LAMP) for cost-effective and quantitative detection of SARS-CoV-2: The change in color in LAMP-based assays quantitatively correlates with viral copy number. Anal. Methods 2020, 13, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Baek, Y.H.; Lloren, K.K.S.; Choi, W.-S.; Jeong, J.H.; Antigua, K.J.C.; Kwon, H.-I.; Park, S.-J.; Kim, E.-H.; Kim, Y.-I.; et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019, 19, 676. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, D.L.; Sullivan, V.; Owen, S.M.; Curtis, K.A. Detection of Acute HIV-1 Infection by RT-LAMP. PLoS ONE 2015, 10, e0126609. [Google Scholar] [CrossRef]
- Bonney, L.C.; Watson, R.J.; Slack, G.S.; Bosworth, A.; Wand, N.I.V.; Hewson, R. A flexible format LAMP assay for rapid detection of Ebola virus. PLOS Neglected Trop. Dis. 2020, 14, e0008496. [Google Scholar] [CrossRef]
- Cao, G.; Qiu, Y.; Long, K.; Xiong, Y.; Shi, M.; Yang, J.; Li, Y.; Nie, F.; Huo, D.; Hou, C. Carbon nanodots combined with loop-mediated isothermal amplification (LAMP) for detection of African swine fever virus (ASFV). Microchim. Acta 2022, 189, 342. [Google Scholar] [CrossRef]
- Wang, S.; Shen, H.; Lin, Q.; Huang, J.; Zhang, C.; Liu, Z.; Sun, M.; Zhang, J.; Liao, M.; Li, Y.; et al. Development of a Cleaved Probe-Based Loop-Mediated Isothermal Amplification Assay for Rapid Detection of African Swine Fever Virus. Front. Cell. Infect. Microbiol. 2022, 12, 884430. [Google Scholar] [CrossRef] [PubMed]
- Areekit, S.; Tangjitrungrot, P.; Khuchareontaworn, S.; Rattanathanawan, K.; Jaratsing, P.; Yasawong, M.; Chansiri, G.; Viseshakul, N.; Chansiri, K. Development of Duplex LAMP Technique for Detection of Porcine Epidemic Diarrhea Virus (PEDV) and Porcine Circovirus Type 2 (PCV 2). Curr. Issues Mol. Biol. 2022, 44, 5427–5439. [Google Scholar] [CrossRef]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, S.; Yang, K.; Liu, X.; Liu, W.; Guo, R.; Liang, W.; Yuan, F.; Liu, Z.; Gao, T.; et al. Rapid and simultaneous detection of Japanese encephalitis virus by real-time nucleic acid sequence-based amplification. Microb. Pathog. 2021, 150, 104724. [Google Scholar] [CrossRef]
- Lu, X.; Shi, X.; Wu, G.; Wu, T.; Qin, R.; Wang, Y. Visual detection and differentiation of Classic Swine Fever Virus strains using nucleic acid sequence-based amplification (NASBA) and G-quadruplex DNAzyme assay. Sci. Rep. 2017, 7, srep44211. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Zhang, J.; Li, N.; Chen, T.; Wang, L.; Zhang, F.; Mi, L.; Zhang, J.; Wang, S.; Wang, Y.; et al. Rapid and Sensitive Recombinase Polymerase Amplification Combined With Lateral Flow Strip for Detecting African Swine Fever Virus. Front. Microbiol. 2019, 10, 1004. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Han, Q.; Wang, K.; Liu, L.; Xu, X.; Yuan, W.; Wang, J. Rapid detection of porcine encephalomyocarditis virus (EMCV) by isothermal reverse transcription recombinase polymerase amplification assays. J. Virol. Methods 2022, 306, 114544. [Google Scholar] [CrossRef]
- Cong, X.; Zhu, Y.; Liu, X.; Lian, Y.; Huang, B.; Luo, Y.; Gu, Y.; Wu, M.; Shi, Y. Establishment of a recombinase polymerase amplification (RPA) fluorescence assay for the detection of swine acute diarrhea syndrome coronavirus (SADS-CoV). BMC Vet. Res. 2022, 18, 369. [Google Scholar] [CrossRef]
- Liu, W.; Dong, D.; Yang, Z.; Zou, D.; Chen, Z.; Yuan, J.; Huang, L. Polymerase Spiral Reaction (PSR): A novel isothermal nucleic acid amplification method. Sci. Rep. 2015, 5, 12723. [Google Scholar] [CrossRef]
- Woźniakowski, G.; Frączyk, M.; Kowalczyk, A.; Pomorska-Mól, M.; Niemczuk, K.; Pejsak, Z. Polymerase cross-linking spiral reaction (PCLSR) for detection of African swine fever virus (ASFV) in pigs and wild boars. Sci. Rep. 2017, 7, srep42903. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Hu, W.; Zuo, K.; Li, Z.; Kan, Y.; Yao, L.; Ji, J.; Bi, Y. Visual detection of porcine epidemic diarrhea virus using a novel reverse transcription poly-merase spiral reaction method. BMC Vet. Res. 2019, 15, 116. [Google Scholar] [CrossRef]
- Woźniakowski, G.; Frączyk, M.; Mazur, N. Comparison of loop-mediated isothermal amplification (LAMP) and cross-priming amplification (CPA) for detection of African swine fever virus. Pol. J. Vet. Sci. 2018, 21, 827–830. [Google Scholar] [CrossRef]
- Gao, M.; Cui, J.; Ren, Y.; Suo, S.; Li, G.; Sun, X.; Su, D.; Opriessnig, T.; Ren, X. Development and evaluation of a novel reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for detection of type II porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2012, 185, 18–23. [Google Scholar] [CrossRef]
- Chen, C.; Cui, S.; Zhang, C.; Li, J.; Wang, J. Development and validation of reverse transcription loop-mediated isothermal amplification for detection of PRRSV. Virus Genes 2009, 40, 76–83. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, P.; Li, J.; Jiang, F.; Li, J.; Wu, W. Development of loop-mediated isothermal amplification method for visuali-zation detection of the highly virulent strains of porcine reproductive and respiratory syndrome virus (PRRSV) in China. Afr. J. Biotechnol. 2013, 10, 13278–13283. [Google Scholar]
- Zhang, L.; Liu, Y.-B.; Chen, L.; Wang, J.-H.; Ning, Y.-B. Rapid and sensitive detection of PRRSV by a reverse transcription-loop-mediated isothermal amplification assay. Virol. Sin. 2011, 26, 252–259. [Google Scholar] [CrossRef]
- Chen, X.-W.; Wang, Q.; Yin, M.; Pu, Z.-H.; Guo, A.-W.; Li, L.; Luo, W.-T.; Wang, X.-Q. Rapid and Simple Detection of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) by Reverse Transcription Loop-Mediated Isothermal Amplification. Pak J Zool. 2018, 50, 55–60. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, Q.-F.; Xue, C.-Y.; Ma, J.-Y.; Zhu, D.-Z.; Cao, Y.-C. Rapid detection of porcine reproductive and respiratory syndrome virus by reverse transcription loop-mediated isothermal amplification assay. J. Virol. Methods 2009, 155, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-T.; Zhang, J.; Sun, D.-H.; Ma, L.-N.; Liu, X.-T.; Quan, K.; Liu, Y.-S. Reverse transcription loop-mediated isothermal amplification for the detection of highly pathogenic porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2008, 153, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.; Abrahante, J.; Murtaugh, M.; Claudia, M.-Z. Reverse Transcription Loop-Mediated Isothermal Amplification for the Detection of Porcine Reproductive and Respiratory Syndrome Virus. J. Vet. Diagn. Investig. 2009, 21, 350–354. [Google Scholar] [CrossRef]
- Park, J.-Y.; Park, S.; Park, Y.-R.; Kang, D.-Y.; Kim, E.-M.; Jeon, H.-S.; Kim, J.-J.; Kim, W.-I.; Lee, K.-T.; Kim, S.-H.; et al. Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the visual detection of European and North American porcine reproductive and respiratory syndrome viruses. J. Virol. Methods 2016, 237, 10–13. [Google Scholar] [CrossRef]
- Wang, C.; Pang, V.F.; Jeng, C.-R.; Lee, F.; Huang, Y.-W.; Lin, Y.-L.; Hsiao, S.-H.; Lai, S.-S. Detection of porcine circovirus type 1 in commercial porcine vaccines by loop-mediated isothermal amplification. Folia Microbiol. 2011, 56, 483–489. [Google Scholar] [CrossRef]
- Wang, H.; Liu, X.; Zeng, F.; Zhang, T.; Lian, Y.; Wu, M.; Xiao, L.; Zhu, Y.; Zhang, Y.; Chen, M.; et al. Development of a real-time loop-mediated isothermal amplification assay for detection of porcine circovirus 3. BMC Vet. Res. 2019, 15, 305. [Google Scholar] [CrossRef]
- Kim, H.; Lim, D.; Chae, H.; Park, J.; Kim, S.; Lee, K.; Lee, C.; Lyoo, Y.S.; Park, C. Advanced target-specific probe-based real-time loop-mediated isothermal amplification assay for the rapid and specific detection of porcine circovirus 3. Transbound. Emerg. Dis. 2020, 67, 2336–2344. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.N.; Nguyen, V.D.; Yamazaki, W.; Okabayashi, T.; Mitoma, S.; Notsu, K.; Sakai, Y.; Yamaguchi, R.; Norimine, J.; Sekiguchi, S. Development of pooled testing system for porcine epidemic diarrhoea using real-time fluorescent reverse-transcription loop-mediated isothermal amplification assay. BMC Vet. Res. 2018, 14, 172. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liang, J.; Yang, D.; Zhang, Q.; Miao, D.; He, X.; Du, Y.; Zhang, W.; Ni, J.; Zhao, K. Visual and Rapid Detection of Porcine Epidemic Diarrhea Virus (PEDV) Using Reverse Transcription Loop-Mediated Isothermal Amplification Method. Animals 2022, 12, 2712. [Google Scholar] [CrossRef] [PubMed]
- Asih, A.U.; Janetanakit, T.; Nasamran, C.; Bunpapong, N.; Boonyapisitsopa, S.; Amonsin, A. Reverse transcription loop-mediated isothermal amplification combined with lateral flow device (RT-LAMP-LFD) for swine influenza virus detection. Thai J. Vet. Med. 2021, 51, 55–67. [Google Scholar] [CrossRef]
- Wongsawat, K.; Dharaku, T.; Narat, P.; Rabablert, J. Detection of nucleic acid of classical swine fever virus by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Health 2011, 03, 447–452. [Google Scholar] [CrossRef]
- Shen, H.; Wang, S.; Huang, J.; Lin, Q.; Zhang, C.; Liu, Z.; Zhang, J.; Liao, M. A Novel, Cleaved Probe-Based Reverse Transcription Loop-Mediated Isothermal Amplifica-tion Method for Specific and Sensitive Detection of Porcine Deltacoronavirus. Front. Vet. Sci. 2022, 9, 747. [Google Scholar] [CrossRef]
- Zhang, F.; Ye, Y.; Song, D.; Guo, N.; Peng, Q.; Li, A.; Zhou, X.; Chen, Y.; Zhang, M.; Huang, D.; et al. A simple and rapid identification method for newly emerged porcine Deltacoronavirus with loop-mediated isothermal amplification. Biol. Res. 2017, 50, 30. [Google Scholar] [CrossRef]
- Qu, G.; Fu, S.; Shen, N.; Wang, J.; Xiao, Y.; Guan, Y.; Tang, N.; Chen, L.; Gao, S.; Shen, Z. Rapid and Sensitive Diagnosis of Porcine Parvovirus by Loop- mediated Isothermal Amplification (LAMP) Method. J. Appl. Anim. Res. 2010, 37, 113–116. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.-X.; Bu, Y.-P.; Liu, Y.; Sun, X.-T.; Shi, N.; Deng, X.-Y.; Lu, R.-G.; Hu, B.; Jin, N.-Y.; et al. Rapid Visual Detection of Getah Virus Using a Loop-Mediated Isothermal Amplification Method. Vector-Borne Zoonotic Dis. 2019, 19, 741–746. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Yang, F.; Liu, P.; Cai, Y.; Huang, J.; Zhu, L.; Xu, Z. Rapid and sensitive detection of porcine torovirus by a reverse transcription loop-mediated isothermal amplification assay (RT-LAMP). J. Virol. Methods 2016, 228, 103–107. [Google Scholar] [CrossRef]
- Blomström, A.-L.; Hakhverdyan, M.; Reid, S.M.; Dukes, J.P.; King, D.P.; Belák, S.; Berg, M. A one-step reverse transcriptase loop-mediated isothermal amplification assay for simple and rapid detection of swine vesicular disease virus. J. Virol. Methods 2008, 147, 188–193. [Google Scholar] [CrossRef]
- Guoshuai, J.; Xiaomeng, X.; Zengdan, G.; Xingxing, H.; Qi, P.; Hanbing, Z.; Yi, W. A rapid and high sensitivity RNA detection based on NASBA and G4-ThT fluorescent biosensor. Sci. Rep. 2022, 12, 10076. [Google Scholar] [CrossRef]
- Fan, X.; Li, L.; Zhao, Y.; Liu, Y.; Liu, C.; Wang, Q.; Dong, Y.; Wang, S.; Chi, T.; Song, F.; et al. Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus. Front. Microbiol. 2020, 11, 1696. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qin, X.; Sun, Y.; Cong, G.; Li, Y.; Zhang, Z. Development of Isothermal Recombinase Polymerase Amplification Assay for Rapid Detection of Porcine Circovirus Type 2. BioMed Res. Int. 2017, 2017, 8403642. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Liu, L.; Li, R.; Yuan, W. Rapid detection of Porcine circovirus 2 by recombinase polymerase amplification. J. Vet. Diagn. Investig. 2016, 28, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zeng, F.; Huang, B.; Zhu, Y.; Wu, M.; Xu, F.; Xiao, L.; Huang, R.; Ma, J.; Cong, F.; et al. Point-of-care diagnostic assay for rapid detection of porcine deltacoronavirus using the recombinase polymerase amplification method. Transbound. Emerg. Dis. 2019, 66, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, X.; Zhang, Y.; Wei, Y.; Wang, Y. Rapid and visual detection of porcine deltacoronavirus by recombinase poly-merase amplification combined with a lateral flow dipstick. BMC Vet. Res. 2020, 16, 130. [Google Scholar] [CrossRef]
- Ma, L.; Lian, K.; Zhu, M.; Tang, Y.; Zhang, M. Visual detection of porcine epidemic diarrhea virus by recombinase polymer-ase amplification combined with lateral flow dipstrip. BMC Vet. Res. 2022, 18, 140. [Google Scholar] [CrossRef]
- Pewlaoo, S.; Phanthong, S.; Kong-Ngoen, T.; Santajit, S.; Tunyong, W.; Buranasinsup, S.; Kaeoket, K.; Thavorasak, T.; Pumirat, P.; Sookrung, N.; et al. Development of a Rapid Reverse Transcription-Recombinase Polymerase Amplification Couple Nucleic Acid Lateral Flow Method for Detecting Porcine Epidemic Diarrhoea Virus. Biology 2022, 11, 1018. [Google Scholar] [CrossRef]
- Tian, X.-X.; Wang, T.; Cui, X.-Y.; Huang, X.-Y.; Sun, Y.; Xia, D.-S.; Yang, Y.-B.; Cai, X.-H.; An, T.-Q. Rapid visual detection of porcine reproductive and respiratory syndrome virus via recombinase polymerase amplification combined with a lateral flow dipstick. Arch. Virol. 2022, 167, 493–499. [Google Scholar] [CrossRef]
- Ji, J.; Xu, X.; Wang, X.; Zuo, K.; Li, Z.; Leng, C.; Kan, Y.; Yao, L.; Bi, Y. Novel polymerase spiral reaction assay for the visible molecular detection of porcine circovirus type 3. BMC Vet. Res. 2019, 15, 322. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Liu, J.; Zhu, W.; Zeng, M.; Xu, K.; Ding, J.; Zhou, H.; Zhu, J.; Ke, Y.; Li, L.Y.; et al. One-Pot Visual Detection of African Swine Fever Virus Using CRISPR-Cas12a. Front. Vet. Sci. 2022, 9, 1032. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tao, D.; Chen, X.; Shen, L.; Zhu, L.; Xu, B.; Liu, H.; Zhao, S.; Li, X.; Liu, X.; et al. Detection of Four Porcine Enteric Coronaviruses Using CRISPR-Cas12a Combined with Multi-plex Reverse Transcriptase Loop-Mediated Isothermal Amplification Assay. Viruses 2022, 14, 833. [Google Scholar] [CrossRef]
- Qian, S.; Chen, Y.; Peng, C.; Wang, X.; Wu, H.; Che, Y.; Wang, H.; Xu, J.; Wu, J. Dipstick-based rapid nucleic acids purification and CRISPR/Cas12a-mediated isothermal amplification for visual detection of African swine fever virus. Talanta 2022, 242, 123294. [Google Scholar] [CrossRef]
- Lei, L.; Liao, F.; Tan, L.; Duan, D.; Zhan, Y.; Wang, N.; Wang, Y.; Peng, X.; Wang, K.; Huang, X.; et al. LAMP Coupled CRISPR-Cas12a Module for Rapid, Sensitive and Visual Detection of Porcine Circovirus 2. Animals 2022, 12, 2413. [Google Scholar] [CrossRef]
- Wang, X.; He, S.; Zhao, N.; Liu, X.; Cao, Y.; Zhang, G.; Wang, G.; Guo, C. Development and clinical application of a novel CRISPR-Cas12a based assay for the detection of African swine fever virus. BMC Microbiol. 2020, 20, 283. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, Y.; Cai, G.; Meng, G.; Shi, S. Rapid and sensitive RPA-Cas12a-fluorescence assay for point-of-care detection of African swine fever virus. PLoS ONE 2021, 16, e0254815. [Google Scholar] [CrossRef]
- Lu, S.; Li, F.; Chen, Q.; Wu, J.; Duan, J.; Lei, X.; Zhang, Y.; Zhao, D.; Bu, Z.; Yin, H. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discov. 2020, 6, 18. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, X.; Yin, D.; Cai, C.; Liu, H.; Yang, Y.; Guo, Z.; Yin, L.; Shen, X.; Dai, Y.; et al. Rapid and Easy-Read Porcine Circovirus Type 4 Detection with CRISPR–Cas13a-Based Lateral Flow Strip. Microorganisms 2023, 11, 354. [Google Scholar] [CrossRef]
- Liu, S.; Tao, D.; Liao, Y.; Yang, Y.; Sun, S.; Zhao, Y.; Yang, P.; Tang, Y.; Chen, B.; Liu, Y.; et al. Highly Sensitive CRISPR/Cas12a-Based Fluorescence Detection of Porcine Reproductive and Res-piratory Syndrome Virus. ACS Synth. Biol. 2021, 10, 2499–2507. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Deng, Y.; Li, T.; Wang, J.; Wang, T.; Tan, F.; Li, X.; Tian, K. Visual detection of porcine reproductive and respiratory syndrome virus using CRISPR-Cas13a. Transbound. Emerg. Dis. 2019, 67, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ji, P.; Fan, H.; Dang, L.; Wan, W.; Liu, S.; Li, Y.; Yu, W.; Li, X.; Ma, X.; et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020, 3, 62. [Google Scholar] [CrossRef]
- Weigl, B.; Domingo, G.; LaBarre, P.; Gerlach, J. Towards non- and minimally instrumented, microfluidics-based diagnostic devices. Lab a Chip 2008, 8, 1999–2014. [Google Scholar] [CrossRef] [PubMed]
- Flores-Contreras, E.A.; González-González, R.B.; Rodríguez-Sánchez, I.P.; León, J.F.Y.-D.; Iqbal, H.M.N.; González-González, E. Microfluidics-Based Biosensing Platforms: Emerging Frontiers in Point-of-Care Testing SARS-CoV-2 and Seroprevalence. Biosensors 2022, 12, 179. [Google Scholar] [CrossRef] [PubMed]
- González-González, E.; Garcia-Ramirez, R.; Díaz-Armas, G.G.; Esparza, M.; Aguilar-Avelar, C.; Flores-Contreras, E.A.; Rodríguez-Sánchez, I.P.; Delgado-Balderas, J.R.; Soto-García, B.; Aráiz-Hernández, D.; et al. Automated ELISA On-Chip for the Detection of Anti-SARS-CoV-2 Antibodies. Sensors 2021, 21, 6785. [Google Scholar] [CrossRef]
- Ratta, B.; Yadav, B.S.; Pokhriyal, M.; Saxena, M.; Sharma, B. Microarray Chip Based Identification of a Mixed Infection of Bovine Herpesvirus 1 and Bovine Viral Diarrhea 2 From Indian Cattle. Curr. Microbiol. 2013, 68, 127–131. [Google Scholar] [CrossRef]
- An, X.; Zuo, P.; Ye, B.-C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 2019, 209, 120571. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, S.; Wu, Y.; Zhou, B.; Wang, K.; Jiang, L.; Long, Y.; Chen, G.; Zeng, D. Multiplex and on-site PCR detection of swine diseases based on the microfluidic chip system. BMC Vet. Res. 2021, 17, 117. [Google Scholar] [CrossRef]
- Chen, L.; Wen, K.; Chen, F.-E.; Trick, A.Y.; Liu, H.; Shao, S.; Yu, W.; Hsieh, K.; Wang, Z.; Shen, J.; et al. Portable Magnetofluidic Device for Point-of-Need Detection of African Swine Fever. Anal. Chem. 2021, 93, 10940–10946. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bai, H.; Mauk, M.G.; Saif, L.; Bau, H.H. A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab. Chip 2021, 21, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Zhang, J.; Zhao, R.; Gao, J. Development of a Handheld Microfluidic Chip for On-Site Multiplex Detection of Four Por-cine Diarrhea-Related Virus. ACS Agric. Sci. Technol. 2022, 2, 805–812. [Google Scholar] [CrossRef]
- Ji, C.; Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Wang, H.; Sun, Y.; et al. Microfluidic-LAMP chip for the point-of-care detection of gene-deleted and wild-type African swine fever viruses and other four swine pathogens. Front. Vet. Sci. 2023, 10, 1116352. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Sun, Y.; Ma, J.; Lan, T. Microfluidic-RT-LAMP chip for the point-of-care detection of emerging and re-emerging enteric coronaviruses in swine. Anal. Chim. Acta 2020, 1125, 57–65. [Google Scholar] [CrossRef]
- Ma, W. Swine influenza virus: Current status and challenge. Virus Res. 2020, 288, 198118. [Google Scholar] [CrossRef] [PubMed]
- Galvis, J.A.; Corzo, C.A.; Prada, J.M.; Machado, G. Modeling between-farm transmission dynamics of porcine epidemic diarrhea virus: Characterizing the dominant transmission routes. Prev. Vet. Med. 2022, 208, 105759. [Google Scholar] [CrossRef]
- Risser, J.; Ackerman, M.; Evelsizer, R.; Wu, S.; Kwon, B.; Hammer, J.M. Porcine reproductive and respiratory syndrome virus genetic variability a management and diagnostic dilemma. Virol. J. 2021, 18, 206. [Google Scholar] [CrossRef]
- Wang, L.; Madera, R.; Li, Y.; McVey, D.S.; Drolet, B.S.; Shi, J. Recent Advances in the Diagnosis of Classical Swine Fever and Future Perspectives. Pathogens 2020, 9, 658. [Google Scholar] [CrossRef]
- Holtkamp, D.; Holtkamp, D.J.; Torremorell, M. Commentary Peer reviewed Proposed modifications to porcine reproduc-tive and respiratory syndrome virus herd classification. J. Swine Health Prod. 2021, 29, 261–270. [Google Scholar] [CrossRef]
- Turlewicz-Podbielska, H.; Włodarek, J.; Pomorska-Mól, M. Noninvasive strategies for surveillance of swine viral diseases: A review. J. Vet. Diagn. Investig. 2020, 32, 503–512. [Google Scholar] [CrossRef]
- Boonbanjong, P.; Treerattrakoon, K.; Waiwinya, W.; Pitikultham, P.; Japrung, D. Isothermal Amplification Technology for Disease Diagnosis. Biosensors 2022, 12, 677. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van Der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Kuupiel, D.; Bawontuo, V.; Mashamba-Thompson, T.P. Improving the Accessibility and Efficiency of Point-of-Care Diagnostics Services in Low- and Middle-Income Countries: Lean and Agile Supply Chain Management. Diagnostics 2017, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Increasing Access to Diagnostics through Technology Transfer and Local Production; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Martins, S.A.M.; Martins, V.C.; Cardoso, F.A.; Germano, J.; Rodrigues, M.; Duarte, C.; Bexiga, R.; Cardoso, S.; Freitas, P.P. Biosensors for On-Farm Diagnosis of Mastitis. Front. Bioeng. Biotechnol. 2019, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-P.; Xia, Y.-J.; Xu, L.; Zhao, J.-J.; Wang, Z.; Zhao, Q.-Z.; Liu, Y.-B.; Zhang, Q.-Y.; Wang, Q. A multiplex real-time PCR assay for simultaneous detection of classical swine fever virus, African swine fever virus, and atypical porcine pestivirus. J. Integr. Agric. 2023, 22, 559–567. [Google Scholar] [CrossRef]
| Type of Multiplex Test | Targets and Reporters (Virus-Gene-Dye) | LOD (Viral Copies) | Reference |
| Quadruplex RT-qPCR | (1) PEDV-N-JUN (2) PDCoV-N-ABY (3) TGEV-N-VIC (4) SADS-CoV-N-FAM | 4–16 | [22] |
| Quadruplex RT-qPCR | (1) PEDV-N-JOE (2) PDCoV-M-FAM (3) TGEV-N-Texas Red (4) SADS-CoV-N-Cy5 | 121 | [36] |
| Quadruplex RT-qPCR | (1) PEDV-ORF1a-FAM (2) PDCoV-ORF1b-Texas Red (3) PtoV-ORF1a-VIC (4) SADS-CoV-ORF1a-Cy5 | 100 | [37] |
| Quadruplex qPCR | ASFV strains (1) B646L-p72-FAM (2) MGF505-2R-CD2v-VIC (3) EP402R-CD2v-Cy5 (4) I177L-CD2v-Texas Red | 3.21–32.1 | [38] |
| Triplex RT-qPCR | (1) ASFV-p72-FAM (2) CSFV-5′ UTR-VIC (3) PRRSV-ORF7-Cy5 | 1.78 | [34] |
| Triplex RT-qPCR | (1) ASFV-p72-Texas Red (2) CSFV-5′ UTR-JOE (3) APPV-5′ UTR-FAM | 2.52 | [35] |
| Triplex qPCR | (1) PCV2-cap-Cy5 (2) PCV3-rep-FAM (3) PCV4-cap-Texas Red | 101 | [39] |
| Duplex qPCR | (1) PCV2-cap-FAM (2) PCV3-cap-ROX | 50 | [40] |
| Duplex qPCR | (1) PCV2-cap-FAM (2) PCV3-rep-HEX | 2.9–22.5 | [41] |
| Duplex qPCR | (1) PCV2-rep/cap-VIC (2) PCV3-rep/ORF3-FAM | 14–17 | [42] |
| Type of Multiplex Test | Targets and Reporter (Virus-Dye) | Detection Time (min) | Company (Web) |
| Duplex qPCR | (1) PCV2-FAM (2) PCV3-Cy5 | 90 | Idexx.com |
| Duplex RT-qPCR | (1) PRRSV1-Cy5 (2) PRRSV2-FAM | 90 | Idexx.com |
| Duplex RT-qPCR | (1) PEDV-FAM (2) PDCoV-Cy5 | 90 | Idexx.com |
| Duplex RT-qPCR | (1) PRRSV1-VIC (2) PRRSV2-FAM | 90 | Thermofisher.com |
| Triplex RT-qPCR | (1) PEDV-LIZ (2) TGEV-FAM (3) PDCoV-VIC | 90 | Thermofisher.com |
| Triplex RT-qPCR | (1) PRRSV-FAM (2) TGEV-VIC (3) PEDV-Cy5 | 90 | Hermes-Bio.com |
| Triplex RT-qPCR | (1) PCV2-FAM (2) PRRSV-VIC (3) PDCoV-Cy5 | 90 | Hermes-Bio.com |
| Triplex RT-qPCR | (1) PoRV-A-FAM (2) PoRV-B-VIC (3) PoRV-C-Cy5 | 90 | Hermes-Bio.com |
| Duplex RT-qPCR | (1) PRRSV-N.A. (2) IAV-N.A. | 140 | Tetracore.com |
| Triplex RT-qPCR | (1) PEDV-FAM (2) TGEV-N.A. (3) PDCoV-Cy5 | 140 | Tetracore.com |
| Method Combined with CRISPR-Cas | Enzymes | Conditions | Target | Detection Signal | LOD | Detection Time (min) | Reference |
| LAMP | Bst, Cas12a |
| PEDV, TGEV, PDCoV, and SADS-CoV | Fluorescence (ROX) | 1 copy/µL | 50 | [97] |
| LAMP | Bst, Cas12a |
| ASFV | Fluorescence (FAM) | 5.8 × 102 copies/µL | 60 | [96] |
| LAMP | Bst, Cas12a |
| ASFV | Fluorescence (SYTO 9) | 1 copy/µL | 40 | [98] |
| LAMP | Bst, Cas12a |
| PCV2 | Fluorescence (FAM) | 1 copy/µL | 30 | [99] |
| RPA | Cas12a, recombinase, DNA polymerase, and SSB |
| ASFV | Fluorescence (FAM) | 7.4 × 104 copies/µL | 40 | [96] |
| RPA | Cas12a, recombinase, DNA polymerase, and SSB |
| ASFV | Fluorescence (HEX) | 1.16 copies/µL | 35 | [100] |
| RPA | Cas12a, recombinase, DNA polymerase, and SSB |
| ASFV | Fluorescence (FAM) | 2 copies of DNA/reaction | 35 min | [101] |
| RPA | Cas12a, recombinase, DNA polymerase, and SSB |
| ASFV | Lateral flow dipstick (LFD) | 2 × 102 copies of viral genome | 120 | [102] |
| RPA | Cas13a, recombinase, DNA polymerase, and SSB |
| PCV4 | Lateral flow dipstick (LFD) | 1 × 100 to 1 × 101 copies/µL | 80 | [103] |
| RPA | Cas12a, recombinase, DNA polymerase, and SSB |
| PRRSV | Fluorescence (FAM) | 1 × 100 copies/µL | 25 | [104] |
| RPA | Cas13a, recombinase, DNA polymerase, and SSB |
| PRRSV | Fluorescence (FAM) | 1.72 × 102 copies/µL | 60 | [105] |
| LFD | Cas12a |
| ASFV | Lateral flow dipstick (LFD) | 2 × 101 copies/reaction | 30 | [106] |
| Microfluidics Platform | Enzymes | Conditions | Target | Detection | Reference |
| Microfluidics Multiplex-PCR | DNA polymerase |
| PRRSV, PEDV, PRV, and PCV2 | Fluorescence (SYBR) | [112] |
| Magnetofluidics Device-qPCR | DNA polymerase |
| ASFV | Fluorescence | [113] |
| 3D-printed Microfluidics Device-LAMP | Bst | 30 min at 65 °C | PEDV, TGEV, and PDCoV | Fluorescence EvaGreen | [114] |
| Handheld Microfluidics Chip-LAMP | Bst | 60 min at 65 °C | PEDV, TGEV, PoRV, and PCV2 | Colorimetric phenol red | [115] |
| Microfluidics Multiplex-LAMP | Bst | 60 min at 63.5 °C | ASFV, PPV, PCV2, PRV, and PRRSV | Fluorescence (SYBR) | [116] |
| Microfluidics Multiplex-LAMP | Bst | 40 min at 63.5 °C | PEDV, PDCoV, and SADS-CoV | Fluorescence (SYBR) | [117] |
| Type of Test | Price | Specificity | Sensibility | Response Time | Available Infrastructure | Detection of Multiple Pathogens | Operation Difficulty |
| Point of Care (POC) | 💲 | 🎯 | 🔍 | ⌛ | ⚙ | 🦠 |  |
| qPCR | 💲💲 | 🎯🎯🎯🎯🎯 | 🔍🔍🔍🔍🔍 | ⌛⌛ | ⚙⚙⚙⚙ | 🦠 |   |
| qPCR Multiplex | 💲💲💲 | 🎯🎯🎯🎯🎯 | 🔍🔍🔍🔍🔍 | ⌛⌛ | ⚙⚙⚙⚙ | 🦠🦠🦠 |   |
| Sequencing | 💲💲💲💲💲 | 🎯🎯🎯🎯🎯 | 🔍🔍🔍🔍🔍 | ⌛⌛⌛⌛⌛ | ⚙ | 🦠🦠🦠🦠🦠 |      |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Contreras, E.A.; Carrasco-González, J.A.; Linhares, D.C.L.; Corzo, C.A.; Campos-Villalobos, J.I.; Henao-Díaz, A.; Melchor-Martínez, E.M.; Iqbal, H.M.N.; González-González, R.B.; Parra-Saldívar, R.; et al. Emergent Molecular Techniques Applied to the Detection of Porcine Viruses. Vet. Sci. 2023, 10, 609. https://doi.org/10.3390/vetsci10100609
Flores-Contreras EA, Carrasco-González JA, Linhares DCL, Corzo CA, Campos-Villalobos JI, Henao-Díaz A, Melchor-Martínez EM, Iqbal HMN, González-González RB, Parra-Saldívar R, et al. Emergent Molecular Techniques Applied to the Detection of Porcine Viruses. Veterinary Sciences. 2023; 10(10):609. https://doi.org/10.3390/vetsci10100609
Chicago/Turabian StyleFlores-Contreras, Elda A., Jorge Alberto Carrasco-González, Daniel C. L. Linhares, Cesar A. Corzo, J. Israel Campos-Villalobos, Alexandra Henao-Díaz, Elda M. Melchor-Martínez, Hafiz M. N. Iqbal, Reyna Berenice González-González, Roberto Parra-Saldívar, and et al. 2023. "Emergent Molecular Techniques Applied to the Detection of Porcine Viruses" Veterinary Sciences 10, no. 10: 609. https://doi.org/10.3390/vetsci10100609
APA StyleFlores-Contreras, E. A., Carrasco-González, J. A., Linhares, D. C. L., Corzo, C. A., Campos-Villalobos, J. I., Henao-Díaz, A., Melchor-Martínez, E. M., Iqbal, H. M. N., González-González, R. B., Parra-Saldívar, R., & González-González, E. (2023). Emergent Molecular Techniques Applied to the Detection of Porcine Viruses. Veterinary Sciences, 10(10), 609. https://doi.org/10.3390/vetsci10100609








