Identification of the Effects of 5-Azacytidine on Porcine Circovirus Type 2 Replication in Porcine Kidney Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and PCV2 Infection
2.2. Cell Activity Detection
2.3. Cell Immunofluorescence Staining
2.4. DNA Extraction
2.5. RNA Extraction and cDNA Synthesis
2.6. qPCR Detection
2.7. Total Protein Extraction from Cell Samples
2.8. Western Blotting
2.9. Reactive Oxygen Species Assessment
2.10. Flow Cytometry Analysis
2.11. RNA Sequencing
2.12. Data Processing and Analyses
3. Results
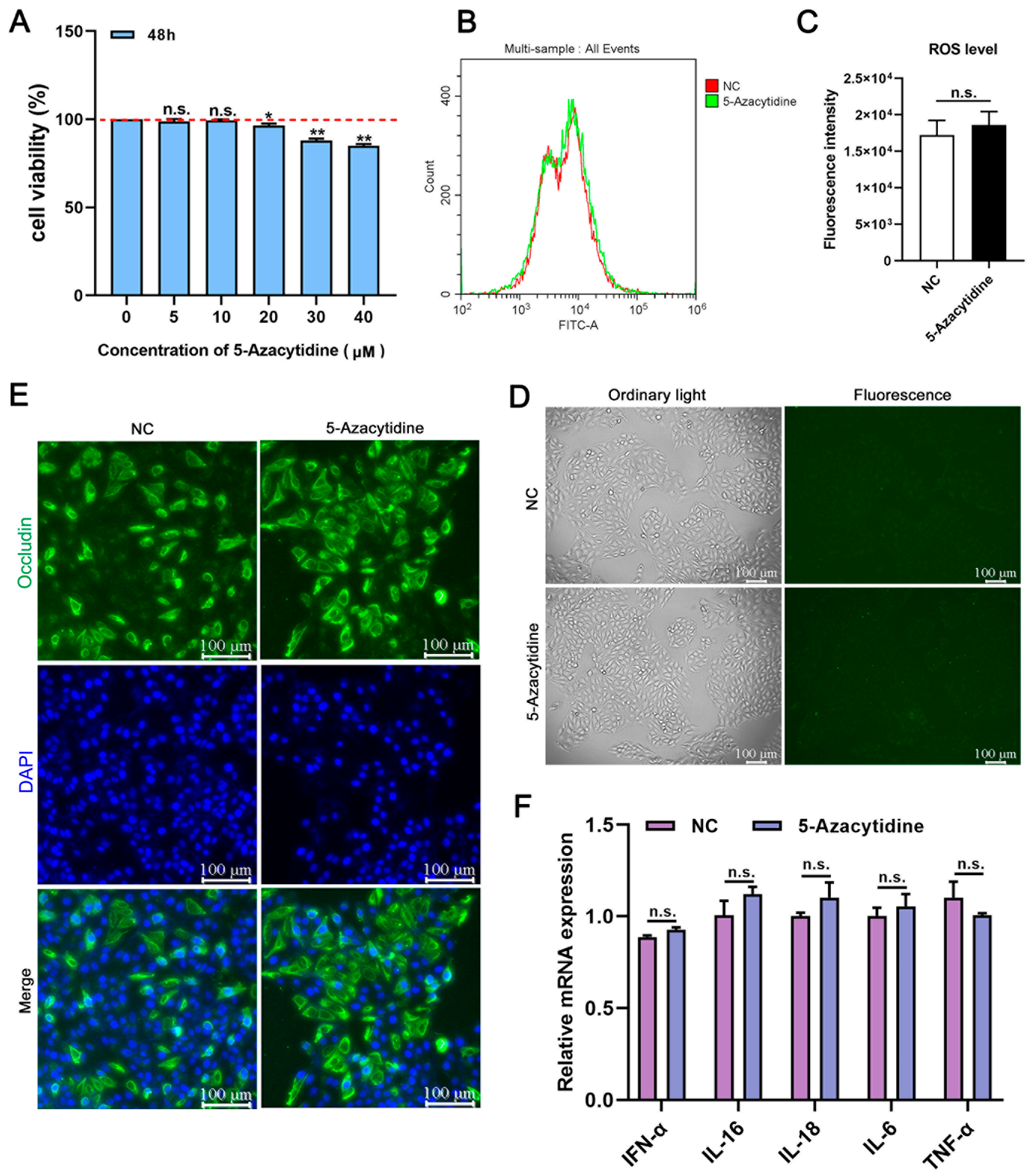
3.1. Working Concentration of 5-Aza and Drug Safety Evaluation
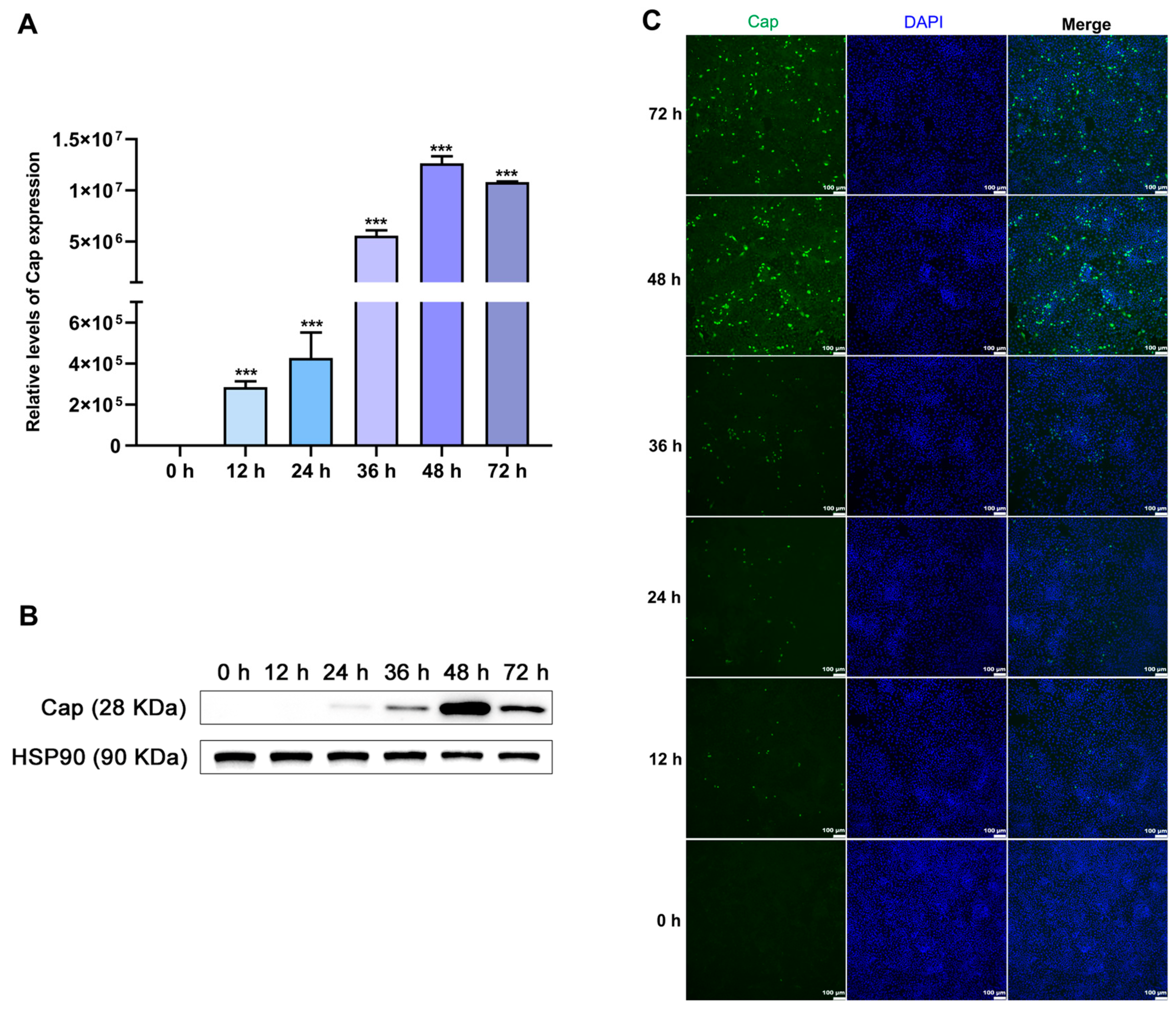
3.2. Establishment of a PCV2-Infected PK15 Cell Model
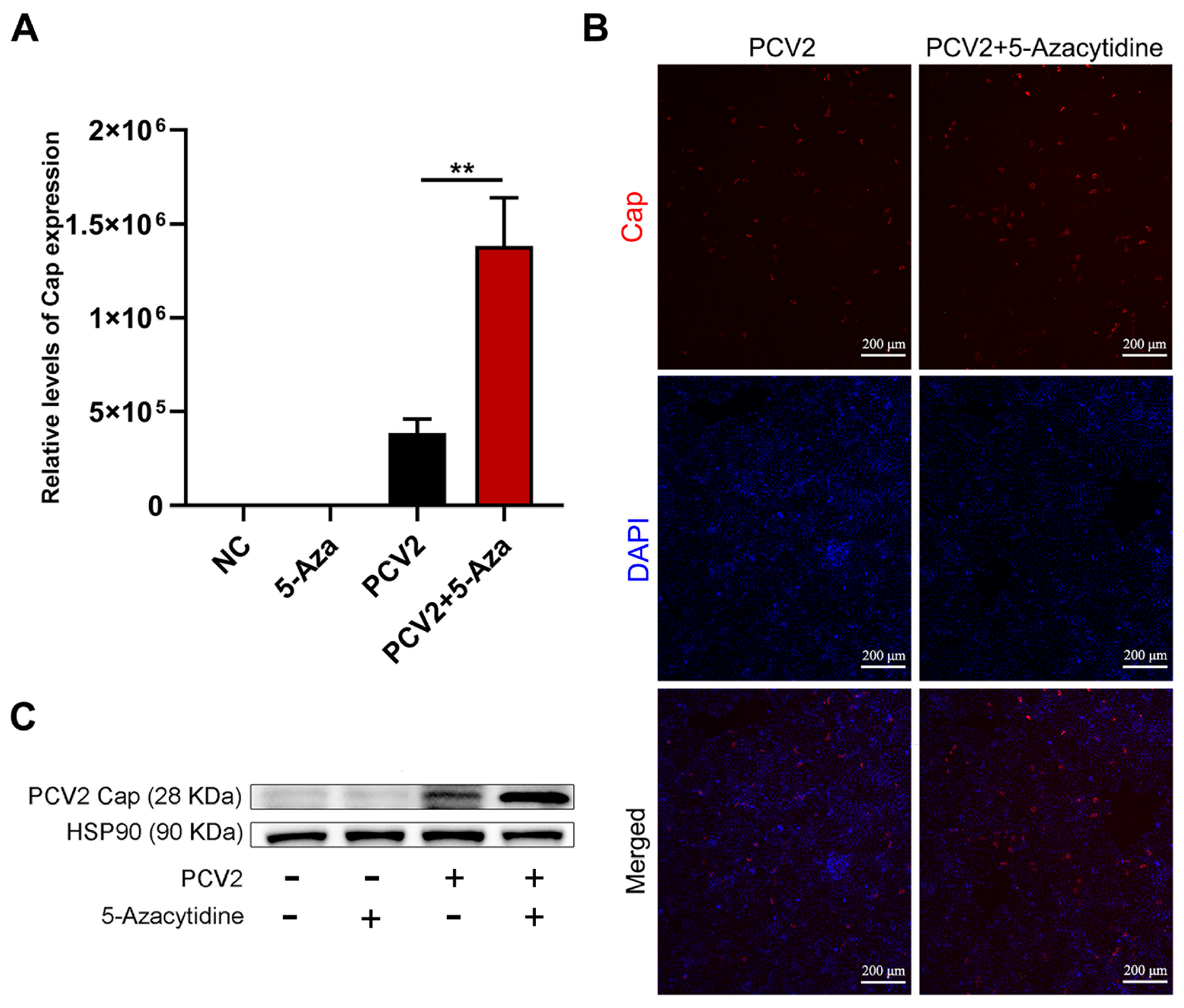
3.3. Effects of 5-Aza on the Replication Level of PCV2
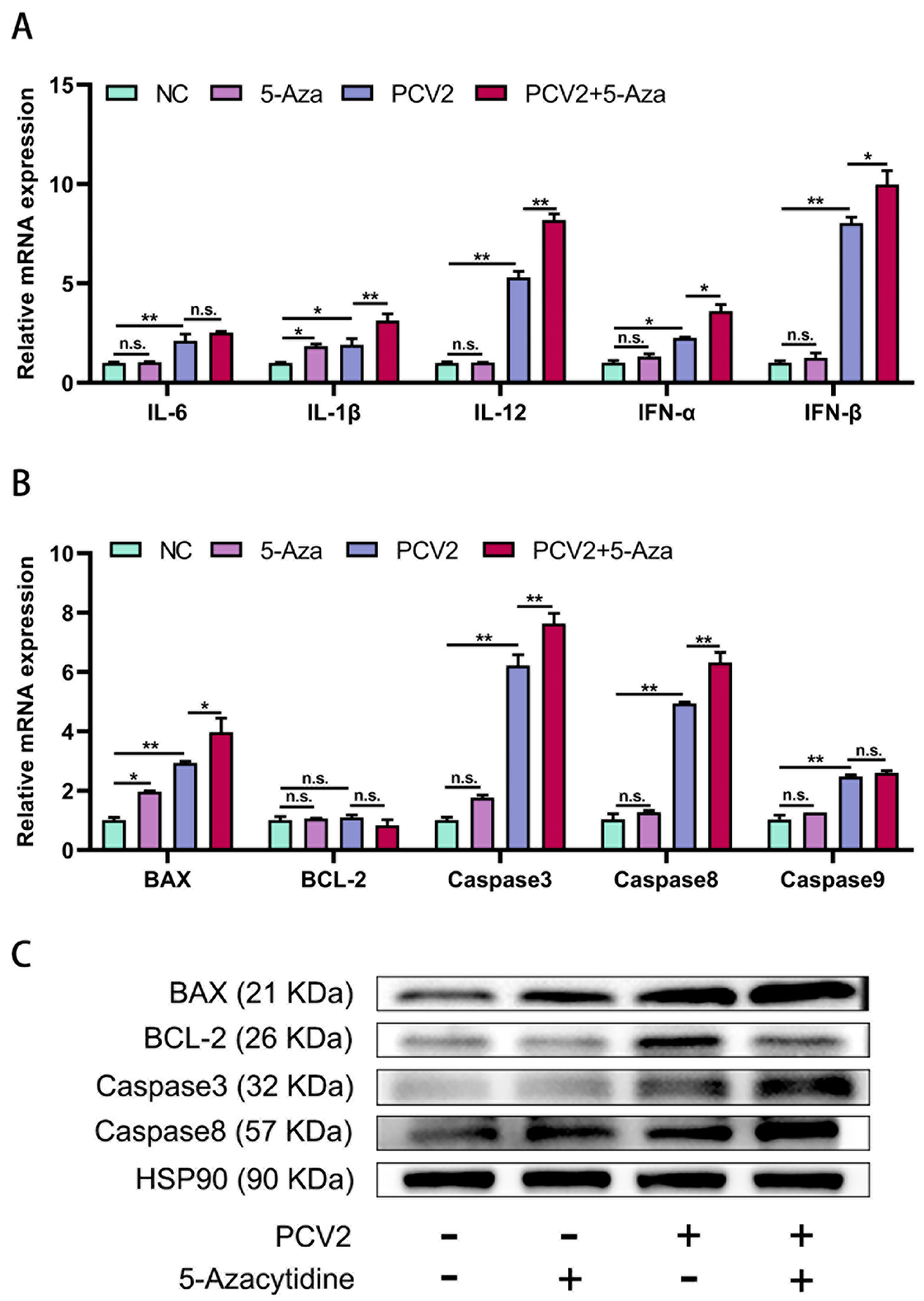
3.4. Effects of 5-Aza on Apoptosis and Cytokine Levels in PCV2 Infection
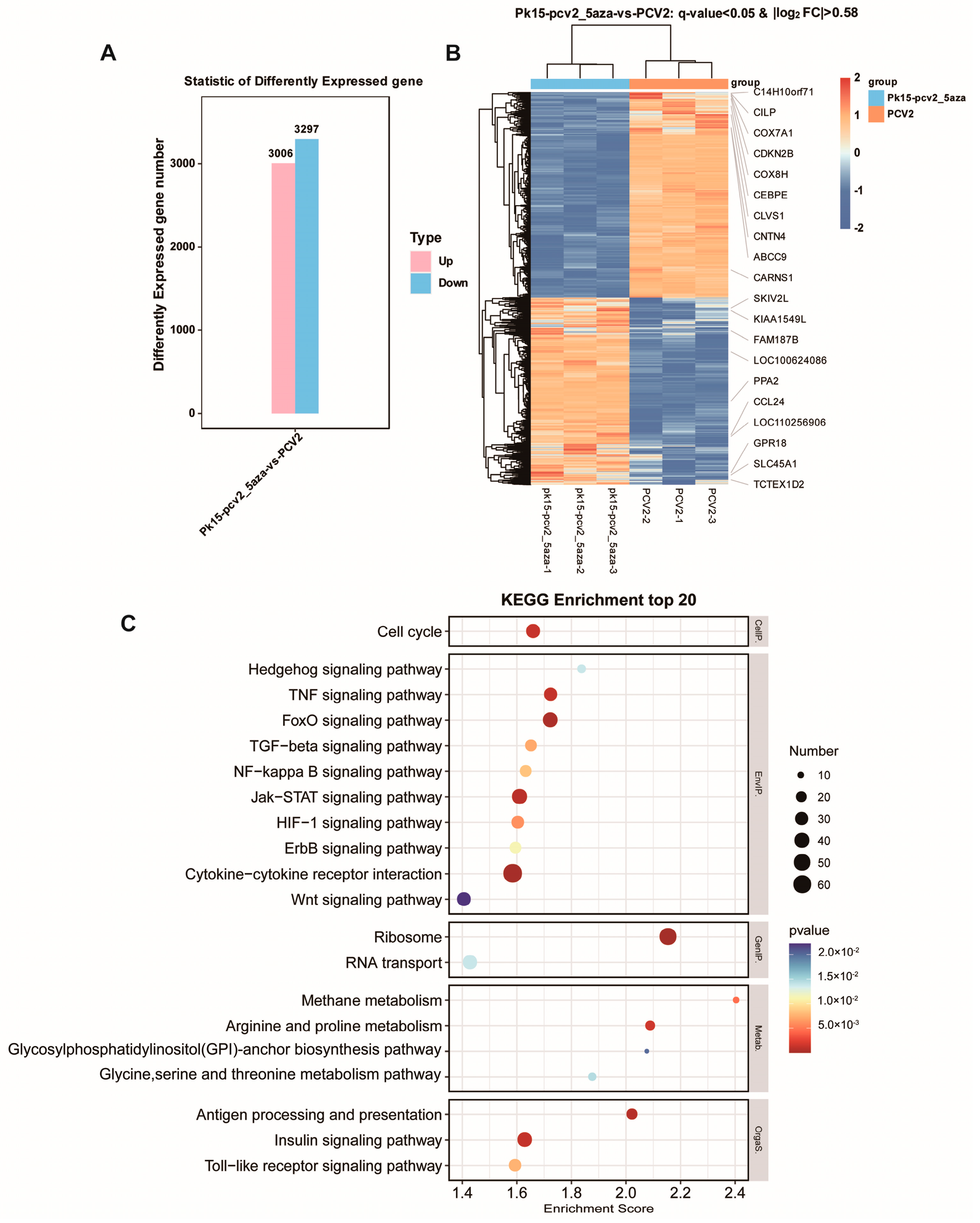
3.5. Transcriptome Sequencing Analysis before and after 5-Aza Treatment and PCV2 Infection
3.5.1. RNA Sequencing of Relevant Genes and Signal Pathways following PCV2 Infection
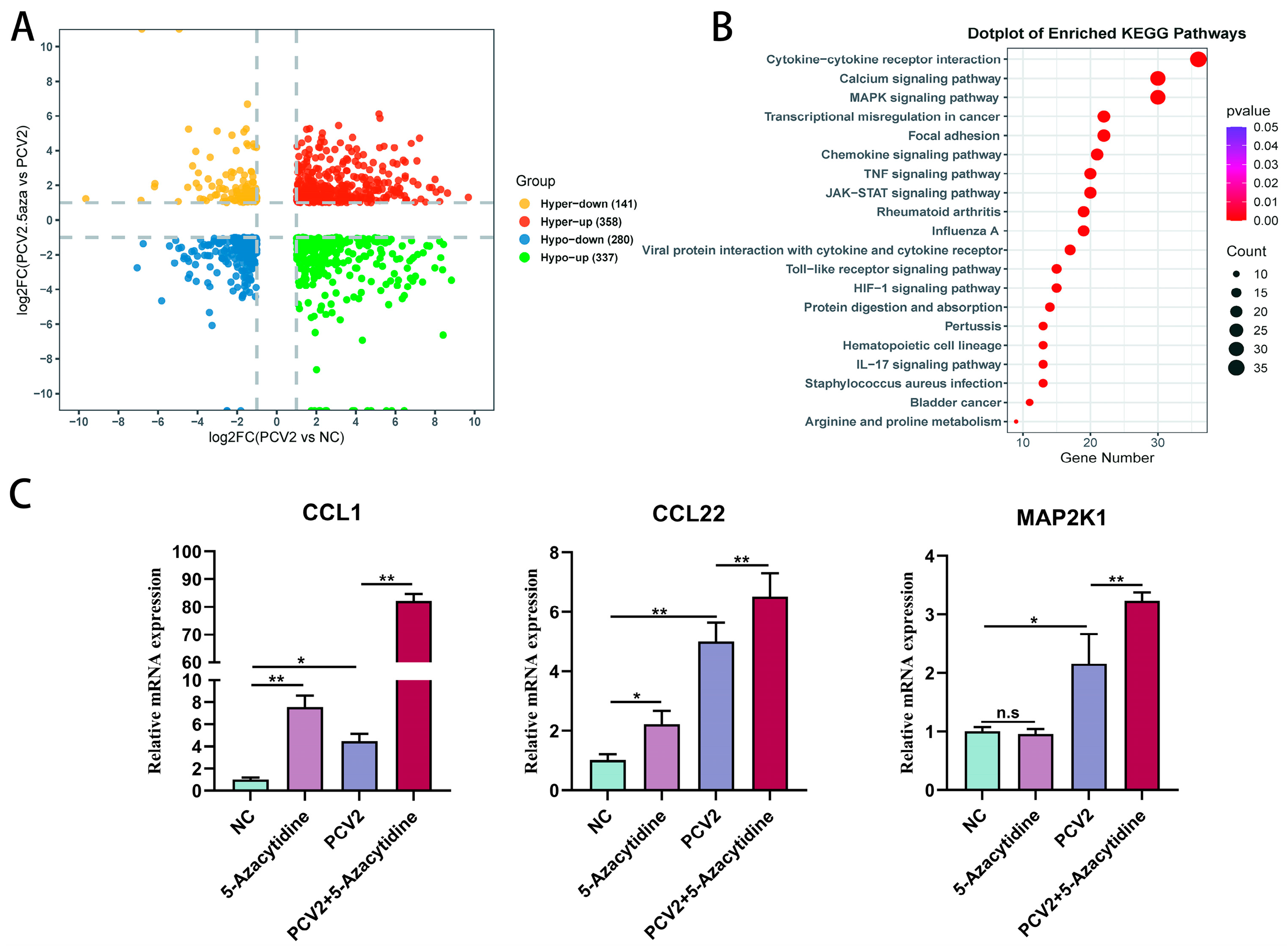
3.5.2. Comparative RNA Sequencing Analysis of PK15 Cells Infected with PCV2 and Cells Treated with PCV2 + 5-Aza
3.5.3. The Addition of 5-Aza Intensifies the Activation of Inflammatory Cytokines and the MAPK Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Finsterbusch, T.; Mankertz, A. Porcine circoviruses—Small but powerful. Virus Res. 2009, 143, 177–183. [Google Scholar] [CrossRef]
- Baekbo, P.; Kristensen, C.S.; Larsen, L.E. Porcine circovirus diseases: A review of PMWS. Transbound. Emerg. Dis. 2012, 59, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81 Pt 9, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhan, Y.; Wang, A.; Zhang, L.; Khayat, R.; Yang, Y. In silico analysis of surface structure variation of PCV2 capsid resulting from loop mutations of its capsid protein (Cap). J. Gen. Virol. 2016, 97, 3331–3344. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Lu, H.; Zhao, K.; Liu, W.; Gao, W.; Lan, Y.; Zhao, J.; Tang, B.; Song, D.; He, W.; et al. Identification and genetic characterization of porcine hemagglutinating encephalomyelitis virus from domestic piglets in China. Arch. Virol. 2014, 159, 2329–2337. [Google Scholar] [CrossRef] [PubMed]
- Nayar, G.P.; Hamel, A.; Lin, L. Detection and characterization of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Can. Vet. J. 1997, 38, 385–386. [Google Scholar] [PubMed]
- Allan, G.M.; McNeilly, F.; Kennedy, S.; Daft, B.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Meehan, B.M.; Adair, B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 1998, 10, 3–10. [Google Scholar] [CrossRef]
- Niagro, F.D.; Forsthoefel, A.N.; Lawther, R.P.; Kamalanathan, L.; Ritchie, B.W.; Latimer, K.S.; Lukert, P.D. Beak and feather disease virus and porcine circovirus genomes: Intermediates between the geminiviruses and plant circoviruses. Arch. Virol. 1998, 143, 1723–1744. [Google Scholar] [CrossRef]
- Khayat, R.; Brunn, N.; Speir, J.A.; Hardham, J.M.; Ankenbauer, R.G.; Schneemann, A.; Johnson, J.E. The 2.3-angstrom structure of porcine circovirus 2. J. Virol. 2011, 85, 7856–7862. [Google Scholar] [CrossRef]
- Meng, X.J. Porcine circovirus type 2 (PCV2): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2013, 1, 43–64. [Google Scholar] [CrossRef]
- Grau-Roma, L.; Segalés, J. Detection of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, swine influenza virus and Aujeszky’s disease virus in cases of porcine proliferative and necrotizing pneumonia (PNP) in Spain. Vet. Microbiol. 2007, 119, 144–151. [Google Scholar] [CrossRef]
- Cheng, C.C.; Lee, Y.F.; Lin, N.N.; Wu, C.L.; Tung, K.C.; Chiu, Y.T. Bronchiolitis obliterans organizing pneumonia in Swine associated with porcine circovirus type 2 infection. J. Biomed. Biotechnol. 2011, 2011, 245728. [Google Scholar] [CrossRef]
- Zhai, S.L.; Chen, S.N.; Xu, Z.H.; Tang, M.H.; Wang, F.G.; Li, X.J.; Sun, B.B.; Deng, S.F.; Hu, J.; Lv, D.H.; et al. Porcine circovirus type 2 in China: An update on and insights to its prevalence and control. Virol. J. 2014, 14, 11–88. [Google Scholar] [CrossRef] [PubMed]
- Krakowka, S.; Allan, G.; Ellis, J.; Hamberg, A.; Charreyre, C.; Kaufmann, E.; Brooks, C.; Meehan, B. A nine-base nucleotide sequence in the porcine circovirus type 2 (PCV2) nucleocapsid gene determines viral replication and virulence. Virus Res. 2012, 164, 90–99. [Google Scholar] [CrossRef]
- Eclercy, J.; Larcher, T.; Andraud, M.; Renson, P.; Bernard, C.; Bigault, L.; Ledevin, M.; Paboeuf, F.; Grasland, B.; Rose, N.; et al. PCV2 co-infection does not impact PRRSV MLV1 safety but enhances virulence of a PRRSV MLV1-like strain in infected SPF pigs. Vet. Microbiol. 2020, 244, 108656. [Google Scholar] [CrossRef]
- Yang, B.C.; Lee, M.S.; Lin, M.K.; Chang, W.T. 5-Azacytidine increases tanshinone production in Salvia miltiorrhiza hairy roots through epigenetic modulation. Sci. Rep. 2022, 12, 9349. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Asada, H.; Tamura, I.; Lee, L.; Maekawa, R.; Taniguchi, K.; Taketani, T.; Matsuoka, A.; Tamura, H.; Sugino, N. DNA methyltransferase expression in the human endometrium: Down-regulation by progesterone and estrogen. Hum. Reprod. 2009, 24, 1126–1132. [Google Scholar] [CrossRef]
- Woellmer, A.; Hammerschmidt, W. Epstein-Barr virus and host cell methylation: Regulation of latency, replication and virus reactivation. Curr. Opin. Virol. 2013, 3, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandan, P.; Darius, H.; Kannangai, R.; Martinez-Murillo, F.; Torbenson, M. Hepatitis B Virus Replication Induces Methylation of both Host and Viral DNA. J. Virol. 2010, 84, 4321–4329. [Google Scholar] [CrossRef]
- Vinokurova, S.; von Knebel Doeberitz, M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE 2011, 6, e24451. [Google Scholar] [CrossRef]
- Barletta, J.; Greer, S.B. Methylation of HSV-1 DNA as a mechanism of viral inhibition: Studies of an analogue of methyldeoxycytidine: Trifluoromethyldeoxycytidine (F3mdCyd). Antivir. Res. 1992, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Sutter, D.; Doerfler, W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl. Acad. Sci. USA 1980, 77, 253–256. [Google Scholar] [CrossRef]
- Cheung, A.K. Porcine circovirus: Transcription and DNA replication. Virus Res. 2012, 164, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Grzybkowska, D.; Morończyk, J.; Wójcikowska, B.; Gaj, M.D. Azacitidine (5-AzaC)-treatment and mutations in DNA methylase genes affect embryogenic response and expression of the genes that are involved in somatic embryogenesis in Arabidopsis. Plant Growth Regul. 2018, 85, 243–256. [Google Scholar] [CrossRef]
- Issa, J.P.; Kantarjian, H.M. Targeting DNA methylation. Clin. Cancer Res. 2009, 15, 3938–3946. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine circovirus 2 genotypes, immunity and vaccines: Multiple genotypes but one single serotype. Pathogens 2020, 9, 1049. [Google Scholar] [CrossRef]
- Kim, S.C.; Nazki, S.; Kwon, S.; Juhng, J.H.; Mun, K.H.; Jeon, D.Y.; Jeong, C.G.; Khatun, A.; Kang, S.J.; Kim, W.I. The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea. BMC Vet. Res. 2018, 26, 294. [Google Scholar] [CrossRef]
- Yao, J.; Qin, Y.; Zeng, Y.; Ouyang, K.; Chen, Y.; Huang, W.; Wei, Z. Genetic analysis of porcine circovirus type 2 (PCV2) strains between 2002 and 2016 reveals PCV2 mutant predominating in porcine population in Guangxi, China. BMC Vet. Res. 2019, 25, 118. [Google Scholar] [CrossRef]
- Hoelzer, K.; Shackelton, L.A.; Parrish, C.R. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res. 2008, 36, 2825–2837. [Google Scholar] [CrossRef]
- Liang, M.; Pan, W.; You, Y.; Qin, X.; Su, H.; Zhan, Z.; Weng, S.; Guo, C.; He, J. Hypermethylated genome of a fish vertebrate iridovirus ISKNV plays important roles in viral infection. Commun. Biol. 2024, 28, 237. [Google Scholar] [CrossRef]
- Cheng, S.; Zhang, M.; Li, W.; Wang, Y.; Liu, Y.; He, Q. Proteomic analysis of porcine alveolar macrophages infected with porcine circovirus type 2. J. Proteom. 2012, 75, 3258–3269. [Google Scholar] [CrossRef]
- Fan, H.; Ye, Y.; Luo, Y.; Tong, T.; Yan, G.; Liao, M. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals protein and pathway regulation in porcine circovirus type 2 infected PK-15 cells. J. Proteome Res. 2012, 11, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, S.; Wang, Y.; Deng, F.; Yan, W.; Yang, K.; Chen, H.; He, Q.; Charreyre, C.; Audoneet, J.C. Transcription analysis of the porcine alveolar macrophage response to porcine circovirus type 2. BMC Genom. 2013, 27, 14–353. [Google Scholar] [CrossRef][Green Version]
- Liu, J.; Bai, J.; Lu, Q.; Zhang, L.; Jiang, Z.; Michal, J.J.; He, Q.; Jiang, P. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine circovirus type 2. J. Proteom. 2013, 79, 72–86. [Google Scholar] [CrossRef]
- Liu, J.; Bai, J.; Zhang, L.; Hou, C.; Li, Y.; Jiang, P. Proteomic alteration of PK-15 cells after infection by porcine circovirus type 2. Virus Genes 2014, 49, 400–416. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, J.; Wu, Y.; Zheng, X.; Ma, G.; Wang, Z.; Jin, Y.; He, J.; Yan, Y. Differential proteome analysis of host cells infected with porcine circovirus type 2. J. Proteome Res. 2009, 8, 5111–5119. [Google Scholar] [CrossRef]
- Mavrommatis, B.; Offord, V.; Patterson, R.; Watson, M.; Kanellos, T.; Steinbach, F.; Grierson, S.; Werling, D. Global gene expression profiling of myeloid immune cell subsets in response to in vitro challenge with porcine circovirus 2b. PLoS ONE 2014, 9, e91081. [Google Scholar] [CrossRef][Green Version]
- Lee, G.; Han, D.; Song, J.Y.; Lee, Y.S.; Kang, K.S.; Yoon, S. Genomic expression profiling in lymph nodes with lymphoid depletion from porcine circovirus 2-infected pigs. J. Gen. Virol. 2010, 91, 2585–2591. [Google Scholar] [CrossRef]
- Klein, A.M.; Zaganjor, E.; Cobb, M.H. Chromatin-tethered MAPKs. Curr. Opin. Cell Biol. 2013, 25, 272–277. [Google Scholar] [CrossRef]
- Kujime, K.; Hashimoto, S.; Gon, Y.; Shimizu, K.; Horie, T. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 2000, 164, 3222–3228. [Google Scholar] [CrossRef]
- Wei, L.; Zhu, S.; Wang, J.; Zhang, C.; Quan, R.; Yan, X.; Liu, J. Regulatory role of ASK1 in porcine circovirus type 2-induced apoptosis. Virology 2013, 447, 285–291. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, L.; Kwang, J.; Wang, J.; Shi, L.; Yang, B.; Li, Y.; Liu, J. Porcine circovirus type 2 induces the activation of nuclear factor κB by IκBα degradation. Virology 2008, 378, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhu, Z.; Wang, J.; Liu, J. JNK and p38 mitogen-activated protein kinase pathways contribute to porcine circovirus type 2 infection. J. Virol. 2009, 83, 6039–6047. [Google Scholar] [CrossRef][Green Version]
- Gu, C.; Gao, X.; Guo, D.; Wang, J.; Wu, Q.; Nepovimova, E.; Wu, W.; Kuca, K. Combined Effect of Deoxynivalenol (DON) and Porcine Circovirus Type 2 (Pcv2) on Inflammatory Cytokine mRNA Expression. Toxins 2021, 13, 422. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, X.; Shi, T.; Luo, L.; Qiao, D.; Wang, Z.; Han, C.; Du, Q.; Tong, D.; Huang, Y. Porcine Circovirus Type 2 Rep Enhances IL-10 Production in Macrophages via Activation of p38-MAPK Pathway. Viruses 2019, 11, 1141. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, Y.; Xiao, Q.; He, K.; Wu, S.; Bao, W.; Wu, Z. Identification of the Effects of 5-Azacytidine on Porcine Circovirus Type 2 Replication in Porcine Kidney Cells. Vet. Sci. 2024, 11, 135. https://doi.org/10.3390/vetsci11030135
Shan Y, Xiao Q, He K, Wu S, Bao W, Wu Z. Identification of the Effects of 5-Azacytidine on Porcine Circovirus Type 2 Replication in Porcine Kidney Cells. Veterinary Sciences. 2024; 11(3):135. https://doi.org/10.3390/vetsci11030135
Chicago/Turabian StyleShan, Yiyi, Qi Xiao, Kongwang He, Shenglong Wu, Wenbin Bao, and Zhengchang Wu. 2024. "Identification of the Effects of 5-Azacytidine on Porcine Circovirus Type 2 Replication in Porcine Kidney Cells" Veterinary Sciences 11, no. 3: 135. https://doi.org/10.3390/vetsci11030135
APA StyleShan, Y., Xiao, Q., He, K., Wu, S., Bao, W., & Wu, Z. (2024). Identification of the Effects of 5-Azacytidine on Porcine Circovirus Type 2 Replication in Porcine Kidney Cells. Veterinary Sciences, 11(3), 135. https://doi.org/10.3390/vetsci11030135








