Characterization of a Novel Infectious Pancreatic Necrosis Virus (IPNV) from Genogroup 6 Identified in Sea Trout (Salmo trutta) from Lake Vänern, Sweden
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Sequencing of the IPNV Isolate
2.1.1. Virus isolation from a Broodstock Farm
2.1.2. RNA Extraction and Preparation of cDNA
2.1.3. Illumina MiSeq Library Preparation and Sequencing
2.1.4. Bioinformatics
2.2. Experimental Trials
2.2.1. Fish
2.2.2. Pre-trial with Rainbow Trout Exposed to Different IPNV Isolates
2.2.3. Infection Trial—Regimens and Doses
2.2.4. Mortality, Sampling, and Virus Detection
2.3. Prevalence Studies
3. Results
3.1. Virus Isolation during the Initial IPNV Outbreak
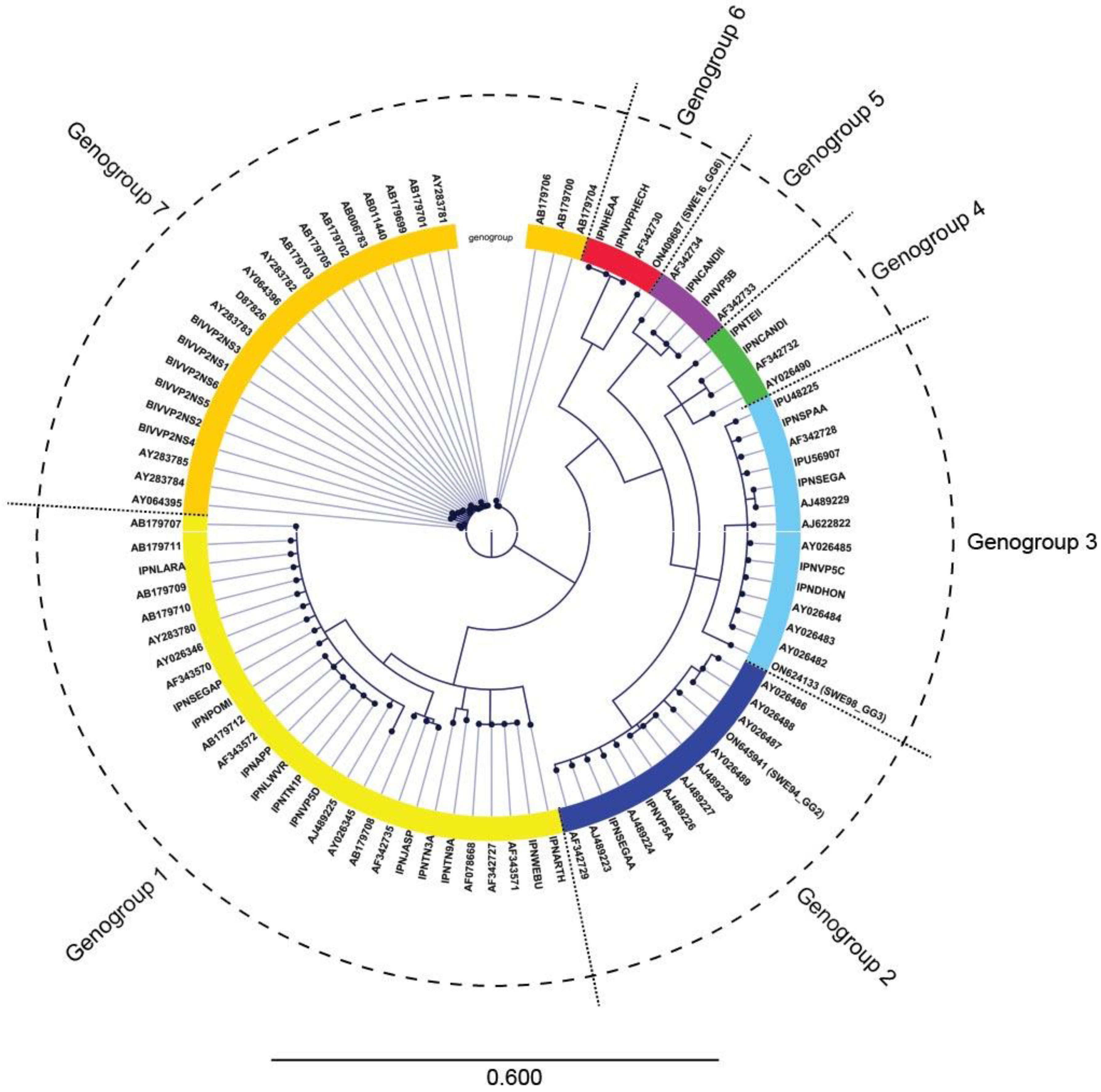
3.2. IPNV Whole Genome Sequencing
3.3. Experimental Infection by IPNV
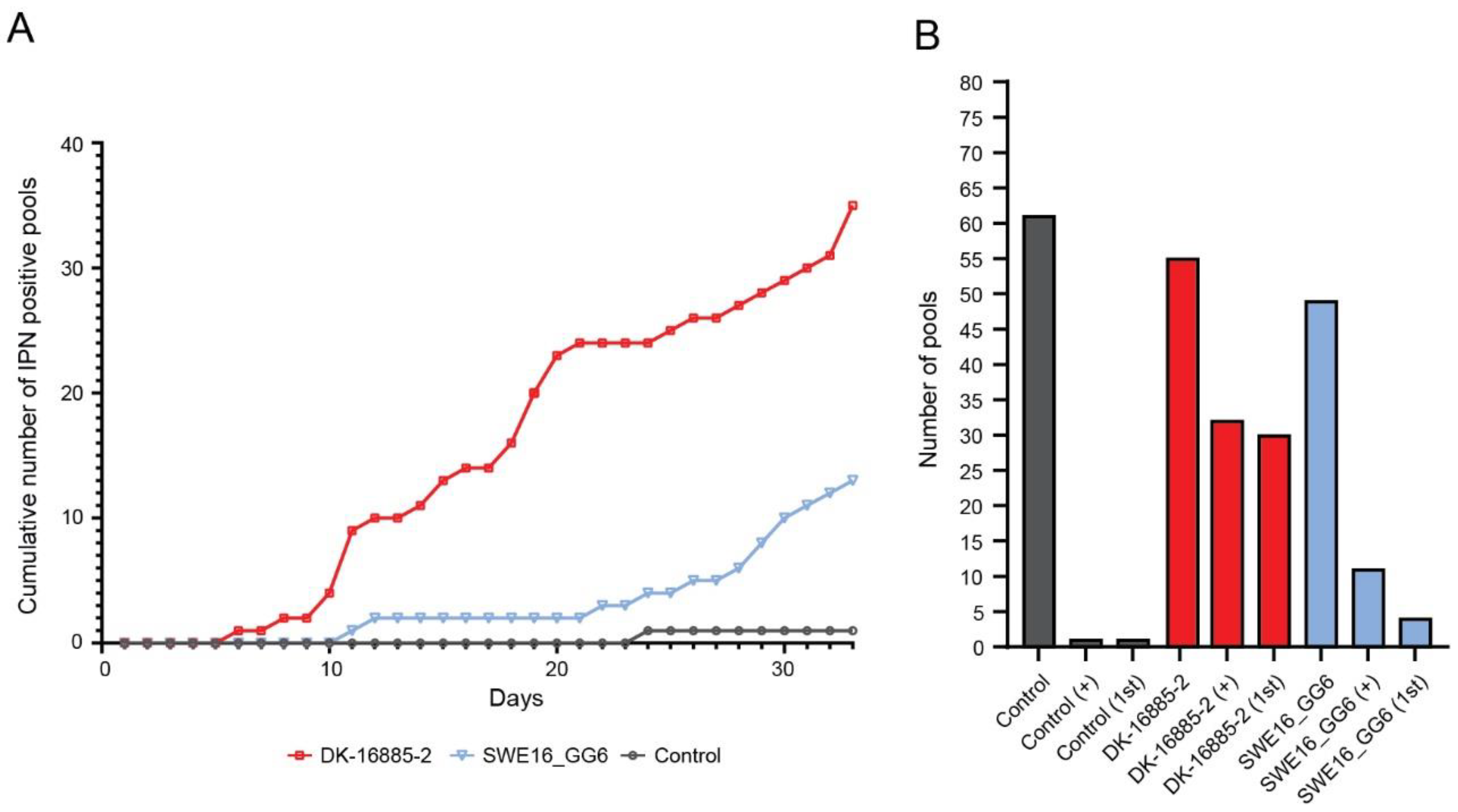
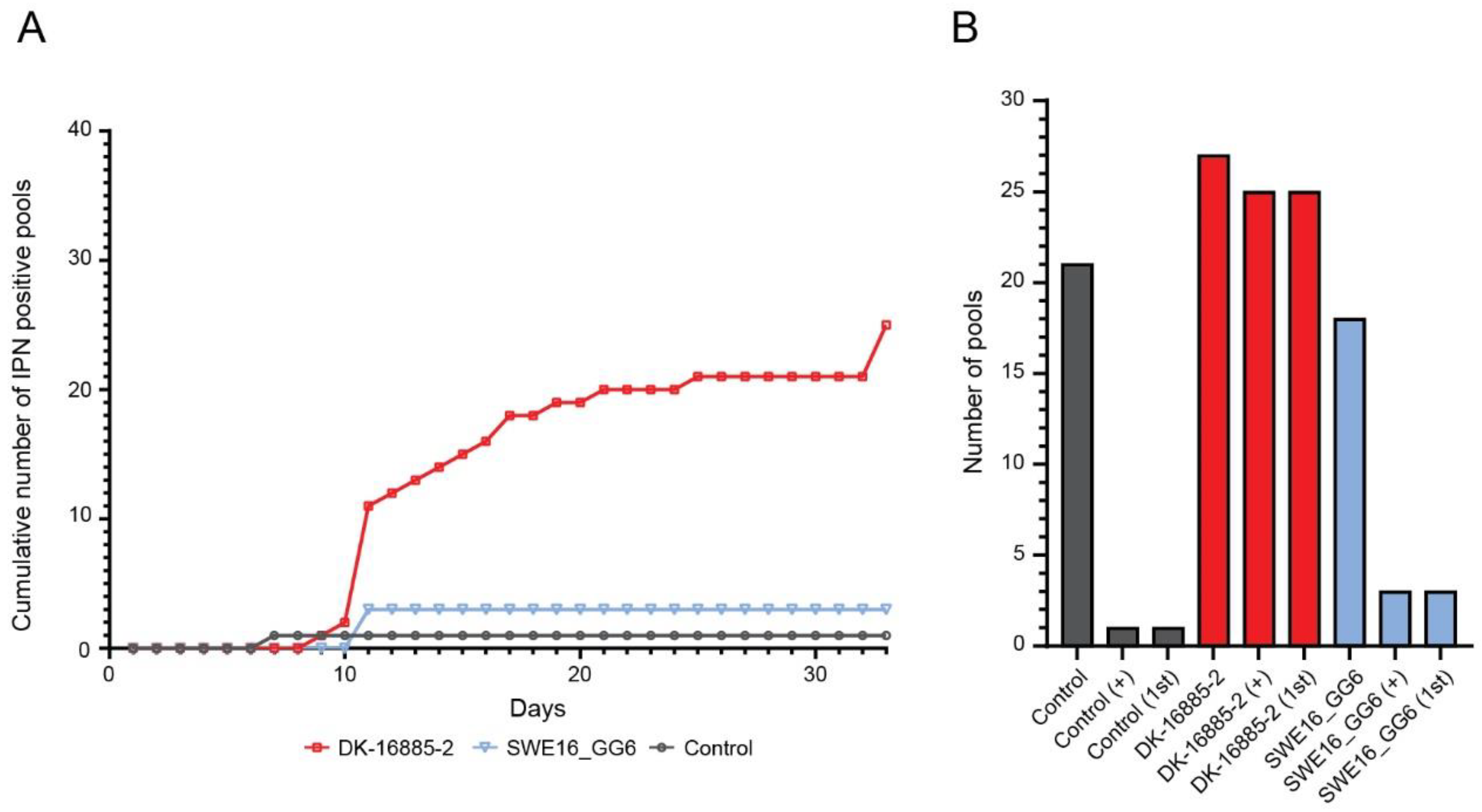
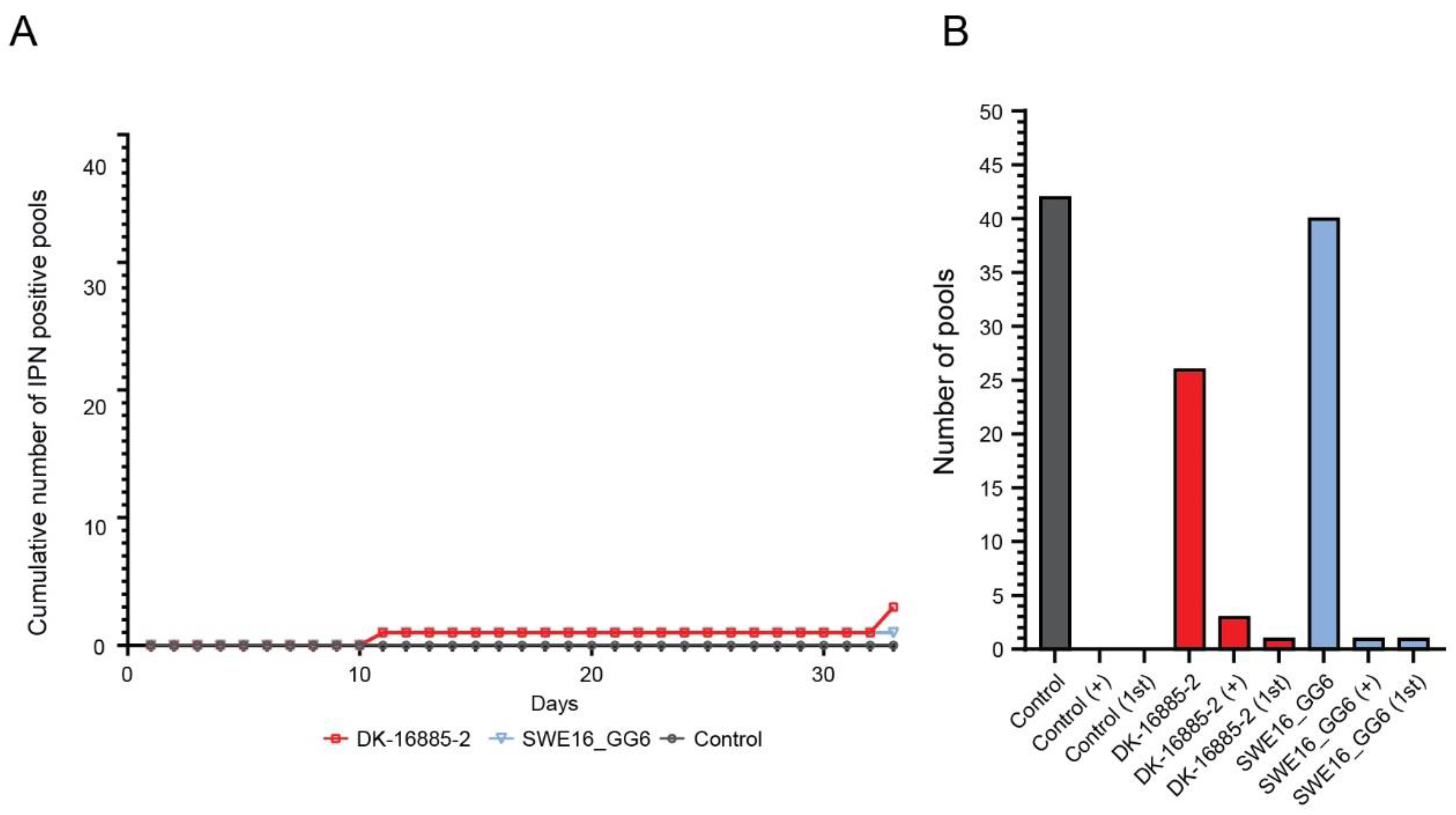
3.3.1. Sea Trout
3.3.2. Rainbow Trout
3.3.3. Atlantic Salmon
3.4. IPNV Prevalence in the Wild
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evensen, Ø.; Santi, N. Infectious Pancreatic Necrosis Virus. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 83–89. [Google Scholar] [CrossRef]
- Delmas, B. Fish viruses. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Elsevier: Oxford, UK, 2008; pp. 321–328. [Google Scholar]
- Dopazo, C.P. The Infectious Pancreatic Necrosis Virus (IPNV) and its Virulence Determinants: What is Known and What Should be Known. Pathogens 2020, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Eriksson-Kallio, A.M.; Holopainen, R.; Koski, P.; Nousiainen, A.; Koskinen, H.; Kause, A.; Gadd, T. Susceptibility of rainbow trout to three different genogroups of infectious pancreatic necrosis virus. Dis. Aquat. Org. 2020, 141, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Ren, G.; Zhao, J.; Lu, T.; Liu, Q.; Xu, L. Dynamic Distribution of Infectious Pancreatic Necrosis Virus (IPNV) Strains of Genogroups 1, 5, and 7 after Intraperitoneal Administration in Rainbow Trout (Oncorhynchus mykiss). Viruses 2022, 14, 2634. [Google Scholar] [CrossRef] [PubMed]
- Santi, N.; Vakharia, V.N.; Evensen, Ø. Identification of putative motifs involved in the virulence of infectious pancreatic necrosis virus. Virology 2004, 322, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Santi, N.; Evensen, O.; Vakharia, V.N. Molecular determinants of infectious pancreatic necrosis virus virulence and cell culture adaptation. J. Virol. 2005, 79, 10289–10299. [Google Scholar] [CrossRef]
- Shivappa, R.B.; Song, H.; Yao, K.; Aas-Eng, A.; Evensen, O.; Vakharia, V.N. Molecular characterization of Sp serotype strains of infectious pancreatic necrosis virus exhibiting differences in virulence. Dis. Aquat. Org. 2004, 61, 23–32. [Google Scholar] [CrossRef]
- Mutoloki, S.; Jossund, T.B.; Ritchie, G.; Munang’andu, H.M.; Evensen, O. Infectious Pancreatic Necrosis Virus Causing Clinical and Subclinical Infections in Atlantic Salmon Have Different Genetic Fingerprints. Front. Microbiol. 2016, 7, 1393. [Google Scholar] [CrossRef]
- Jensen, I.; Overrein, M.C.; Fredriksen, B.N.; Strandskog, G.; Seternes, T. Differences in smolt status affect the resistance of Atlantic salmon (Salmo salar L.) against infectious pancreatic necrosis, while vaccine-mediated protection is unaffected. J. Fish Dis. 2019, 42, 1271–1282. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, X.; Wang, K.; Yang, Q.; He, J.; Qin, Z.; Geng, Y.; Ouyang, P.; Huang, X. Outbreak of infectious pancreatic necrosis virus (IPNV) in farmed rainbow trout in China. Acta Trop. 2017, 170, 63–69. [Google Scholar] [CrossRef]
- Kristoffersen, A.B.; Devold, M.; Aspehaug, V.; Gjelstenli, O.; Breck, O.; Bang Jensen, B. Molecular tracing confirms that infection with infectious pancreatic necrosis virus follows the smolt from hatchery to grow-out farm. J. Fish Dis. 2018, 41, 1601–1607. [Google Scholar] [CrossRef]
- Julin, K.; Johansen, L.H.; Sommer, A.I.; Jorgensen, J.B. Persistent infections with infectious pancreatic necrosis virus (IPNV) of different virulence in Atlantic salmon, Salmo salar L. J. Fish Dis. 2015, 38, 1005–1019. [Google Scholar] [CrossRef]
- Ahne, W. Isolation and characterization of infectious pancreatic necrosis virus from pike (Esox lucius). Arch. Virol. 1978, 58, 65–69. [Google Scholar] [CrossRef]
- Hill, B.J.; Way, K. Serological classification of infectious pancreatic necrosis (IPN) virus and other aquatic birnaviruses. Annu. Rev. Fish Dis. 1995, 5, 55–77. [Google Scholar] [CrossRef]
- Blomstrom, A.L.; Widen, F.; Hammer, A.S.; Belak, S.; Berg, M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010, 48, 4392–4396. [Google Scholar] [CrossRef]
- Allander, T.; Tammi, M.T.; Eriksson, M.; Bjerkner, A.; Tiveljung-Lindell, A.; Andersson, B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 2005, 102, 12891–12896. [Google Scholar] [CrossRef]
- Axen, C.; Hakhverdyan, M.; Boutrup, T.S.; Blomkvist, E.; Ljunghager, F.; Alfjorden, A.; Hagstrom, A.; Olesen, N.J.; Juremalm, M.; Leijon, M.; et al. Emergence of a new rhabdovirus associated with mass mortalities in eelpout (Zoarces viviparous) in the Baltic Sea. J. Fish Dis. 2017, 40, 219–229. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Nishizawa, T.; Kinoshita, S.; Yoshimizu, M. An approach for genogrouping of Japanese isolates of aquabirnaviruses in a new genogroup, VII, based on the VP2/NS junction region. J. Gen. Virol. 2005, 86 Pt 7, 1973–1978. [Google Scholar] [CrossRef]
- Simon, R.C.; Schill, W.B. Tables of sample size requirements for detection of fish infected by pathogens: Three confidence levels for different infection prevalence and various population sizes. J. Fish Dis. 1984, 7, 515–520. [Google Scholar] [CrossRef]
- Coulibaly, F.; Chevalier, C.; Delmas, B.; Rey, F.A. Crystal structure of an Aquabirnavirus particle: Insights into antigenic diversity and virulence determinism. J. Virol. 2010, 84, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
| Group | Virus Strain | Infection Route | Number of fish | Infection Dose (TCID50/mL) * | Infection Dose/Fish | ||
|---|---|---|---|---|---|---|---|
| Injected | Cohabitants | Immersion | |||||
| NC | Negative control | i.p + cohab | 29 | 30 | - | 0 | 0 |
| StR | 16619-3 | i.p + cohab | 30 | 30 | - | 5.9 × 104 | 1.48 × 103 |
| C | 15512-2 | i.p + cohab | 30 | 30 | - | 8.6 × 105 | 2.15 × 104 |
| H | 17692-1 | i.p + cohab | 30 | 30 | - | 1.9 × 105 | 4.75 × 103 |
| R-inj | 16885-2 | i.p + cohab | 30 | 30 | - | 1.3 × 108 | 3.25 × 105 |
| R-imm | 16885-2 | immersion | - | - | 60 | 2.6 × 105 | |
| Tank 4 | Tank 21 | Tank 5 | Tank 20 | Tank 6 | Tank 19 | Total | |
|---|---|---|---|---|---|---|---|
| Treatment | Control | Control | DK-16885-2 | DK-16885-2 | SWE16_GG6 | SWE16_GG6 | |
| Total number | 80 | 73 | 75 | 89 | 49 | 88 | 454 |
| Survivors | 56 | 20 | 41 | 2 | 20 | 25 | 164 |
| Moribund | 24 | 53 | 34 | 87 | 29 | 63 | 290 |
| Tank 10 | Tank 15 | Tank 11 | Tank 14 | Tank 12 | Tank 13 | Total | |
|---|---|---|---|---|---|---|---|
| Treatment | Control | Control | DK-16885-2 | DK-16885-2 | SWE16_GG6 | SWE16_GG6 | |
| Total number | 85 | 104 | 98 | 99 | 100 | 97 | 583 |
| Survivors | 84 | 103 | 93 | 90 | 98 | 96 | 564 |
| Moribund | 1 | 1 | 5 | 9 | 2 | 1 | 19 |
| Tank 7 | Tank 18 | Tank 8 | Tank 17 | Tank 9 | Tank 16 | Total | |
|---|---|---|---|---|---|---|---|
| Treatment | Control | Control | DK-16885-2 | DK-16885-2 | SWE16_GG6 | SWE16_GG6 | |
| Total number | 82 | 93 | 94 | 75 | 84 | 67 | 495 |
| Survivors | 75 | 43 | 83 | 67 | 29 | 53 | 350 |
| Moribund | 7 | 50 | 11 | 8 | 55 | 14 | 145 |
| Species | Population | Upstream 1 | Lake Vänern 2 | Total |
|---|---|---|---|---|
| Atlantic salmon | Klarälven | 131 | 98 | 229 |
| Atlantic salmon | Gullspång | 153 | 46 | 199 |
| Atlantic salmon | Unknown | 0 | 124 | 124 |
| Sea trout | Klarälven | 155 | 26 | 181 |
| Sea trout | Gullspång | 152 | 14 | 166 |
| Sea trout | Unknown | 0 | 73 | 73 |
| Mixed | Unknown | 0 | 7 | 7 |
| Total | 591 | 388 | 979 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Persson, B.D.; Schmidt, J.G.; Hakhverdyan, M.; Leijon, M.; Olesen, N.J.; Axén, C. Characterization of a Novel Infectious Pancreatic Necrosis Virus (IPNV) from Genogroup 6 Identified in Sea Trout (Salmo trutta) from Lake Vänern, Sweden. Vet. Sci. 2023, 10, 58. https://doi.org/10.3390/vetsci10010058
Persson BD, Schmidt JG, Hakhverdyan M, Leijon M, Olesen NJ, Axén C. Characterization of a Novel Infectious Pancreatic Necrosis Virus (IPNV) from Genogroup 6 Identified in Sea Trout (Salmo trutta) from Lake Vänern, Sweden. Veterinary Sciences. 2023; 10(1):58. https://doi.org/10.3390/vetsci10010058
Chicago/Turabian StylePersson, B. David, Jacob Günther Schmidt, Mikhayil Hakhverdyan, Mikael Leijon, Niels Jørgen Olesen, and Charlotte Axén. 2023. "Characterization of a Novel Infectious Pancreatic Necrosis Virus (IPNV) from Genogroup 6 Identified in Sea Trout (Salmo trutta) from Lake Vänern, Sweden" Veterinary Sciences 10, no. 1: 58. https://doi.org/10.3390/vetsci10010058
APA StylePersson, B. D., Schmidt, J. G., Hakhverdyan, M., Leijon, M., Olesen, N. J., & Axén, C. (2023). Characterization of a Novel Infectious Pancreatic Necrosis Virus (IPNV) from Genogroup 6 Identified in Sea Trout (Salmo trutta) from Lake Vänern, Sweden. Veterinary Sciences, 10(1), 58. https://doi.org/10.3390/vetsci10010058






