Abstract
High-quality, openly accessible clinical datasets remain a significant bottleneck in advancing both research and clinical applications within medical artificial intelligence. Case reports, often rich in multimodal clinical data, represent an underutilized resource for developing medical AI applications. We present an enhanced version of MultiCaRe, a dataset derived from open-access case reports on PubMed Central. This new version addresses the limitations identified in the previous release and incorporates newly added clinical cases and images (totaling 93,816 and 130,791, respectively), along with a refined hierarchical taxonomy featuring over 140 categories. Image labels have been meticulously curated using a combination of manual and machine learning-based label generation and validation, ensuring a higher quality for image classification tasks and the fine-tuning of multimodal models. To facilitate its use, we also provide a Python package for dataset manipulation, pretrained models for medical image classification, and two dedicated websites. The updated MultiCaRe dataset expands the resources available for multimodal AI research in medicine. Its scale, quality, and accessibility make it a valuable tool for developing medical AI systems, as well as for educational purposes in clinical and computational fields.
Dataset: https://doi.org/10.5281/zenodo.10079369.
Dataset License: CC-BY-NC-SA
1. Summary
Acquiring high-quality, open-access datasets is a persistent challenge in the clinical domain. In medical image classification, datasets are typically limited to a single type of medical image, such as chest x-rays [1,2], gastrointestinal endoscopies [3,4], skin photographs [5,6,7], or histopathological images [8,9,10,11]. Only exceptionally do medical image classification datasets include a broader variety of image types [12]. When it comes to multimodal datasets that include both visual and textual data, they typically feature a broader range of image types [13,14,15,16,17,18,19,20,21,22,23,24]. However, the text provided in these datasets usually focuses on the image content, and only a few exceptions refer to the image context (the actual clinical case) [17,21,23].
This paper introduces a new version of MultiCaRe, a dataset consisting of clinical cases, labeled images, and captions, extracted from 72,581 open-access case reports from PubMed Central (see Figure 1 for an example) [25,26]. This version addresses the limitations and errors identified in the original release, and includes an updated collection of clinical cases (93,816 in total) and images (130,791), and a richer taxonomy (145 categories, with a hierarchical structure). The dataset is optimized for image classification, and can also be used for other tasks, such as fine-tuning large multimodal models, training image captioning or image generation models, and for data annotation (for example, for Natural Language Processing or image segmentation tasks). Differences between this updated version and the initial release are detailed in Appendix A. Citation data for each article used in MultiCaRe is available in the metadata.parquet file. When compared to other datasets sourced from PubMed Central articles, MultiCaRe offers two main advantages: its large number of image classes and the inclusion of clinical cases associated with the images (see Table 1). Apart from the dataset, this release includes: (a) a Python package containing all the code needed to create and use the dataset; (b) medical image classification models; (c) the corresponding taxonomy with documentation; and (d) two websites with clinical cases intended for healthcare professionals and medical students (all links are available in Section 4). The project’s code is available in the GitHub repository [27].
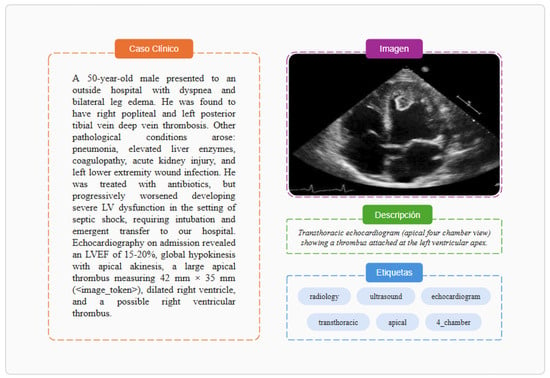
Figure 1.
Example of data elements from the MultiCaRe dataset, adapted from patient PMC7992397_01 [28].

Table 1.
Comparison of datasets derived from PubMed Central data.
Before describing the dataset and its creation, it is important to address some key points regarding case report articles. Case reports are a type of medical publication that describe and discuss the clinical presentation of one or more patients. Compared to other forms of clinical research, this article type has both advantages and limitations: on the one hand, it enables the detection of clinical novelties, the generation of hypotheses, and the study of rare diseases; on the other hand, it does not allow for generalizations or causal inferences [34]. Case reports are a relatively underutilized resource for model training and AI application development, despite being a rich resource of multimodal clinical data. Key differences between case report datasets and data extracted from medical records are detailed in Appendix B.
2. Data Description
This section outlines the contents and structure of the entire dataset, which is available for download on Zenodo [25]. However, for specific use cases (such as training image classification models for CT scans), the recommended approach is to create customizable subsets using the MedicalDatasetCreator class from Python’s multiversity library, rather than accessing the data directly from the repository (see Section 4) [35].
These are the files included in the dataset:
- data_dictionary.csv: This file describes the dataset’s contents, providing definitions for all data fields in each file.
- metadata.parquet: This file contains metadata for case report articles, including authors, year, journal, keywords (extracted from the article’s content), MeSH terms, DOI, PMCID, license, and link.
- abstracts.parquet: This file stores the abstracts of the articles.
- cases.parquet: This file contains the text of the clinical cases, along with the age of the patient (in years), and the gender (Female, Male, Transgender or Unknown).
- case_images.parquet: This file contains metadata from the raw images, such as the file name and the tag present in the EuropePMC API, captions, and text references (substrings from the clinical case text that refer to the images).
- captions_and_labels.csv: This file contains the metadata of all the images of the dataset. It includes two types of columns:
- –
- file information columns: they include details such as the image name, the source image (original name in EuropePMC’s API), license, size, image caption, and substrings from the case text referencing the image.
- –
- label columns:
- ∗
- image_type: multiclass classification column with seven possible classes (‘radiology’, ‘pathology’, ‘chart’, ‘endoscopy’, ‘medical_photograph’, ‘ophthalmic_imaging’ and ‘electrography’).
- ∗
- image_subtype: multiclass classification column with 40 possible classes (including ‘x_ray’, ‘ct’, ‘mri’, ‘ekg’, ‘immunostaining’, etc).
- ∗
- radiology_region: multiclass classification column, including nine possible anatomical classes that are applicable only for ‘radiology’ images (‘thorax’, ‘abdomen’, ‘head’, ‘lower_limb’, etc).
- ∗
- radiology_region_granular: multiclass classification column, including 19 possible anatomical classes that are applicable only for ‘radiology’ images (the same classes as in radiology_region, but ‘upper_limb’ and ‘lower_limb’ are replaced with more detailed classes, such as ‘hip’ and ‘knee’).
- ∗
- radiology_view: multiclass classification column, including 12 possible classes that are only applicable for ‘radiology’ images (e.g., ‘axial’, ‘frontal’, ‘transabdominal’, etc).
- ∗
- ml_labels_for_supervised_classification: multilabel classification column (there can be more than one label per image). This column includes all the labels assigned to images by machine learning (ML) models, including the ones present in the multiclass columns.
- ∗
- gt_labels_for_semisupervised_classification: multilabel classification column (there can be more than one label per image). Labels were primarily assigned based on the image captions, except when they conflicted with the ML-generated labels. In such cases, the caption-based label was removed—unless the ML label had a confidence score above 0.95, in which case it replaced the caption-based label. Compared to the labels from the ml_labels_for_supervised_classification, the ones from gt_labels_for_semisupervised_classification have a higher quality and a wider taxonomy, but they are not complete (for example, not all the CT scans have a ‘ct’ label, because some of their captions do not mention words that are specific to that class). Coincidences between both columns mean that the ML models predicted the same labels as extracted using the caption-based approach. This column is well-suited for semi-supervised learning, as it includes a mix of labeled and unlabeled data. Additionally, it can be used to sample images from specific classes when higher label quality is required—even if the total number of labeled examples is lower than in ml_labels_for_supervised_classification.
- Image files: This dataset contains 130,791 images in .webp format, with an average resolution of 430 × 365 pixels (this average varies depending on the image class). The image files are located in 90 folders (from ‘\PMC1\PMC10\’ to ‘\PMC9\PMC99\’), based on the first digits of the PMCID of the article they were sourced from. The file names encode relevant information about each image. For example, in PMC10000728_fmed-09-985235-g001_A_1_3.webp, ‘PMC10000728’ is the PMCID of the corresponding case report article, ‘fmed-09-985235-g001’ is the name of the file present in EuropePMC’s API, ‘A’ is the reference used in the caption to refer to this part of the original file, and ‘1’ and ‘3’ mean that the file is the first component from a compound image with three components. The names of images that were not split during post-processing end with ‘_undivided_1_1.webp’.
3. Methods
3.1. Dataset Creation
This section provides an explanation of the dataset creation process, which involved multiple steps: downloading data from PubMed, preprocessing texts and images, developing a hierarchical taxonomy for image labels, and assigning labels to images—initially based on their captions, and later refined using machine learning models. The entire process can be replicated by running the cell from Figure 2 (for more details on the code, please refer to the multiversity folder from the GitHub repository).
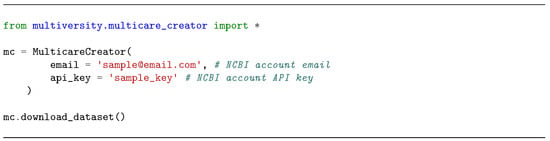
Figure 2.
Python code used to replicate the creation of the dataset.
3.2. Data Download
The IDs of the relevant case report articles were retrieved from PubMed using Biopython, a bioinformatics library that provides access to the NCBI Entrez API [36]. The query string was designed with the following criteria: (a) the “Publication Type” field had to be “case report,” or the “Title/Abstract” field needed to include specific terms such as “clinical case,” “case report,” or “case series"; (b) only full-text articles freely available were retrieved using the “ffrft” filter; (c) articles categorized as “animal” were excluded; and (d) only articles published after 1990—and before the search date (19 February 2025)—were considered. Although language was not explicitly filtered during the query process, non-English articles were excluded later due to the reliance on English-specific regex patterns for text processing.
Article metadata and abstracts were retrieved from PubMed’s website using the requests [37] and BeautifulSoup [38] packages. The BioC API for PMC was used to extract the remaining textual contents of the articles, including image captions and file names [39]. Figures were downloaded via the Europe PMC RESTful API [40]. The articles used to create this dataset are licensed under CC BY, CC BY-NC, CC BY-NC-SA, or CC0. Other licenses included in PMC Open Access Subset, such as no-derivatives licenses, were filtered out.
3.3. Data Preprocessing
Case report articles include information beyond the actual case presentations (such as introduction or conclusion sections), and may describe more than one patient. Therefore, it was necessary to identify the boundaries of each case. To achieve this, HTML tags, section headers, and mentions of patient ages—commonly found at the beginning of clinical cases—were considered. Additionally, text processing tasks included extracting and normalizing mentions of age and gender to populate demographic metadata fields, identifying figure mentions in text to map image files to their corresponding textual references, and splitting compound image captions into individual captions (e.g., converting ‘Chest X-rays (A) AP view and (B) lateral view’ into ‘Chest X-rays AP view’ and ‘Chest X-rays lateral view’). Demographic factors other than age and gender, such as ethnicity or socioeconomic status were not extracted because they are typically not explicitly mentioned in clinical cases.
Articles often include compound images, which are single files containing multiple image components. Using OpenCV [41], these figures were split into individual components, as illustrated in the example in Figure 3. This process involved the following steps: (1) edge detection, achieved by blurring the image with a Gaussian filter followed by applying a Canny edge detector; (2) limit detection, based on identifying rows and columns of pixels with a low density of edges; (3) image slicing, using the detected limits; and (4) order sorting, where image components were arranged in the original file’s layout from left to right and top to bottom (ensuring, for example, that the second image is the one directly to the right of the first). Borders present in the input images were removed throughout this procedure. The results from the image preprocessing were cross-checked with those from caption splitting (i.e., only the images that were split into the same number of pieces as their corresponding captions were included in the dataset).
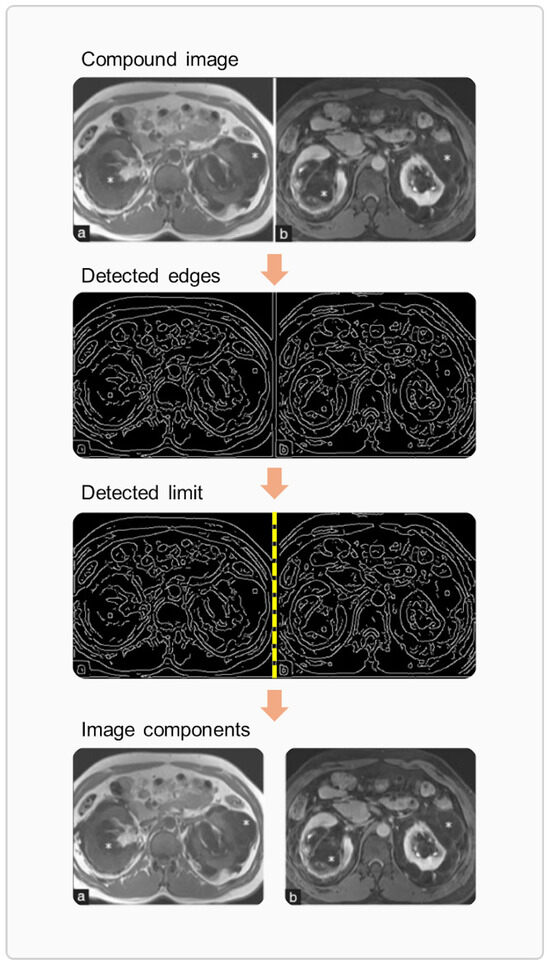
Figure 3.
An example of image preprocessing applied to a compound image adapted from a case report article [42].
3.4. Programmatic Image Labeling
The image labeling process comprised three main steps: taxonomy development, caption-to-label conversion, and machine learning-based annotation.
3.4.1. Taxonomy Development
The taxonomy of a classification dataset consists of its classes, their hierarchical relationships, and the logical constraints among them (such as mutual exclusivity). In this version of the dataset, the taxonomy was developed using the previous version as a foundation, considering the terms present in image captions, and referencing different domain ontologies accessed via BioPortal: SNOMED CT, MeSH, the Foundational Model of Anatomy (FMA) and RadLex [43,44,45,46,47]. The structure of the taxonomy follows the principles of the Multiplex Classification Framework, a methodology that enables complex classification problems to be addressed as a set of simpler binary or multiclass tasks, organized either in parallel or sequentially [48]. The full taxonomy was published in Bioportal as MCR_TX, and includes 163 classes (including the root_class, 17 negative classes such as no_doppler, and 145 positive classes) [49]. A reduced version with 81 classes (80 positive ones and the root_class) was developed by excluding the least important ones in order to simplify the training of machine learning (ML) models (see the ML_MCR_TX taxonomy in BioPortal) [50]. Figure 4 displays multiple classes from the MCR_TX taxonomy, each accompanied by an example image. In the MultiCaRe dataset, an image can have multiple associated labels, provided that the logical constraints among them are respected (for more details, refer to the taxonomy documentation available in the GitHub repository).
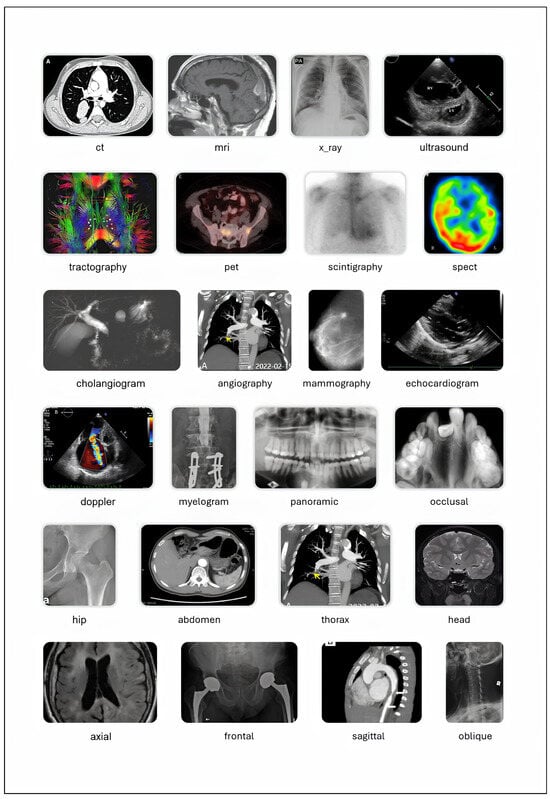
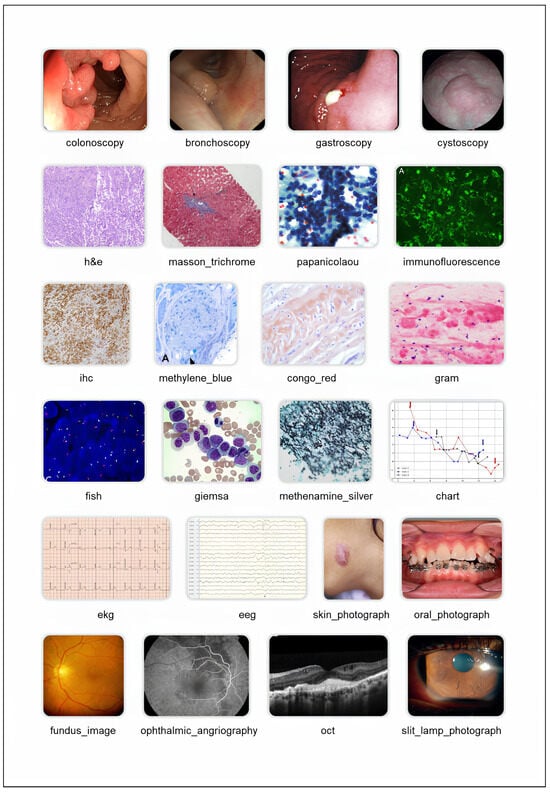
Figure 4.
Selected classes from the medical image taxonomy. For details on the class structure, please refer to the taxonomy documentation. ‘ct’: Computed Tomography, ‘ekg’: Electrocardiogram, ‘eeg’: Electroencephalogram, ‘fish’: Fluorescence In Situ Hybridization, ‘h&e’: Hematoxylin and Eosin, ‘ihc’: Immunohistochemistry, ‘mri’: Magnetic Resonance Imaging, ‘oct’: Optical Coherence Tomography, ‘pet’: Positron Emission Tomography, ‘spect’: Single Photon Emission Computed Tomography. Image credits: ct [51], mri [52], x_ray [53], ultrasound [54], tractography [55], pet [52], scintigraphy [56], spect [57], cholangiogram [58], angiography [59], mammography [60], echocardiogram [61], doppler [54], myelogram [62], panoramic [63], occlusal [64], hip [65], abdomen [66], thorax [59], head [67], axial [68], frontal [69], sagittal [70], oblique [71]. All images are licensed under CC BY. Selected classes from the medical image taxonomy. Image credits: colonoscopy [72], bronchoscopy [73], gastroscopy [74], cystoscopy [75], h&e [76], masson_trichrome [77], papanicolaou [78], immunofluorescence [79], ihc [80], methylene_blue [81], congo_red [82], gram [83], fish [84], giemsa [85], methenamine_silver [86], chart [87], ekg [88], eeg [89], oral_photograph [90], skin_photograph [91], fundus_photograph [92], ophthalmic_angiography [93], oct [94], slit_lamp_photograph [95]. All images are licensed under CC BY.
3.4.2. Caption-to-Label Conversion
At this step of the process, each image was assigned labels based on the content of its caption. To do this, first a list was created including all the unique sequences of words or tokens (n-grams) present in captions. Sequences longer than five tokens were excluded (no important class was lost in the process because relevant long n-grams generally consist of a combination of multiple shorter n-grams). The list was manually annotated using a spreadsheet, assigning corresponding labels to each n-gram based on the dataset taxonomy. The annotated spreadsheet was then used to assign labels to each caption based on the n-grams it contained (e.g., the label ‘ct’ was assigned to captions containing the n-gram ‘computed tomography’). When captions included overlapping n-grams (e.g., ‘right’ and ‘right lobe’), only the longest n-gram was considered. Polysemic n-grams, such as ‘left’ (which could indicate laterality or the past tense of ‘to leave’), were annotated disregarding such polysemy, and then the n-grams associated with irrelevant meanings (e.g., ‘nothing left’) were tagged with a dummy label, which was removed during post-processing to prevent the assignment of incorrect labels. Finally, any pairs of incompatible labels assigned to the same caption (e.g., ‘mri’ and ‘medical_photograph’) were also removed during post-processing. The outcome labels are included in the gt_labels_for_semisupervised_classification column of captions_and_labels.csv, except for those incompatible with machine learning predictions, which were filtered out.
This type of annotation is effective for image captions because they rarely include negated or hypothetical terms (unlike other medical texts), and therefore there is no need for additional resources like negation detection or assertion status models. The main advantage of this approach is its efficiency, as it requires fewer working hours to annotate larger datasets. However, it also has limitations. First of all, it demands annotators with significant expertise both in data annotation and medical text handling. Second of all, for negative classes (such as ‘no_doppler’), manual annotations are generally required because captions do not typically mention absent facts. Finally, this approach tends to produce a high number of false negatives because sometimes captions do not explicitly reference the image classes (for example, not all CT scan images contain n-grams related to the ‘ct’ class in their captions). This limitation can be addressed using ML-based annotations (as detailed in the following section).
3.4.3. Machine Learning-Based Annotation
After converting captions into image labels, ML was used to improve label quality for the two most important sets of classes: (a) main image types, such as ‘radiology’ or ‘endoscopy’; and (b) radiology subclasses, such as ‘ultrasound’ or ‘mri’. This process consisted of three steps: (1) image classification models were trained on pre-labeled images; (2) cases with the highest loss values were manually reviewed and analyzed to establish a quality threshold; (3) labels with loss values exceeding this threshold were replaced with the model’s predictions. Following this initial quality improvement, thirteen ML models were trained on the entire image dataset for all classes within the reduced taxonomy to predict missing labels and complete the dataset (details of model training and metrics of each model can be found in MultiCaReClassifier folder from the GitHub repository). Finally, the quality improvement process was reapplied, yielding more robust labels and the final set of thirteen image classification models. The models, along with the code for model ensembling and data post-processing, have been published on HuggingFace [96]. The predictions of these models have been used to populate the supervised learning columns from captions_and_labels.csv (ml_labels_for_supervised_classification and all the multiclass columns, such as image_type). The distribution of image types, along with the subtypes of the main class (’radiology’), is shown in Figure 5. Additional information—such as the number of images per class, average image height, average image width, and average file size—is provided in the file image_data_per_class.csv available in the GitHub repository.
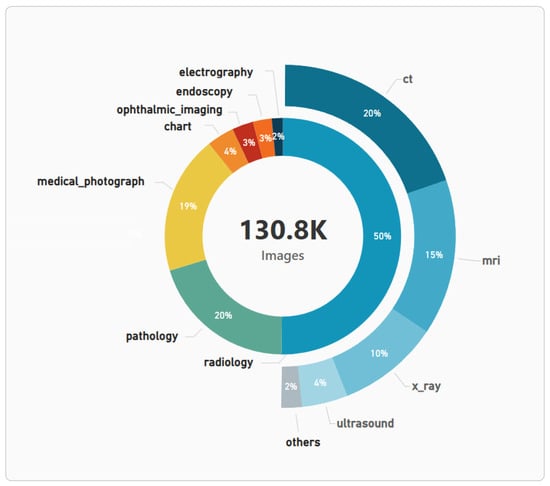
Figure 5.
Distribution of image types, and the subtypes within the ‘radiology’ class.
All the models mentioned in this work used the ResNet50 architecture, with the implementation provided by the Torchvision library [97,98]. An intensive data augmentation pipeline was applied to prevent overfitting, consisting of the HorizontalFlip, VerticalFlip, Rotate, Sharpen, ColorJitter, RGBShift, GaussianBlur, GaussNoise and RandomSizedCrop methods from the Albumentations library [99]. Oversampling was also used, meaning that the probability of sampling each image depends on the relative frequency of its class (images from underrepresented classes are more likely to be sampled). Models were trained using an image size of 224, an 80/20 split for training and validation data, a batch size of 16, and a learning rate of 0.0003. The cross entropy loss function with label smoothing was employed with the Adam optimizer over 30 epochs. The training process utilized the ‘fit_one_cycle’ method from the FastAI Python library [100]. The performance of the models was evaluated using accuracy and F1 score as metrics. The confusion matrices of each model in the ensemble can be found in the MultiCaReClassifier folder from the GitHub repository. However, it’s important to note that these are not traditional confusion matrices, as ML-generated labels are compared to caption-based labels (which are incomplete and somewhat inaccurate), instead of manually annotated ground truth labels.
3.5. Data Validation and Quality Control
Data quality assessment was conducted at three levels: text quality, image quality, and label quality. The first two aspects were evaluated in the previous version of the dataset, whereas the assessment of image label quality was introduced in this version [26].
For textual data, 100 case reports were reviewed, with 84% found to have no problems. The identified problems were: (a) incorrect article type (3%); (b) errors in demographic extraction (4%); (c) extra splits (3%), which means that the text of one patient was split into multiple cases; (d) missing splits (3%), which means that multiple patients were combined into a single case; and (e) incorrect case content (3%), for example if part of the introduction of an article was included in the content of a case.
Regarding image quality, 153 images (corresponding to 100 article figures) were examined, and 86% were verified as correct. The problems found at this point were: (a) missing splits in compound images (4%), generally related to errors in the image caption; (b) incorrect image order (2%); (c) removal of relevant parts of the image (2%); and (e) incorrect patient assignment (1%). Although errors related to image splitting are relatively infrequent, if complete avoidance is necessary, users can filter out split images by selecting only those that contain the term ‘undivided’ in their names.
Finally, regarding label quality, two types of tests were performed by reviewing labels manually:
- Random subset review: Six random samples of 50 labels each were reviewed for quality assessment—three from the gt_labels_for_semisupervised_classification column and three from the ml_labels_for_supervised_classification column. In the first group, 96%, 90%, and 96% of the labels were correct, corresponding to a mean error rate of 6% with a standard deviation of 3.46%. In the second group, 70%, 78%, and 76% of the labels were correct, resulting in a mean error rate of 25.3% and a standard deviation of 4.16%.
- Stratified manual review: To account for class imbalance, another manual review was conducted on 200 caption-based labels and 200 ML-based labels. This time, the sampling ensured a minimum number of examples per class (at least one per class for caption-based labels and two per class for ML-based labels). The review found that 90% of the caption-based labels and 70% of the ML-based labels were correct. Notably, nearly half of the errors in the ML-based labels stemmed from incorrect parent class assignments—for instance, when a skin image is misclassified as ‘immunostaining’, the root cause is often an upstream misclassification under the broader ‘pathology’ class instead of ‘medical_photograph’.
Given this, we recommend using caption-based labels when higher label quality is needed, ideally with review by a clinical data annotator. ML-generated labels are preferable when a larger dataset is required, provided that the labels are also reviewed to ensure quality.
4. Usage Notes
4.1. Creation of Customized Subsets
As mentioned before, rather than accessing the whole dataset through Zenodo, we recommend creating subsets based on the specific use case using the MedicalDatasetCreator class from Python’s multiversity library. This can be accomplished through a few simple steps. First, the library needs to be installed in order to import and instantiate the MedicalDatasetCreator class (see Figure 6). At this step, which may take around five minutes, the MultiCaRe Dataset is imported from Zenodo into a folder called ‘whole_multicare_dataset’ in the main directory (‘/sample/directory/’ in this example).

Figure 6.
Python code used to import and instantiate the MedicalDatasetCreator class.
The part of the whole dataset that will be included in the customized subset depends on the list of filters that are used. Some filters work at an article level (e.g., ‘min_year’ filters by the year of the case report article), others work at a patient level (e.g., ‘gender’) and some others work at an image level (e.g., ‘caption’). The contents of the subset also depend on the selected subset type (text, image, multimodal, and case series). For additional examples and details, please refer to the Demos folder from the GitHub repository. In all cases, the subsets include these three files:
- readme.txt: a short description of the subset, including its name, type, creation date, and the filters that were used during its creation.
- case_report_citations.json: file including all the metadata necessary to cite the case report articles, such as title, authors, and year.
- article_metadata.json: file including the rest of the metadata (license, keywords and mesh terms).
4.1.1. Text Subsets
This type of subsets contains textual data without annotations, which can be used for text labeling, language modeling or text mining. All the clinical cases are included in the cases.csv file, along with the patient’s age and gender. In the example from Figure 7, the created text subset includes cases after 2015 involving patients older than 18 years diagnosed that mention either ‘lung cancer’ or ‘lung carcinoma’. First the corresponding filters are defined, and then the subset is created by using the .create_dataset() method. In this case, the dataset_type parameter is set to ‘text’.

Figure 7.
Python code used to create a text subset.
4.1.2. Image Classification Subsets
This subset type is intended for training image classification models. The data are organized into folders: one folder per image class (containing image files and the image_metadata.json file) and an additional folder with the image_classification.csv file.
- image_metadata.json: it includes the patient ID, file path, license, caption, and labels for each image.
- image_classification.csv: it contains one column with the file path, and another one with the corresponding class.
It is possible to create a subset of the captions_and_labels.csv file by using the .create_dataset() method with the dataset_type parameter set to ‘image’ (this works the same way as for ‘text’ subsets). However, users may sometimes need to create multiple customized subsets to train models that classify images into one of these subsets. For example, a user might want to create two subsets of head MRIs showing metastases—one for female patients and one for male patients—in order to train a gender classifier. This can be achieved using the .create_image_classification_dataset() method, which allows the application of a general filter list for all the subsets, as well as specific filter lists for each of them. The output is a CSV file with a ‘class’ column for the classes included in the dataset_dict (‘female’ and ‘male’ in the example below), along with the same label columns as captions_and_labels.csv if the keep_label_columns parameter is set to True. The filtered images will have exactly one label in the ‘class’ column (instances with multiple labels are excluded). In the example from Figure 8, the created subset is named ‘gender_classifier’, it includes head MRI images, and it has two possible classes based on the patient’s gender: male and female.
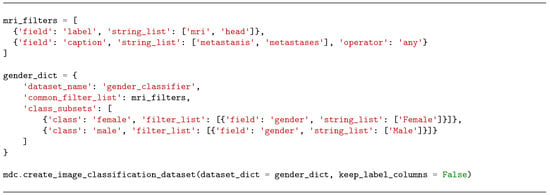
Figure 8.
Python code used to create an image classification subset.
The .create_image_classification_dataset() method allows to create datasets for specific use cases based on the contents of image captions, which can be useful in cases where specific image classes are not present in the dataset taxonomy. If negative classes are needed, ‘none’ can be used as a logical operator to include in the negative subset any image whose caption does not contain relevant terms associated with the positive class. In the example from Figure 9, a thorax X-ray dataset is created with the classes ‘fracture’ and ‘no_fracture’ using this approach. It is important to note that any dataset containing negative classes defined by the absence of terms in captions should be manually reviewed. This helps prevent the negative subset from including samples of the positive class—which can occur if the image class is not explicitly mentioned in the caption.

Figure 9.
Example of image subset creation with a negative class (no_fracture).
4.1.3. Multimodal Subsets
This subset type is intended for the development of multimodal AI applications. It contains image files organized in folders, the same image_metadata.json file as in image classification subsets, and the same case.csv file as in text subsets, with one key difference: image references (e.g., “Figure 1”) in the clinical case text are replaced by either image tokens (e.g., <image_token>) or the file path to the corresponding image (see example in Figure 1).
In the example from Figure 10, the created multimodal subset includes patients diagnosed with lung cancer and with a head MRI showing the presence of metastases. First the corresponding filters are defined, and then the subset is created by using the .create_dataset() method, with ‘multimodal’ as the dataset_type.

Figure 10.
Python code used to create a multimodal subset.
4.2. Use of Image Classification Models
The model ensemble for medical image classification is available on HuggingFace [96]. These models can be used by following the steps from Figure 11.

Figure 11.
Python code used to get image classes.
4.3. Resources for Healthcare Professionals
All the resources mentioned so far are intended specially for data scientists and related roles. However, this release also includes resources specially intended for healthcare professionals and students.
- Case series subsets: If ‘case_series’ is used as the dataset_type for the .create_dataset() method, then a subset with multimodal patient data is created, with the following distinctions: (a) the image organization differs from other subset types, with each patient having their own folder, and (b) the cases.csv file is very similar to the one from text subsets, except for some additional columns (including link to the case report article, number of images per patient, and path to the corresponding image folder).
- Clinical Case Hub [101]: This specialized platform is designed for medical students and healthcare professionals to explore the dataset’s contents, including clinical case texts and medical images. The hub’s user-friendly interface enables users to apply filters without requiring programming knowledge.
- Digital Twin Retriever [102]: This AI-powered tool with a chatbot interface can be used to search clinical cases and answer questions about them, helping healthcare professionals make more informed decisions. The current version operates only on textual data.
4.4. Dataset Limitations
The following limitations should be considered when using this dataset:
- Differences from real-life data: As noted in Appendix B, data from case reports differs from real clinical data in several ways. The text is shorter, rare cases tend to be overrepresented, images are generally smaller and lower in resolution, and they may contain burned-in annotations such as arrows or asterisks.
- Label quality: This is a silver standard dataset, meaning that image labels were generated semi-automatically and may contain a relatively high number of errors. As a result, label quality is lower than in datasets annotated manually by domain experts.
- Data imbalance: There is substantial variation in both the number and resolution of images across classes. This imbalance reflects the nature of the original data sources.
- Data loss: Not all content from PMC case reports was included. Two major causes of data loss are: (a) exclusion of articles with non-derivative licenses, and (b) removal of images when the number of cropped subimages did not match the number of caption parts, as explained in Section 3.3.
Author Contributions
Conceptualization, M.N.O.; methodology, F.R. and M.N.O.; software, M.C.G.G., M.M. and M.N.O.; data curation, M.N.O.; writing—original draft preparation, M.N.O.; writing—review and editing, C.D., F.R., M.M. and M.C.G.G.; visualization, M.C.G.G.; supervision, C.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
https://doi.org/10.5281/zenodo.10079369, accessed on 29 July 2025.
Acknowledgments
The authors are thankful to each and every patient, medical doctor, researcher, editor and any other person involved in the creation of the open access case reports included in this dataset. The citation information for each article used in this dataset is available in the metadata.parquet file. F.R. and M.M. receive financial support from the National Scientific and Technical Research Council (CONICET). No other specific grants from funding agencies in the public, commercial, or non-profit sectors were received for this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | artificial intelligence |
| API | application programming interface |
| CC0 | Creative Commons Zero |
| CC BY | Creative Commons Attribution |
| CC BY-NC | Creative Commons Attribution-NonCommercial |
| CC BY-NC-SA | Creative Commons Attribution-NonCommercial-ShareAlike |
| CSV | comma-separated values |
| CT | computed tomography |
| DOI | digital object identifier |
| EEG | electroencephalogram |
| EKG | electrocardiogram |
| FISH | fluorescence in situ hybridization |
| FMA | Foundational Model of Anatomy |
| H&E | hematoxylin and eosin |
| HTML | HyperText Markup Language |
| IHC | immunohistochemistry |
| MeSH | Medical Subject Headings |
| ML | machine learning |
| MRI | magnetic resonance imaging |
| NCBI | National Center for Biotechnology Information |
| OCT | optical coherence tomography |
| PET | positron emission tomography |
| PHI | protected health information |
| PMC | PubMed Central |
| PMCID | PubMed Central ID |
| SNOMED CT | Systematized Nomenclature of Medicine Clinical Terms |
| SPECT | single-photon emission computed tomography |
Appendix A. Differences Between the Updated and Initial Versions of the MultiCaRe Dataset
This updated version includes several improvements, as detailed in Table A1. Two main changes are particularly noteworthy:
- Data Licenses: Data with non-derivative license types were excluded from the updated version, as derivatives based on such data cannot be redistributed. This change ensures full compliance with licensing terms, although it resulted in a slight reduction in the number of cases and images. However, if needed, these excluded data can still be downloaded using the MulticareCreator class with the parameter license_types = [] (see Section 3.1).
- Image Format: The image format was changed from .jpg to .webp to reduce the overall dataset size. This modification does not affect the resolution, quality, or visual fidelity of the images.

Table A1.
Differences between the initial and the updated versions of the MultiCaRe dataset.
Table A1.
Differences between the initial and the updated versions of the MultiCaRe dataset.
| Initial Version | Updated Version | |
|---|---|---|
| update date | August 2023 | February 2025 |
| excluded license types | none | non-derivatives |
| amount of cases | 96,428 | 93,816 |
| amount of images | 135,596 | 130,791 |
| image classes | 138 | 150 |
| taxonomy quality | low | high |
| taxonomy structure | flat | hierarchical, with logical relations |
| documented taxonomy | no | yes |
| image labeling | caption-based | caption- and ml-based |
| file format | .jpg | .webp |
| dataset size | 8.8gb | 2.9gb |
| associated python package | no | yes |
| other associated products | none | image classifiers, clinical case hub, digital twin retriever |
Appendix B. Differences Between a Dataset of Case Reports and a Dataset of Medical Records
There are key differences between case report datasets and data extracted from medical records [26]. These differences are summarized in Table A2 and include:
- Healthy cases: Patients with no significant clinical problems are typically excluded from case report datasets, but healthy cases are sometimes included in medical record datasets (e.g., routine checkups or screening visits).
- Rare cases: These may be overrepresented in a case report dataset, and this can be considered as something beneficial because it increases the variability and diversity of the dataset. However, it is important to mention that normal disease presentations are still well represented since most case reports are published for educational purposes rather than because of their rarity [103].
- Text extension: Case reports typically consist of one or a few paragraphs, whereas medical records are generally much longer.
- Irrelevant information: Phrases like “patient denies any chest pain” are common in medical records. In contrast, case reports usually exclude absent symptoms, and normal test results may also be omitted.
- Text quality: Although text quality in medical records is high, when it comes to case reports it is much higher, as they undergo a rigorous review process before being published in a medical journal.
- Personal Health Information (PHI): Medical records are not de-identified because they are confidential documents maintained within a healthcare institution. In contrast, case reports are intended to be shared publicly, so they do not include any personal information of the patient. This ensures the protection of patient privacy, which is a fundamental ethical requirement for open clinical datasets.
- Images: Images from case reports are much smaller in size, they may contain burned drawings (such as arrows, circles or asterisks) and they do not contain any personal information of the patient.

Table A2.
Differences between a dataset of case reports and a dataset of medical records.
Table A2.
Differences between a dataset of case reports and a dataset of medical records.
| Case Reports | Medical Records | |
|---|---|---|
| Normal Cases | Underrepresented | Proportionally Represented |
| Rare Cases | Overrepresented | Proportionally Represented |
| Text Extension | Paragraphs | Pages |
| Irrelevant Information | Absent | Present |
| Text Quality | Very High | High |
| Personal Health Information | Absent | Present |
| Images | Small Size (<200 kb) | Large Size |
References
- Luo, L.; Yu, L.; Chen, H.; Liu, Q.; Wang, X.; Xu, J.; Heng, P.A. Deep Mining External Imperfect Data for Chest X-ray Disease Screening. arXiv 2020, arXiv:2006.03796. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- Borgli, H.; Thambawita, V.; Smedsrud, P.H.; Hicks, S.; Jha, D.; Eskeland, S.L.; Randel, K.R.; Pogorelov, K.; Lux, M.; Nguyen, D.T.D.; et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 2020, 7, 283. [Google Scholar] [CrossRef]
- Polat, G.; Kani, H.T.; Ergenc, I.; Ozen Alahdab, Y.; Temizel, A.; Atug, O. Improving the Computer-Aided Estimation of Ulcerative Colitis Severity According to Mayo Endoscopic Score by Using Regression-Based Deep Learning. Inflamm. Bowel Dis. 2023, 29, 1431–1439. [Google Scholar] [CrossRef]
- Hernández-Pérez, C.; Combalia, M.; Podlipnik, S.; Codella, N.C.F.; Rotemberg, V.; Halpern, A.C.; Reiter, O.; Carrera, C.; Barreiro, A.; Helba, B.; et al. BCN20000: Dermoscopic Lesions in the Wild. Sci. Data 2024, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Yap, M.H.; Cassidy, B.; Pappachan, J.M.; O’Shea, C.; Gillespie, D.; Reeves, N. Analysis Towards Classification of Infection and Ischaemia of Diabetic Foot Ulcers. In Proceedings of the 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), Athens, Greece, 27–30 July 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Da, Q.; Huang, X.; Li, Z.; Zuo, Y.; Zhang, C.; Liu, J.; Chen, W.; Li, J.; Xu, D.; Hu, Z.; et al. DigestPath: A benchmark dataset with challenge review for the pathological detection and segmentation of digestive-system. Med. Image Anal. 2022, 80, 102485. [Google Scholar] [CrossRef]
- Koziarski, M.; Cyganek, B.; Niedziela, P.; Olborski, B.; Antosz, Z.; Żydak, M.; Kwolek, B.; Wąsowicz, P.; Bukała, A.; Swadźba, J.; et al. DiagSet: A dataset for prostate cancer histopathological image classification. Sci. Rep. 2024, 14, 6780. [Google Scholar] [CrossRef]
- Maksoud, S.; Zhao, K.; Hobson, P.; Jennings, A.; Lovell, B. SOS: Selective Objective Switch for Rapid Immunofluorescence Whole Slide Image Classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 3861–3870. [Google Scholar] [CrossRef]
- Phoulady, H.A.; Mouton, P.R. A New Cervical Cytology Dataset for Nucleus Detection and Image Classification (Cervix93) and Methods for Cervical Nucleus Detection. arXiv 2018, arXiv:1811.09651. [Google Scholar]
- Yang, J.; Shi, R.; Wei, D.; Liu, Z.; Zhao, L.; Ke, B.; Pfister, H.; Ni, B. MedMNIST v2 - A large-scale lightweight benchmark for 2D and 3D biomedical image classification. Sci. Data 2023, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Abacha, A.B.; Sarrouti, M.; Demner-Fushman, D.; Hasan, S.A.; Müller, H. Overview of the VQA-Med Task at ImageCLEF 2021: Visual Question Answering and Generation in the Medical Domain. In Proceedings of the CLEF 2021 Conference and Labs of the Evaluation Forum-Working Notes, Bucharest, Romania, 21–24 September 2021. [Google Scholar] [CrossRef]
- Chen, J.; Gui, C.; Ouyang, R.; Gao, A.; Chen, S.; Chen, G.H.; Wang, X.; Zhang, R.; Cai, Z.; Ji, K.; et al. HuatuoGPT-Vision, Towards Injecting Medical Visual Knowledge into Multimodal LLMs at Scale. arXiv 2024, arXiv:2406.19280. [Google Scholar]
- He, X.; Zhang, Y.; Mou, L.; Xing, E.; Xie, P. PathVQA: 30000+ Questions for Medical Visual Question Answering. arXiv 2020, arXiv:2003.10286. [Google Scholar]
- Ikezogwo, W.O.; Seyfioglu, M.S.; Ghezloo, F.; Geva, D.S.C.; Mohammed, F.S.; Anand, P.K.; Krishna, R.; Shapiro, L. Quilt-1M: One Million Image-Text Pairs for Histopathology. arXiv 2023, arXiv:2306.11207. [Google Scholar]
- Jin, Q.; Chen, F.; Zhou, Y.; Xu, Z.; Cheung, J.M.; Chen, R.; Summers, R.M.; Rousseau, J.F.; Ni, P.; Landsman, M.J.; et al. Hidden flaws behind expert-level accuracy of multimodal GPT-4 vision in medicine. NPJ Digit. Med. 2024, 7, 190. [Google Scholar] [CrossRef]
- Lau, J.J.; Gayen, S.; Ben Abacha, A.; Demner-Fushman, D. A dataset of clinically generated visual questions and answers about radiology images. Sci. Data 2018, 5, 180251. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhao, Z.; Zhang, X.; Wu, C.; Zhang, Y.; Wang, Y.; Xie, W. PMC-CLIP: Contrastive Language-Image Pre-training using Biomedical Documents. arXiv 2023, arXiv:2303.07240. [Google Scholar]
- Liu, B.; Zhan, L.M.; Xu, L.; Ma, L.; Yang, Y.; Wu, X.M. SLAKE: A Semantically-Labeled Knowledge-Enhanced Dataset for Medical Visual Question Answering. arXiv 2021, arXiv:2102.09542. [Google Scholar]
- Matos, J.; Chen, S.; Placino, S.; Li, Y.; Pardo, J.C.C.; Idan, D.; Tohyama, T.; Restrepo, D.; Nakayama, L.F.; Pascual-Leone, J.M.M.; et al. WorldMedQA-V: A multilingual, multimodal medical examination dataset for multimodal language models evaluation. arXiv 2024, arXiv:2410.12722. [Google Scholar]
- Rückert, J.; Bloch, L.; Brüngel, R.; Idrissi-Yaghir, A.; Schäfer, H.; Schmidt, C.S.; Koitka, S.; Pelka, O.; Abacha, A.B.; Herrera, A.G.S.d.; et al. ROCOv2: Radiology Objects in COntext Version 2, an Updated Multimodal Image Dataset. Sci. Data 2024, 11, 688. [Google Scholar] [CrossRef]
- Siragusa, I.; Contino, S.; Ciura, M.L.; Alicata, R.; Pirrone, R. MedPix 2.0: A Comprehensive Multimodal Biomedical Dataset for Advanced AI Applications. arXiv 2024, arXiv:2407.02994. [Google Scholar]
- Xie, Y.; Zhou, C.; Gao, L.; Wu, J.; Li, X.; Zhou, H.Y.; Liu, S.; Xing, L.; Zou, J.; Xie, C.; et al. MedTrinity-25M: A Large-scale Multimodal Dataset with Multigranular Annotations for Medicine. arXiv 2024, arXiv:2408.02900. [Google Scholar]
- Nievas Offidani, M. MultiCaRe: An Open-Source Clinical Case Dataset for Medical Image Classification and Multimodal AI Applications. 2025. Available online: https://zenodo.org/records/14994046 (accessed on 29 July 2025). [CrossRef]
- Nievas Offidani, M.A.; Delrieux, C.A. Dataset of clinical cases, images, image labels and captions from open access case reports from PubMed Central (1990–2023). Data Brief 2024, 52, 110008. [Google Scholar] [CrossRef]
- Nievas Offidani, M.A. MultiCaRe_Dataset. 2025. Available online: https://github.com/mauro-nievoff/MultiCaRe_Dataset (accessed on 29 July 2025).
- Belharty, N.; Azouzi, R.E.; Chafai, Y.; Mouine, N.; Benyass, A. Concomitant in situ and in transit right heart thrombi: A case report. Pan Afr. Med J. 2020, 37, 355. [Google Scholar] [CrossRef]
- Li, C.; Wong, C.; Zhang, S.; Usuyama, N.; Liu, H.; Yang, J.; Naumann, T.; Poon, H.; Gao, J. LLaVA-Med: Training a Large Language-and-Vision Assistant for Biomedicine in One Day. arXiv 2023, arXiv:2306.00890. [Google Scholar]
- Zhang, S.; Xu, Y.; Usuyama, N.; Xu, H.; Bagga, J.; Tinn, R.; Preston, S.; Rao, R.; Wei, M.; Valluri, N.; et al. BiomedCLIP: A multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. arXiv 2024, arXiv:2303.00915. [Google Scholar]
- Zhang, X.; Wu, C.; Zhao, Z.; Lin, W.; Zhang, Y.; Wang, Y.; Xie, W. PMC-VQA: Visual Instruction Tuning for Medical Visual Question Answering. arXiv 2024, arXiv:2305.10415. [Google Scholar]
- Zhao, Z.; Jin, Q.; Chen, F.; Peng, T.; Yu, S. A large-scale dataset of patient summaries for retrieval-based clinical decision support systems. Sci. Data 2023, 10, 909. [Google Scholar] [CrossRef]
- Subramanian, S.; Wang, L.L.; Mehta, S.; Bogin, B.; Zuylen, M.v.; Parasa, S.; Singh, S.; Gardner, M.; Hajishirzi, H. MedICaT: A Dataset of Medical Images, Captions, and Textual References. arXiv 2020, arXiv:2010.06000. [Google Scholar]
- Nissen, T.; Wynn, R. The clinical case report: A review of its merits and limitations. BMC Res. Notes 2014, 7, 264. [Google Scholar] [CrossRef]
- Nievas Offidani, M. Multiversity Package. Available online: https://github.com/mauro-nievoff/MultiCaRe_Dataset/tree/main/multiversity_library/multiversity/multiversity (accessed on 29 July 2025).
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Reitz, K. Requests: HTTP for Humans. Available online: https://docs.python-requests.org/en/latest/ (accessed on 29 July 2025).
- Richardson, L. Beautiful Soup Documentation. Available online: https://www.crummy.com/software/BeautifulSoup/bs4/doc/ (accessed on 29 July 2025).
- Comeau, D.C.; Wei, C.H.; Islamaj Doğan, R.; Lu, Z. PMC text mining subset in BioC: About three million full-text articles and growing. Bioinformatics 2019, 35, 3533–3535. [Google Scholar] [CrossRef] [PubMed]
- Europe PMC Consortium. Europe PMC: A full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015, 43, D1042–D1048. [Google Scholar] [CrossRef] [PubMed]
- Bradski, G. The OpenCV Library. J. Softw. Tools 2000, 25, 120–123. [Google Scholar]
- Elbanna, K.Y.; Almutairi, B.M.; Zidan, A.T. Bilateral renal lymphangiectasia: Radiological findings by ultrasound, computed tomography, and magnetic resonance imaging. J. Clin. Imaging Sci. 2015, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Structural Informatics Group. Foundational Model of Anatomy. 2019. Available online: http://sig.biostr.washington.edu/projects/fm/AboutFM.html (accessed on 29 July 2025).
- National Library of Medicine Medical. Medical Subject Headings; U.S. National Library of Medicine: Bethesda, MD, USA, 2024. [Google Scholar]
- Radiological Society of North America. RadLex Radiology Lexicon. 2020. Available online: https://www.rsna.org/practice-tools/data-tools-and-standards/radlex-radiology-lexicon (accessed on 29 July 2025).
- National Center for Biomedical Ontology. BioPortal. 2024. Available online: https://bioportal.bioontology.org/ (accessed on 29 July 2025).
- Snomed International. SNOMED CT. 2024. Available online: https://www.snomed.org/ (accessed on 29 July 2025).
- Nievas Offidani, M.A.; Roffet, F.; Delrieux, C.A.; González Galtier, M.C.; Zarate, M.D. The Multiplex Classification Framework: Optimizing Multi-Label Classifiers Through Problem Transformation, Ontology Engineering, and Model Ensembling. Appl. Ontol. 2025. [Google Scholar] [CrossRef]
- Nievas Offidani, M. MCR_TX Taxonomy. Available online: https://bioportal.bioontology.org/ontologies/MCR_TX (accessed on 29 July 2025).
- Nievas Offidani, M. ML_MCR_TX Taxonomy. Available online: https://bioportal.bioontology.org/ontologies/ML_MCR_TX (accessed on 29 July 2025).
- Zarfati, A.; Martucci, C.; Crocoli, A.; Serra, A.; Persano, G.; Inserra, A. Chemotherapy-induced cavitating Wilms’ tumor pulmonary metastasis: Active disease or scarring? A case report and literature review. Front. Pediatr. 2023, 11, 1083168. [Google Scholar] [CrossRef]
- Bailey, D.D.; Montgomery, E.Y.; Garzon-Muvdi, T. Metastatic high-grade meningioma: A case report and review of risk factors for metastasis. Neuro-Oncol. Adv. 2023, 5, vdad014. [Google Scholar] [CrossRef]
- Rouientan, H.; Gilani, A.; Sarmadian, R.; Rukerd, M.R.Z. A rare case of an immunocompetent patient with isolated pulmonary mucormycosis. IDCases 2023, 31, e01726. [Google Scholar] [CrossRef]
- Mohanty, V.; Sharma, S.K.; Goswami, S.; Gudhage, R.; Deora, S. Idiopathic Isolated Right Ventricular Cardiomyopathy: A Rare Case Report. Avicenna J. Med. 2023, 13, 56–59. [Google Scholar] [CrossRef]
- Idris, Z.; Ghazali, F.H.; Abdullah, J.M. Fibromyalgia and arachnoiditis presented as an acute spinal disorder. Surg. Neurol. Int. 2014, 5, 151. [Google Scholar] [CrossRef]
- Nishiki, E.; Honda, S.; Yamano, M.; Kawasaki, T. Midventricular Obstruction With Diastolic Paradoxic Jet Flow in Transthyretin Cardiac Amyloidosis: A Case Report. Cureus 2025, 17, e77097. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Fujisawa, M.; Hozumi, H.; Tsuboi, T.; Kuwashima, S.; Hirao, J.i.; Sugita, K.; Arisaka, O. Systemic inflammatory response syndrome and prolonged hypoperfusion lesions in an infant with respiratory syncytial virus encephalopathy. J. Infect. Chemother. 2013, 19, 978–982. [Google Scholar] [CrossRef]
- Jiang, C.; Tang, W.; Yang, X.; Li, H. Immune checkpoint inhibitor-related pancreatitis: What is known and what is not. Open Med. 2023, 18, 20230713. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Feng, L.; Huang, X.; Hu, B.; Yuan, Y. Case report: An elderly woman with recurrent syncope after pacemaker implantation. Front. Cardiovasc. Med. 2023, 10, 1117244. [Google Scholar] [CrossRef] [PubMed]
- Caputo, R.; Pagliuca, M.; Pensabene, M.; Parola, S.; De Laurentiis, M. Long-term complete response with third-line PARP inhibitor after immunotherapy in a patient with triple-negative breast cancer: A case report. Front. Oncol. 2023, 13, 1214660. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, H.; Chen, J.; He, X. Case report: Echocardiographic diagnosis of cardiac involvement caused by congenital generalized lipodystrophy in an infant. Front. Pediatr. 2023, 11, 1087833. [Google Scholar] [CrossRef]
- Derman, P.B.; Rogers-LaVanne, M.P.; Satin, A.M. Minimally Invasive Revision of Luque Plate Instrumentation: A Case Report. Cureus 2024, 16, e53120. [Google Scholar] [CrossRef] [PubMed]
- Handschel, J.G.; Depprich, R.A.; Zimmermann, A.C.; Braunstein, S.; Kübler, N.R. Adenomatoid odontogenic tumor of the mandible: Review of the literature and report of a rare case. Head Face Med. 2005, 1, 3. [Google Scholar] [CrossRef]
- Adlakha, V.K.; Chandna, P.; Gandhi, S.; Chopra, S.; Singh, N.; Singh, S. Surgical Repositioning of a Dilacerated Impacted Incisor. Int. J. Clin. Pediatr. Dent. 2011, 4, 55–58. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, J.; Chen, Y.; Ding, X.; Chen, F.; Feng, K.; Chen, K. Cryptococcal Osteomyelitis of the Left Acetabulum: A Case Report. Curr. Med Imaging Former. Curr. Med Imaging Rev. 2023, 19, e251122211248. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Wang, Y.W.; Zhang, W.; Zhang, B.X. Case Report: Solitary metastasis to the appendix after curative treatment of HCC. Front. Surg. 2023, 10, 1081326. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, B.; Xue, Y.; Wang, Y.; Yan, J.; Xu, L. Case report: A case of Culler-Jones syndrome caused by a novel mutation of GLI2 gene and literature review. Front. Endocrinol. 2023, 14, 1133492. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.T.; Torres, D.; Robinson, A.; Malone, C.; Vera, J.C.; Daghighi, S.; Dunn-Pirio, A.; Khoromi, S.; Nowell, J.; Léger, G.C.; et al. Case report: Neuronal intranuclear inclusion disease presenting with acute encephalopathy. Front. Neurol. 2023, 14, 1184612. [Google Scholar] [CrossRef]
- Singh, A.; Gandavaram, S.; Patel, K.; Herlekar, D. Use of a Trephine to Extract a Fractured Corail Femoral Stem During Revision Total Hip Arthroplasty: Tips From Our Case Report. Cureus 2024, 16, e52996. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Wang, Z.; Lin, J.; Zou, L.; Xu, X.; Yang, X.; Xu, Y.; Zhang, Y.; Zheng, Q.; Xu, X.; et al. Case report: Aggressive progression of acute heart failure due to juvenile tuberculosis-associated Takayasu arteritis with aortic stenosis and thrombosis. Front. Cardiovasc. Med. 2023, 10, 1076118. [Google Scholar] [CrossRef]
- Cupler, Z.A.; Anderson, M.T.; Stefanowicz, E.T.; Warshel, C.D. Post-infectious ankylosis of the cervical spine in an army veteran: A case report. Chiropr. Man. Ther. 2018, 26, 40. [Google Scholar] [CrossRef]
- Pang, S.; Song, J.; Zhang, K.; Wang, J.; Zhao, H.; Wang, Y.; Li, P.; Zong, Y.; Wu, Y. Case report: Coexistence of sigmoid tumor with unusual pathological features and multiple colorectal neuroendocrine tumors with lymph node metastases. Front. Oncol. 2023, 13, 1073234. [Google Scholar] [CrossRef] [PubMed]
- Sivapalan, P.; Gottlieb, M.; Christensen, M.; Clementsen, P.F. An obstructing endobronchial lipoma simulating COPD. Eur. Clin. Respir. J. 2014, 1, 25664. [Google Scholar] [CrossRef]
- Koizumi, M.; Sata, N.; Yoshizawa, K.; Tsukahara, M.; Kurihara, K.; Yasuda, Y.; Nagai, H. Post-ERCP pancreatogastric fistula associated with an intraductal papillary-mucinous neoplasm of the pancreas—A case report and literature review. World J. Surg. Oncol. 2005, 3, 70. [Google Scholar] [CrossRef][Green Version]
- Sekino, Y.; Mochizuki, H.; Kuniyasu, H. A 49-year-old woman presenting with hepatoid adenocarcinoma of the urinary bladder: A case report. J. Med Case Rep. 2013, 7, 12. [Google Scholar] [CrossRef]
- Bohbot, Y.; Garot, J.; Danjon, I.; Thébert, D.; Nahory, L.; Gros, P.; Salerno, F.; Garot, P. Case report: Diagnosis, management and evolution of a bulky and invasive cardiac mass complicated by complete atrioventricular block. Front. Cardiovasc. Med. 2023, 10, 1135233. [Google Scholar] [CrossRef]
- Tan, Y.; Zheng, S. Clinicopathological characteristics and diagnosis of hepatic sinusoidal obstruction syndrome caused by Tusanqi–Case report and literature review. Open Med. 2023, 18, 20230737. [Google Scholar] [CrossRef]
- Pailoor, K.; Kini, H.; Rau, A.R.; Kumar, Y. Cytological diagnosis of a rare case of solid pseudopapillary neoplasm of the pancreas. J. Cytol./Indian Acad. Cytol. 2010, 27, 32–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Dong, L.; Yu, L.; Huang, J. Autoimmune glial fibrillary acidic protein astrocytopathy coexistent with reversible splenial lesion syndrome: A case report and literature review. Front. Neurol. 2023, 14, 1192118. [Google Scholar] [CrossRef]
- Ge, W.; Qu, Y.; Hou, T.; Zhang, J.; Li, Q.; Yang, L.; Cao, L.; Li, J.; Zhang, S. Case report: Surgical treatment of a primary giant epithelioid hemangioendothelioma of the spine with total en-bloc spondylectomy. Front. Oncol. 2023, 13, 1109643. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.M.; Tran, N.T.; Patel, S.S.; Hou, J.; Jhaveri, K.D.; Parikh, R.; Selamet, U.; Ghobry, L.; Wassef, O.; Barsoum, M.; et al. Thrombotic Microangiopathy and Acute Kidney Injury Induced After Intravitreal Injection of Vascular Endothelial Growth Factor Inhibitors VEGF Blockade-Related TMA After Intravitreal Use. Front. Med. 2020, 7, 579603. [Google Scholar] [CrossRef]
- Orchard, G.E. Gout With Associated Cutaneous AA Amyloidosis: A Case Report and Review of the Literature. Br. J. Biomed. Sci. 2023, 80, 11442. [Google Scholar] [CrossRef]
- Yang, K.; Kruse, R.L.; Lin, W.V.; Musher, D.M. Corynebacteria as a cause of pulmonary infection: A case series and literature review. Pneumonia 2018, 10, 10. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, Y.; Zhang, W.; Wang, K. Multimodal imaging features of primary pericardial synovial sarcoma: A case report. Front. Oncol. 2023, 13, 1181778. [Google Scholar] [CrossRef]
- Juthani, R.; Singh, A.R.; Basu, D. A case series of therapy-related leukemias: A deadly ricochet. Leuk. Res. Rep. 2023, 20, 100382. [Google Scholar] [CrossRef] [PubMed]
- Olobatoke, A.O.; David, D.; Hafeez, W.; Van, T.; Saleh, H.A. Pulmonary carcinosarcoma initially presenting as invasive aspergillosis: A case report of previously unreported combination. Diagn. Pathol. 2010, 5, 11. [Google Scholar] [CrossRef]
- Liu, S.C.; Suresh, M.; Jaber, M.; Mercado Munoz, Y.; Sarafoglou, K. Case Report: Anastrozole as a monotherapy for pre-pubertal children with non-classic congenital adrenal hyperplasia. Front. Endocrinol. 2023, 14, 1101843. [Google Scholar] [CrossRef]
- Jin, J.; Xia, G.; Luo, Y.; Cai, Y.; Huang, Y.; Yang, Z.; Yang, Q.; Yang, B. Case report: Implantable cardioverter-defibrillator implantation with optimal medical treatment for lethal ventricular arrhythmia caused by recurrent coronary artery spasm due to tyrosine kinase inhibitors. Front. Cardiovasc. Med. 2023, 10, 1145075. [Google Scholar] [CrossRef]
- Xiang, Y.; Li, F.; Song, Z.; Yi, Z.; Yang, C.; Xue, J.; Zhang, Y. Two pediatric patients with hemiplegic migraine presenting as acute encephalopathy: Case reports and a literature review. Front. Pediatr. 2023, 11, 1214837. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Tanna, N. Maxillary protraction and vertical control utilizing skeletal anchorage for midfacial-maxillary deficiency. Dent. Press J. Orthod. 2021, 26, e2120114. [Google Scholar] [CrossRef]
- Al-Qattan, M.M.; Almotairi, M.I. Facial cutaneous lesions of dental origin: A case series emphasizing the awareness of the entity and its medico-legal consequences. Int. J. Surg. Case Rep. 2018, 53, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Al Kandari, J.; Al Sabti, K.; Wani, V.B. Partial-thickness macular hole in vitreomacular traction syndrome: A case report and review of the literature. J. Med Case Rep. 2010, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Koufakis, D.; Konstantopoulos, D.; Koufakis, T. An uncommon cause of progressive visual loss in a heavy smoker. Pan Afr. Med J. 2015, 22, 47. [Google Scholar] [CrossRef]
- Makri, O.E.; Tsapardoni, F.N.; Plotas, P.; Pallikari, A.; Georgakopoulos, C.D. Intravitreal aflibercept for choroidal neovascularization secondary to angioid streaks in a non-responder to intravitreal ranibizumab. Int. Med Case Rep. J. 2018, 11, 229–231. [Google Scholar] [CrossRef]
- Babu, K.; Murthy, V.; Akki, V.; Prabhakaran, V.; Murthy, K. Coin-shaped epithelial lesions following an acute attack of erythema multiforme minor with confocal microscopy findings. Indian J. Ophthalmol. 2010, 58, 64. [Google Scholar] [CrossRef]
- Nievas Offidani, M.; Roffet, F. MultiCaReClassifier. 2024. Available online: https://huggingface.co/mauro-nievoff/MultiCaReClassifier (accessed on 29 July 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Torchvision Maintainers and Contributors. M.N.O.: The Correct Name Is Torchvision Maintainers and Contributors. TorchVision: PyTorch’s Computer Vision Library. 2016. Available online: https://github.com/pytorch/vision (accessed on 29 July 2025).
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Howard, J.; Gugger, S. Fastai: A Layered API for Deep Learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Gonzalez Galtier, M.C.; Nievas Offidani, M.A.; Massiris, M. Clinical Case Hub. Available online: https://clinical-cases.streamlit.app/ (accessed on 29 July 2025).
- González Galtier, M.C.; Nievas Offidani, M.A. Digital Twin Retriever. Available online: https://digital-twin-retriever.streamlit.app/ (accessed on 29 July 2025).
- Lundh, A.; Christensen, M.; Jørgensen, A. International or national publication of case reports. Dan. Med Bull. 2011, 58, A4242. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).