Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strain, Inoculum Standardisation, and Cell Viability
2.2. Glass Surface
2.3. Wort Preparation
2.4. Biofilm Formation Parameters
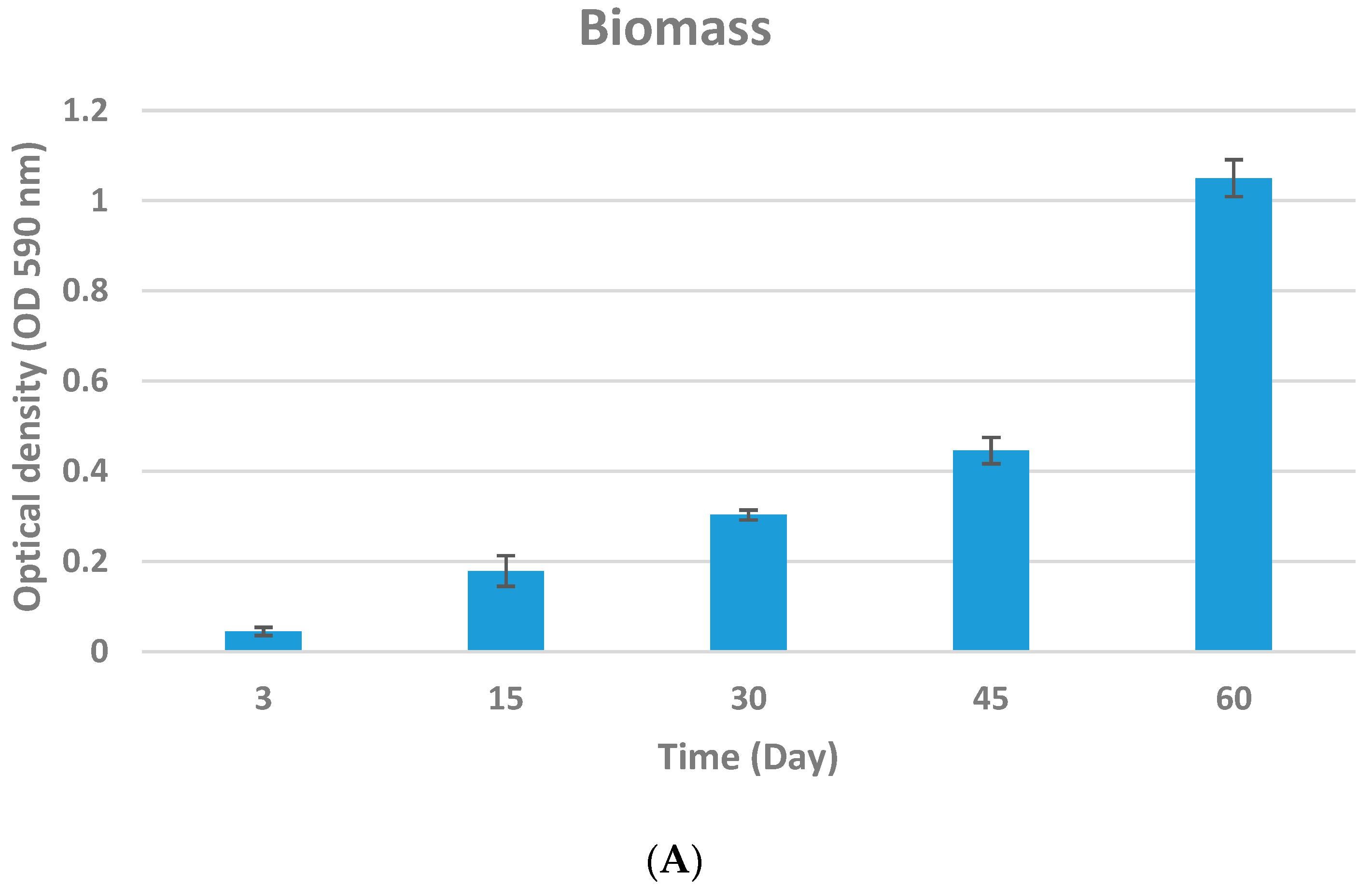
2.4.1. Biomass
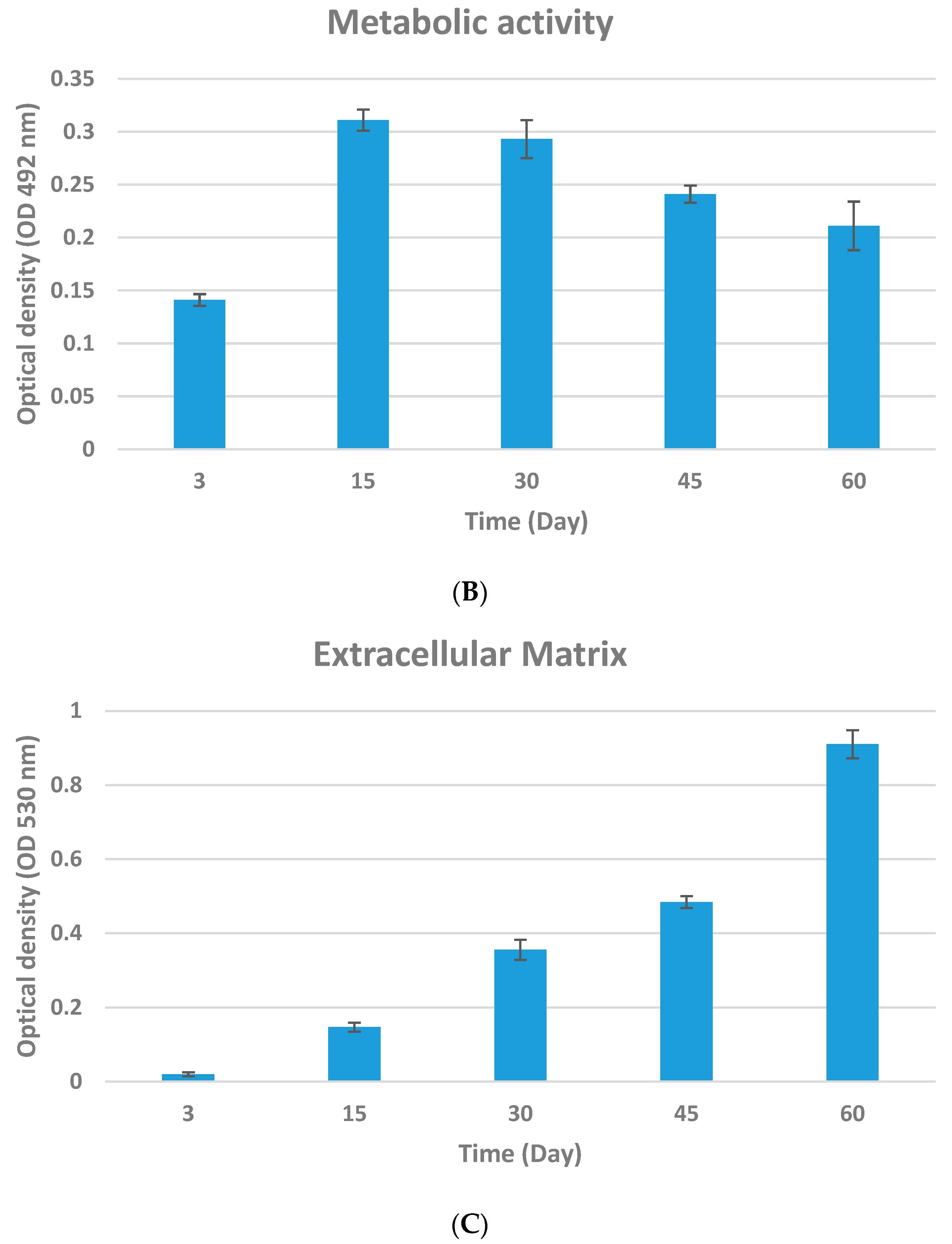
2.4.2. Metabolic Activity
2.4.3. Extracellular Matrix
2.5. Microscopic Visualisation of Biofilm S. boulardii Cells
2.6. HPLC Analysis
2.7. Transcription Analysis Using Real-Time Quantitative PCR (RT-qPCR)
2.8. Statistical Analysis
3. Results
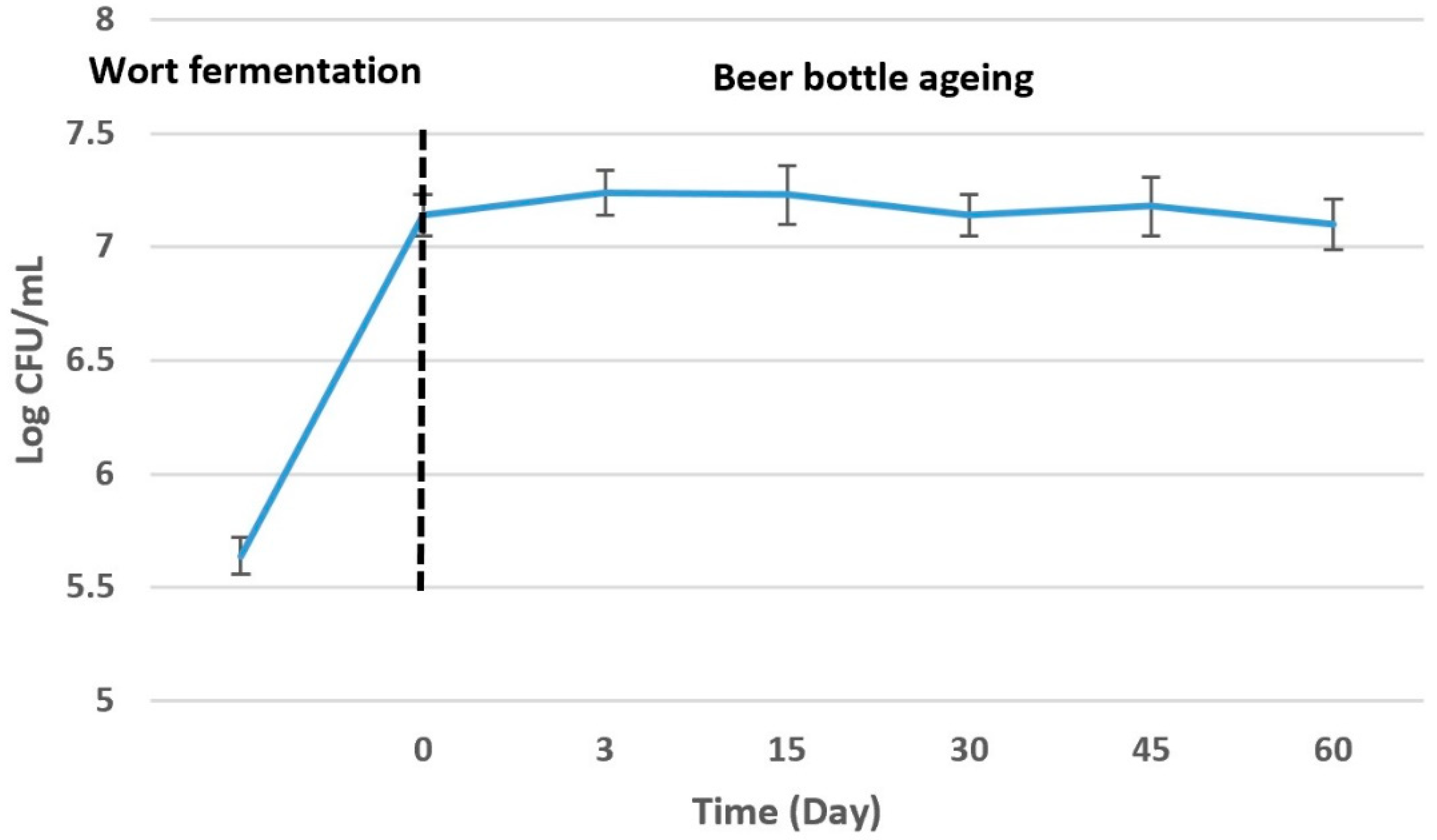
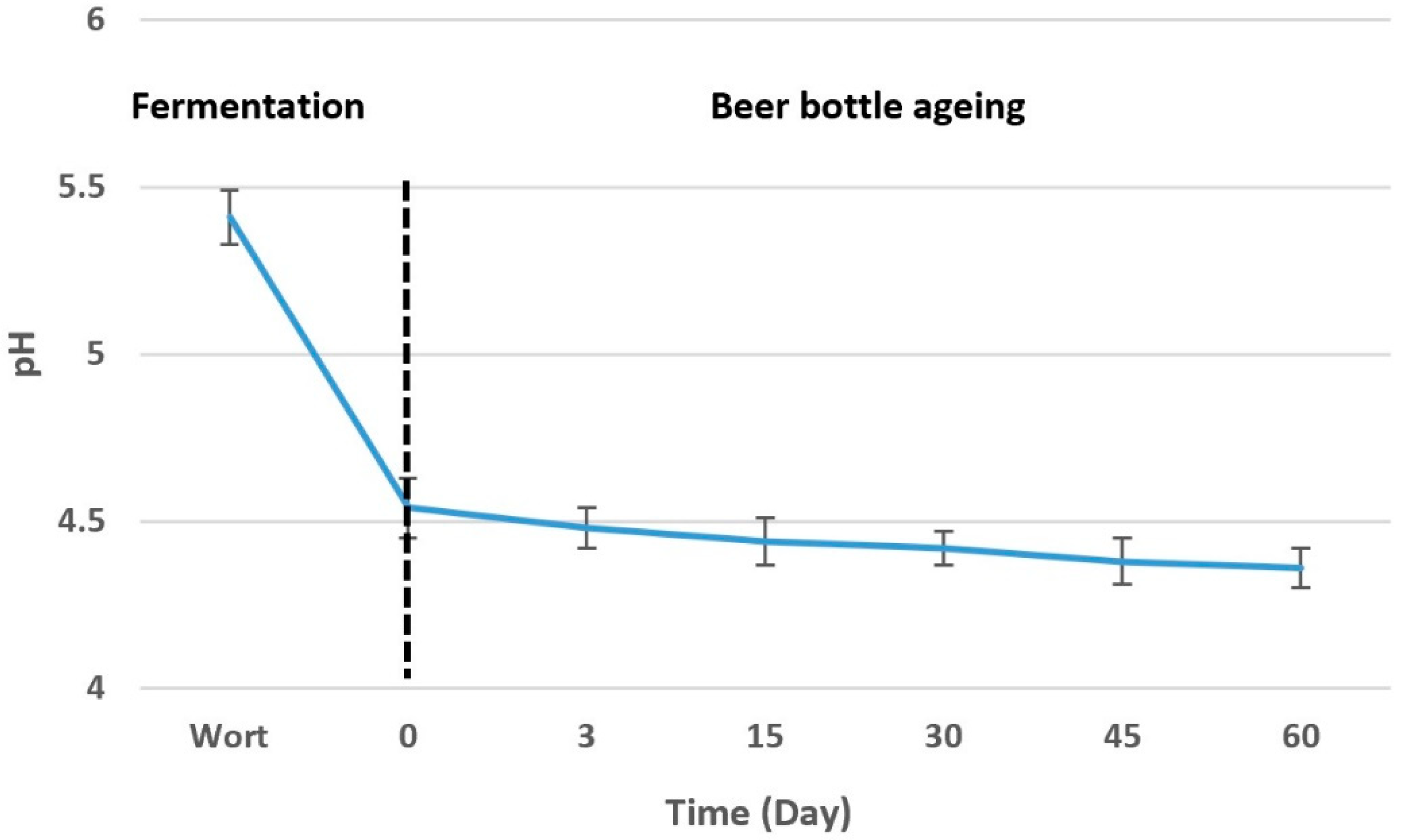
3.1. Metabolic Activity of S. boulardii
3.2. Biofilm Quantification
3.3. Biofilm Morphology
3.4. Expression Level of FLO11 Gene in Planktonic and Biofilm Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Riedl, R.M.A.F. Systems for Validation of Brewing Equipment Hygiene on the Basis of Characteristic Biofilm Organisms; mediaTUM, Universitätsbibliothek, Technische Universität München: Munich, Germany, 2020. [Google Scholar]
- Galiè, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Lolis, N.; Veldekis, D.; Moraitou, H.; Kanavaki, S.; Velegraki, A.; Triandafyllidis, C.; Tasioudis, C.; Pefanis, A.; Pneumatikos, I. Saccharomyces boulardii fungaemia in an intensive care unit patient treated with caspofungin. Crit. Care 2008, 12, 1–2. [Google Scholar]
- Richard, M.L.; Nobile, C.J.; Bruno, V.M.; Mitchell, A.P. Candida albicans Biofilm-Defective Mutants. Eukaryot. Cell 2005, 4, 1493–1502. [Google Scholar] [CrossRef]
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef]
- Yang, L.; Zheng, C.; Chen, Y.; Ying, H. FLO genes family and transcription factor MIG1 regulate Saccharomyces cerevisiae biofilm formation during immobilized fermentation. Front. Microbiol. 2018, 9, 1860. [Google Scholar] [CrossRef] [PubMed]
- Stringer, J.R.; Keely, S.P. Genetics of Surface Antigen Expression in Pneumocystis carinii. Infect. Immun. 2001, 69, 627–639. [Google Scholar] [CrossRef]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—A model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef]
- Teunissen, A.W.R.H.; Steensma, H.Y. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 1995, 11, 1001–1013. [Google Scholar] [CrossRef]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11 -Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef]
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 2007, 6, 2214–2221. [Google Scholar] [CrossRef]
- Silva, A.P.; Jager, G.; van Bommel, R.; van Zyl, H.; Voss, H.-P.; Hogg, T.; Pintado, M.; de Graaf, C. Functional or emotional? How Dutch and Portuguese conceptualise beer, wine and non-alcoholic beer consumption. Food Qual. Prefer. 2016, 49, 54–65. [Google Scholar] [CrossRef]
- Barnett, A.; Velasco, C.; Spence, C. Bottled vs. canned beer: Do they really taste different? Beverages 2016, 2, 25. [Google Scholar] [CrossRef]
- Back, W. Biofilme in der Brauerei und Getrankeindustrie-15 Jahre Praxiserfahrung. Brauwelt 2003, 24, 766–777. [Google Scholar]
- Flemming, H. Wingender: The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Vávrová, A.; Matoulková, D.; Balážová, T.; Šedo, O. MALDI-TOF MS Analysis of Anaerobic Bacteria Isolated from Biofilm-Covered Surfaces in Brewery Bottling Halls. J. Am. Soc. Brew. Chem. 2014, 72, 95–101. [Google Scholar] [CrossRef]
- Riedl, R.; Fütterer, J.; Goderbauer, P.; Michel, M.; Jacob, F.; Hutzler, M. Combined Yeast Biofilm Screening—Characterization and Validation of Yeast Related Biofilms in a Brewing Environment with Combined Cultivation and Specific Real-time PCR Screening of Selected Indicator Species. J. Am. Soc. Brew. Chem. 2019, 77, 99–112. [Google Scholar] [CrossRef]
- Wagner, E.M.; Thalguter, S.; Wagner, M.; Rychli, K. Presence of Microbial Contamination and Biofilms at a Beer Can Filling Production Line. J. Food Prot. 2021, 84, 896–902. [Google Scholar] [CrossRef]
- de Paula, B.P.; de Souza Lago, H.; Firmino, L.; Lemos Júnior, W.J.F.; Dutra Corrêa, M.F.; Guerra, A.F.; Pereira, K.S.; Zarur Coelho, M.A. Technological features of Saccharomyces cerevisiae var. boulardii for potential probiotic wheat beer development. LWT 2021, 135, 110233. [Google Scholar]
- Senkarcinova, B.; Dias, I.A.G.; Nespor, J.; Branyik, T. Probiotic alcohol-free beer made with Saccharomyces cerevisiae var. boulardii. LWT 2019, 100, 362–367. [Google Scholar] [CrossRef]
- Reitenbach, A.F.; Iwassa, I.J.; Barros, B.C.B. Production of functional beer with the addition of probiotic: Saccharomyces boulardii. Res. Soc. Dev. 2021, 10. [Google Scholar] [CrossRef]
- Li, R.; Yassami, S.; Kiviniemi, E.A.; Qiao, W.; Takala, T.M.; Saris, P.E. Listeria decontamination of chicken meat with beer brewed with bacteriocin producing Saccharomyces boulardii. LWT 2021, 152, 112323. [Google Scholar] [CrossRef]
- Rodriguez, E.; Martínez-Flores, H.; Rodiles-Lopez, O.; Zamora-Vega, R.; Salgado-Garciglia, R.; Pérez-Sánchez, E. Survival rate of Saccharomyces boulardii adapted to a functional freeze-dried yogurt: Experimental study related to processing, storage and digestion by Wistar rats. Funct. Foods Health Dis. 2017, 7, 98–114. [Google Scholar]
- Speranza, B.; Corbo, M.R.; Campaniello, D.; Altieri, C.; Sinigaglia, M.; Bevilacqua, A. Biofilm formation by potentially probiotic Saccharomyces cerevisiae strains. Food Microbiol. 2020, 87, 103393. [Google Scholar] [CrossRef] [PubMed]
- Muller, R. The production of low-alcohol and alcohol-free beers by limited fermentations. Ferment 1990, 3, 224–230. [Google Scholar]
- Muller, R. The effects of mashing temperature and mash thickness on wort carbohydrate composition. J. Inst. Brew. 1991, 97, 85–92. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Butcher, J.; Lang, S.; Williams, C.; Ramage, G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med Microbiol. 2007, 56, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Mello, T.P.; Aor, A.C.; Gonçalves, D.S.; Seabra, S.H.; Branquinha, M.H.; Santos, A.L.S. Assessment of biofilm formation by Scedosporium apiospermum, S. aurantiacum, S. minutisporum and Lomentospora prolificans. Biofouling 2016, 32, 737–749. [Google Scholar] [CrossRef]
- Mello, T.P.; Oliveira, S.S.C.; Frasés, S.; Branquinha, M.H.; Santos, A.L.S. Surface properties, adhesion and biofilm formation on different surfaces by Scedosporium spp. and Lomentospora prolificans. Biofouling 2018, 34, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.; Kačar, D.; Mali, T.; Lundell, T. Lignocellulose bioconversion to ethanol by a fungal single-step consolidated method tested with waste substrates and co-culture experiments. AIMS Energy 2018, 6, 866–879. [Google Scholar] [CrossRef]
- Purevdorj-Gage, B.; Sheehan, K.B.; Hyman, L.E. Effects of Low-Shear Modeled Microgravity on Cell Function, Gene Expression, and Phenotype in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006, 72, 4569–4575. [Google Scholar] [CrossRef] [PubMed]
- Narendranath, N.V.; Power, R. Relationship between pH and Medium Dissolved Solids in Terms of Growth and Metabolism of Lactobacilli and Saccharomyces cerevisiae during Ethanol Production. Appl. Environ. Microbiol. 2005, 71, 2239–2243. [Google Scholar] [CrossRef]
- Forehand, A.L.; Myagmarsuren, D.; Chen, Z.; Murphy, H.A. Variation in pH gradients and FLO11 expression in mat biofilms from environmental isolates of the yeast Saccharomyces cerevisiae. MicrobiologyOpen 2022, 11, e1277. [Google Scholar] [CrossRef] [PubMed]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Liu, D.; Zhao, N.; Cheng, H.; Ren, H.; Guo, T.; Niu, H.; Zhuang, W.; Wu, J.; et al. Involvement of glycolysis/gluconeogenesis and signaling regulatory pathways in Saccharomyces cerevisiae biofilms during fermentation. Front. Microbiol. 2015, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, P.; Plocek, V.; Váchová, L.; Palková, Z. Glucose, Cyc8p and Tup1p regulate biofilm formation and dispersal in wild Saccharomyces cerevisiae. npj Biofilms Microbiomes 2020, 6, 1–10. [Google Scholar] [CrossRef]
- Lo, W.-S.; Dranginis, A.M. The Cell Surface Flocculin Flo11 Is Required for Pseudohyphae Formation and Invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 161–171. [Google Scholar] [CrossRef]
- Kumamoto, C.A.; Vinces, M.D. Alternative Candida albicans lifestyles: Growth on Surfaces. Annu. Rev. Microbiol. 2005, 59, 113–133. [Google Scholar] [CrossRef]
- Livas, D.; Almering, M.J.; Daran, J.-M.; Pronk, J.T.; Gancedo, J.M. Transcriptional responses to glucose in Saccharomyces cerevisiae strains lacking a functional protein kinase A. BMC Genom. 2011, 12, 405. [Google Scholar] [CrossRef]
- Boulton, C.; Quain, D. Brewing Yeast and Fermentation; John Wiley & Sons: Hoboken, NJ, USA, 2008; p. 210. [Google Scholar]
- Stewart, G.; Zheng, X.; Russell, I. Wort Sugar Uptake and Metabolism—The Influence of Genetic and Environmental Factors. In Poceedings of Congress- European Brewery Convention; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Griffin, S.R. Maltase activity in Saccharomyces cerevisiae during fermentation of synthetic media. J. Inst. Brew. 1969, 75, 342–346. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Lee, W.G.; Chang, Y.K.; Chang, H.N. Modelling of ethanol production by Saccharomyces cerevisiae from a glucose and maltose mixture. Biotechnol. Lett. 1995, 17, 791–796. [Google Scholar] [CrossRef]
- Henriques, M.; Azeredo, J.; Oliveira, R. Candida albicans and Candida dubliniensis: Comparison of biofilm formation in terms of biomass and activity. Br. J. Biomed. Sci. 2006, 63, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Honraet, K.; Goetghebeur, E.; Nelis, H.J. Comparison of three assays for the quantification of Candida biomass in suspension and CDC reactor grown biofilms. J. Microbiol. Methods 2005, 63, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Balkis, M.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Uses and Limitations of the XTT Assay in Studies of Candida Growth and Metabolism. J. Clin. Microbiol. 2003, 41, 506–508. [Google Scholar] [CrossRef]
- Rollin-Pinheiro, R.; de Meirelles, J.V.; Vila, T.V.M.; Fonseca, B.B.; Alves, V.; Frases, S.; Rozental, S.; Barreto-Bergter, E. Biofilm Formation by Pseudallescheria/Scedosporium Species: A Comparative Study. Front. Microbiol. 2017, 8, 1568. [Google Scholar] [CrossRef]
- Beauvais, A.; Loussert, C.; Prevost, M.C.; Verstrepen, K.; Latgé, J.P. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 2009, 9, 411–419. [Google Scholar] [CrossRef]
- Santos, A.L.S.d.; Galdino, A.C.M.; de Mello, T.P.; Ramos, L.d.S.; Branquinha, M.H.; Bolognese, A.M.; Neto, J.C.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Memórias Do Inst. Oswaldo Cruz 2018, 113. [Google Scholar] [CrossRef]
- Abdo, H.; Catacchio, C.R.; Ventura, M.; D’Addabbo, P.; Calabrese, F.M.; Laurent, J.; David-Vaizant, V.; Alexandre, H.; Guilloux-Bénatier, M.; Rousseaux, S. Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages 2020, 6, 9. [Google Scholar] [CrossRef]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. The Extracellular Matrix of Fungal Biofilms. Fungal Biofilms Relat. Infect. 2016, 931, 21–35. [Google Scholar] [CrossRef]
- Mercier-Bonin, M.; Ouazzani, K.; Schmitz, P.; Lorthois, S. Study of bioadhesion on a flat plate with a yeast/glass model system. J. Colloid Interface Sci. 2004, 271, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Bayly, J.C.; Douglas, L.M.; Pretorius, I.; Bauer, F.F.; Dranginis, A.M. Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 1151–1156. [Google Scholar] [CrossRef]
- Kleyn, J.; Hough, J. The microbiology of brewing. Annu. Rev. Microbiol. 1971, 25, 583–608. [Google Scholar] [CrossRef]
- Blair, M.G.; Pigman, W. Fermentability of maltose. Arch. Biochem. Biophys. 1954, 48, 17–22. [Google Scholar] [CrossRef]
- Vincent, M.; Johnny, Q.; Adeni, D.S.A.; Suhaili, N. Potential of Candida glabrata from ragi as a bioethanol producer using selected carbohydrate substrates. Nusant. Biosci. 2020, 13. [Google Scholar] [CrossRef]
- Liu, F. Changes in organic acids during beer fermentation. J. Am. Soc. Brew. Chem. 2015, 73, 275–279. [Google Scholar] [CrossRef]
- Monošík, R.; Magdolen, P.; Streďanský, M.; Šturdík, E. Monitoring of monosaccharides, oligosaccharides, ethanol and glycerol during wort fermentation by biosensors, HPLC and spectrophotometry. Food Chem. 2013, 138, 220–226. [Google Scholar] [CrossRef]
- Zhao, X.; Procopio, S.; Becker, T. Flavor impacts of glycerol in the processing of yeast fermented beverages: A review. J. Food Sci. Technol. 2015, 52, 7588–7598. [Google Scholar] [CrossRef]

| Gene | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) | Length (bp) |
|---|---|---|---|
| FLO11 | AACCAAGTCCATCCCAACC | GCGAGTAGCAACCACATAAAG | 300 |
| 18S | TTAATGACCCACTCGGCAC | CACCACCACCCACAAAATC | 215 |
| Process | Time | Sugar Composition (g/L) | Metabolites Production (g/L) | |||
|---|---|---|---|---|---|---|
| Glucose | Maltose | Ethanol | Glycerol | Acetate | ||
| Wort fermentation | DF0 * | 15.29 ± 0.16 a | 55.64 ± 0.22 a | – | – | – |
| Beer bottle ageing | D0 ** | 6.36 ± 0.12 b | 33.31 ± 0.13 b | 17.35 ± 0.13 a | 1.68 ± 0.07 a | 0.34 ± 0.06 a |
| D3 | 2.52 ± 0.09 c | 32.93 ± 0. 09 b | 18.88 ± 0.16 a | 1.58 ± 0.07 a | 0.36 ± 0.09 a | |
| D15 | 1.99 ± 0.12 c | 30.06 ± 0.19 b | 17.23 ±0.11 a | 1.76 ± 0.08 a | 0.34 ± 0.07 a | |
| D30 | 1.94 ± 0.11 c | 23.61 ± 0.14 c | 19.21 ± 0.08 a | 1.85 ± 0.04 a | 0.38 ± 0.07 a | |
| D45 | 1.92 ± 0.07 c | 15.68 ± 0.08 d | 33.27 ± 0.14 b | 2.30 ±0.09 a | 0.34 ± 0.08 a | |
| D60 | 1.21 ± 0.08 c | – | 40.36 ± 0.12 c | 2.28 ± 0.06 a | 0.38 ± 0.08 a | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, K.; Saris, P.E.J. Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing. Beverages 2022, 8, 77. https://doi.org/10.3390/beverages8040077
Mohammadi K, Saris PEJ. Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing. Beverages. 2022; 8(4):77. https://doi.org/10.3390/beverages8040077
Chicago/Turabian StyleMohammadi, Khosrow, and Per Erik Joakim Saris. 2022. "Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing" Beverages 8, no. 4: 77. https://doi.org/10.3390/beverages8040077
APA StyleMohammadi, K., & Saris, P. E. J. (2022). Biofilm Formation of Probiotic Saccharomyces cerevisiae var. boulardii on Glass Surface during Beer Bottle Ageing. Beverages, 8(4), 77. https://doi.org/10.3390/beverages8040077







