Abstract
Nemea and Mantinia are famous wine regions in Greece known for two indigenous grape varieties, Agiorgitiko and Moschofilero, which produce high quality PDO wines. In the present study, indigenous Saccharomyces cerevisiae yeast strains were isolated and identified from spontaneous alcoholic fermentation of Agiorgitiko and Moschofilero musts in order to evaluate their oenological potential. Random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) recovered the presence of five distinct profiles from a total of 430 yeast isolates. The five obtained strains were evaluated at microvinifications trials and tested for basic oenological and biochemical parameters including sulphur dioxide and ethanol tolerance as well as H2S production in sterile grape must. The selected autochthonous yeast strains named, Soi2 (Agiorgitiko wine) and L2M (Moschofilero wine), were evaluated also in industrial (4000L) fermentations to assess their sensorial and oenological characteristics. The volatile compounds of the produced wines were determined by GC-FID. Our results demonstrated the feasibility of using Soi2 and L2M strains in industrial fermentations for Agiorgitiko and Moschofilero grape musts, respectively.
1. Introduction
Wine production, the process of gradually transforming grape sugars and other grape compounds to alcohol, carbon dioxide and other metabolites is mainly regulated by a wide range of microorganisms of different taxonomic orders, the most important of them, the yeast Saccharomyces cerevisiae is almost exclusively responsible for the final outcome of the fermentation [1]. S. cerevisiae is most commonly found in nature as a minor resident on the surface of grapes, usually being carried by insect vectors [2,3,4,5,6]. Additionally, the yeast species has an increased capacity to colonize the cellar and thus being part of the microflora of the winery [7]. Consequently, there is often the question if the strain that dominates the fermentation process comes from the vineyard or the winery [8].
Wine yeasts affect wine sensory profile by various mechanisms, such as the metabolism of the grape’s juice sugar and nitrogen components, the enzymatic hydrolysis of grape aroma precursors which affect wine aroma, flavour, and finally by the yeast’s cell autolysis and the bio-absorption [9,10]. More precisely the metabolism of yeast leads to a vast array of secondary fermentation by-products which act as flavor-impacting substances such as organic acids, glycerol, higher alcohols, esters, aldehydes, ketones, amines and sulphur volatiles [9,10,11,12]. Such complex volatile profiles are highly dependent not only on the yeast species but also on the yeast strains. Therefore, an extensive strain screening is necessary to select those with positive oenological attributes (e.g., enhanced ester formation, enhanced glycerol production) and reject those producing off-flavours (e.g., overproduction of hydrogen sulphide, acetic acid, etc.) [9,13].
S. cerevisiae affects a wine’s character by several biochemical reactions including: (a) the liberation of terpenes, C13-nor-isoprenoids and volatile phenols by its glycosidases [12,14,15,16], (b) the transformation of non-volatile thiols into volatile ones such as 4-mercapto-4-methylpentan-2-one (4MMP), 3-mercaptohexan-1-ol (3MH) and 3-mercaptohexyl acetate (3MHA) which result in desirable passion fruit, grapefruit, citrus and other aromas [17,18], (c) the bio-absorption of grape components (e.g., mycotoxins and microbial metabolites) by its cell’s wall glucan polysaccharides and mannoproteins [19], which also affects the colloidal stability of wines by interaction with the grape’s tartrates and proteins [20], and (d) the stabilization and the extraction of the anthocyanin pigments from the grape skins [20,21,22]. All previous characteristics vary within the strain of S. cerevisiae and need to be examined when selecting yeasts for starter culture development [9].
Unique strains of yeasts are associated with particular grape varieties in specific geographical locations and the significant diversity and regional character, or ‘terroir’, are introduced into the winemaking process via this association [23,24,25,26,27]. Terroir can be defined as an interactive ecosystem, in a given place, including climate, soil the vine as well as the native microflora [28,29]. In the past, wine fermentation was spontaneously carried out by indigenous yeasts. This method is still applied by several wineries in order to preserve the typicity (or regional character) of the wines and for marketing purposes. Previous studies showed that indigenous S. cerevisiae strains strengthen the terroir expression in the resulting wines [27,30].
The grapes of a region represent an important source of yeasts for starter culture development when trying to preserve both yeast biodiversity and the regional influence on the characteristics of a wine [9,13,27]. Selection of indigenous S. cerevisiae strains from their natural environments is a developing and important practice for wine industry in order to express the regional characteristics and to improve the quality of the resulting wines, or just simply differentiate the organoleptic character of the wines. Since the overall genetic variation between yeast strains has been estimated to be between 0.1 and 0.5% based on approaches using multilocus sequence typing, multilocus microsatellite analysis, genome sequencing and whole genome tiling arrays [31,32,33,34,35], molecular methodologies that allow the characterization of the yeast populations at the species and strain level have been widely developed. These methods include DNA fingerprinting methodologies commonly used in yeast population studies like, restriction fragment length polymorphism analysis of mitochondrial DNA (RFLP-mtDNA), pulsed field gel electrophoresis (PFGE) and RAPD-PCR analyses [26,36,37,38,39,40,41].
The purpose of this study was to isolate and characterize indigenous S. cerevisiae yeast strains from two protected designations of origin located in Greece. The isolated yeast strains were assessed as potential fermentation starters in order to enhance the expression of the regional characteristics of the produced wines. Indigenous S. cerevisiae yeast strains were isolated from spontaneously fermented grapes of cv. Agiorgitiko (a region of Nemea) and of cv. Moschofilero (a region of Mantinia). 201 and 229 S. cerevisiae yeasts were isolated from Agiorgitiko and Moschofilero, respectively, spontaneously fermented wines and two of them (one from each fermentation) were selected according to their unique RAPD-PCR profiles and their advanced sensorial and oenological characteristics. The aforementioned strains, symbolised as L2M (Moschofilero) and Soi2 (Agiorgitiko) were used in industrial fermentations of Agiorgitiko and Moschofilero grape musts. The isolation, selection and characterization of yeast strains, as well as their evaluation concerning their impact to wine quality based on laboratory to industrial fermentations, are also discussed in this study.
2. Materials and Methods
2.1. Growth Media, Yeast Storage and Industrial Inoculations
Yeasts isolates were routinely being kept in YPD liquid and YPD agar plates (1% w/v Yeast extract, 2% w/v Peptone, 2% w/v Glucose and 1.8% w/v Bacteriological agar). For preserving yeasts, 500 μL of overnight liquid cultures were being transferred in 1.5 mL eppendorf microcentrifuge tubes and supplied with 500 μL of 30% v/v glycerol. The mixtures were stored at −80 °C as biological banks.
For supplying indigenous yeasts as liquid cultures for industrial fermentations, S. cerevisiae strains were serially grown from 1 L Erlenmeyer Flasks-YPD cultures to 100 L of sterile rectified concentrated grape must of Savvatiano grape variety (Bé = 12 at 20 °C, pH = 3.45, total acidity 4.95 g/L). The yeasts were grown in 100 L of the sterilized water-diluted must in sterile 120 L Brupak’s brewing plastic vessels and were continuously supplied with O2 with a vacuum air-pump fixed with a microbiological filter membrane Millipore, Filter type HA, 0.45 μm pore size (Μerck Μillipore, 400 Summit Drive Burlington MA 1803, USA) at room temperature. The liquid yeast cultures were grown until concentration of 107 cells/ ml was achieved. At this concentration, 50 L of the liquid culture was used to ferment 5000 L of each grape must variety.
2.2. Isolation of Indigenous S. cerevisiae Yeast Strains from Spontaneous Fermentations of Agiorgitiko and Moschofilero Grape Musts
Two different spontaneous fermentations were performed in 5000 L stainless-steel fermenters, which contained 2000 L and 3000 L of Agiorgitiko (Nemea PDO, Domaine Zafeiri, Nemea, Korinthia, Greece, 205 00, 37.82025845094733/22.66105504910692) and Moschofilero (Mantinia PDO, Domaine Papadopoulos, Arkadia, Greece, 221 50, 37.617141117629984/22.38346160985462) musts, respectively, during the 2013 vintage. Fermentations were carried out until Bé was equal to 0, then density was measured down to 0.991 and the achievement of fermentation was verified by enzymatic analysis of glucose/fructose. After completion of fermentations, 50 mL sample from the mid-zone of each fermenter was taken out and stored into 50 mL sterile falcon conical tubes. The samples were placed at 20 °C for temporary storage. Serial dilutions of wine samples (10−1–10−10) were performed in 2% w/v of peptone solutions. A total of 100 μL of each dilution was used to inoculate YPD agar following incubation for 2 days at 28 °C. The dilution on which yeasts were clearly distinguishable was used for further analyses. All yeasts were first observed under an optical microscope in order to establish their cellular morphology following re-streaking into fresh YPD agar plates and analysed using the API 20 C Aux and ID 32 C strips (bioMérieux Hellas S.A., Athens, Greece, 152 32). Results were submitted into the Apiweb identification system [42] and according to this software all 201 and 229 yeasts isolates from Agiorgitiko and Moschofilero wines respectively belonged to the Saccharomyces cerevisiae species. Additional morphological analyses of the isolated yeast strains has also been performed after streaking quantities of glycerol stocks on Wallerstein agar (WLN agar) plates and incubating for 4–5 days at 28 °C.
2.3. DNA Extraction, RAPD-PCR Genotyping, rDNA-ITS Sequencing and Genetic Analysis of Indigenous Yeasts
For DNA isolation eight single colonies of freshly streaked glycerol stocks were diluted in 250 μL of 1 M sorbitol, 0.1 M EDTA, pH 7.5 and were sonicated for 4 min at 20% power, 50% pulser. The mixture was centrifuged for 15 min at 16,300× g, resuspended in 350 μL of 50 mM Tris-HCl, 20 mM EDTA, pH 7.4 and supplied with 25 μL of 20% w/v SDS following incubation at 65 °C for 30 min. An amount of 200 μL of ice-cold CH3COOK 5M were added and the resulting mixture was vortexed and transferred on ice for 30 min following centrifugation at 4 °C for 15 min at the maximum speed. The aqueous phase was transferred into a clean 1.5 mL microcentrifuge tube and after mixing it up with 1 mL of isopropanol it was incubated at room temperature for 5 min. This was then centrifuged for 15 min at maximum speed at 4 °C. Finally, the pellet was washed-up with 500 μL of 70% v/v ethanol, centrifuged for 15 min at maximum speed at 4 °C and followed by ethanol removal the pellet was resuspended in 50 μL ddH2O [43].
Two kinds of genotyping assays were performed. In the first assay, RAPD-PCR was used to analyse 201 and 229 S. cerevisiae yeast colonies from Agiorgitiko and Moschofilero wines, respectively. For this analysis, genomic DNA was isolated and yeasts were screened with the RAPD-12 primer (5′-ACGGCGATGA-3′). After the completion of this screen, the total number of the analyzed colonies was reduced to 5. In the second RAPD-PCR assay, the OPA-12 (5′-TCGGCGATAG-3′) and OPC-06 (5′-GAACGGACTC-3′) primers (Operon. Technologies, Alameda, CA, USA) were used to fingerprint the selected strains and to increase band polymorphism for genetic analyses.
RAPD-PCR reactions contained 200 ng of genomic DNA, 2 mM of MgCl2, 0.6 μM of each RAPD-PCR primer, 0.2 mM of each dNTP and 1U of the DreamTaq DNA polymerase, (Thermo Fisher Scientific Inc., Waltham, MA, USA) in 50 µL of final volume. The amplification reactions were carried out in a MJ research PTC-200 thermal cycler (MJ Research Inc., St. Bruno, QC, Canada). The reaction’s conditions are described as follows: initial denaturation at 94.0 °C for 2:00 min, 2 cycles at 94.0 °C for 0:30 min, 33.0 °C for 1:00 min, 72.0 °C for 2:00 min, followed by 6 cycles at 94.0 °C for 0:30 min, 35.0 °C for 1:00 min, 72.0 °C for 2:00 min, followed by 41 cycles at 94.0 °C for 0:30 min, 37.0 °C for 1:00 min, 72.0 °C for 2:00 min and a final extension at 72 °C for 10 min. RAPD-PCR profiles were analysed on 1% w/v agarose gel electrophoresis.
Images of DNA banding patterns obtained after agarose gel electrophoresis were analysed using the PyElph version 1.2 software [44] in order to prepare the phylogenetic dendrograms using the neighbour-joining method. Reproducibility of banding patterns for all isolates was also evaluated.
The universal primer set, (ITS1: 5′-TCCGTAGGTGAACCTGCGG-3′/ITS4: 5′-TCCTCCGCTTATTGATATGC-3′) was used to amplify the rDNA ITS regions from selected S. cerevisiae indigenous strains [45]. PCR-amplified ITS fragments were purified using the QIAquickGel Extraction kit, according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Each ITS purified fragment was sequenced in both directions with the ITS1 and ITS4 primers (Macrogen, Europe). rDNA-ITS sequences were deposited in GenBank as follows: (L2M, KP723678), (L26A, KP723679), (M9, KP723680), (Soi2, KP723681), (Soi103, KP723682).
Amplified ITS sequences were compared with the GenBank Nucleotide Database [46] using the algorithm BLAST N [47]. Putative rDNA-ITS sequences from various S. cerevisiae strains were retrieved from Genbank. The retrieved sequences were aligned with the CLUSTALX program by the multiple alignment method [48]. The multiple alignment resulting from CLUSTALX analysis was used as input for the GeneDoc version 2.7.000 program [49] to indicate nucleotide similarity. A phylogenetic tree was constructed using the neighbour-joining method. The constructed tree was analysed using TreeView version 1 software [50].
2.4. Sulphur Dioxide (SO2) and Ethanol Tolerance Tests
SO2 content was adjusted by adding potassium metabisulphite, to the rectified concentrated grape must of Savvatiano grape variety (Bé = 12 at 20 °C, pH = 3.45, total acidity 4.95 g/L). Strains were pre-cultured in sterile rectified concentrated grape must at 0.01 optical density measured at 660 nm (O.D.660). After reaching the desirable optical density, different amounts of total SO2 (0, 150, 300 ppm) were added. The cultures were then incubated for 24 h at 28 °C and optical densities were read. Yeast ethanol tolerance was assayed according to the maximum ethanol production method [51]. Aliquots of S. cerevisiae yeast strains were used to inoculate 100 mL of sterile rectified concentrated grape must of Savvatiano grape variety (Bé = 14.20 at 20 °C, pH = 3.45, total acidity 4.95 g/L) in 250 mL Erlenmeyer flasks at a concentration of 1 × 107 cells/mL. Fermentation lasted 10 days at 28 °C under static culture conditions. During fermentation, sucrose was added to maintain 14.20 Βé in the mid-stages of the fermentation and 4.20 Βé in the final stages [51].
All experiments were performed in triplicates and data were expressed as mean ± SD of three experimental replicates.
2.5. H2S Production Tests
For BiGGY (Bismuth Glucose Glycine Yeast) agar screen assays, cells were directly streaked from −80 °C glycerol stocks on BiGGY agar plates and then incubated for 36 h at 28 °C. Additionally a second one relative H2S production assay was performed using the membrane overlay method, which was developed by [52]. Strains were pre-cultured in sterile rectified concentrated grape must at 0.01 optical density measured at 660 nm (O.D.660). After reaching the desirable optical density, 250 μL of each culture was added in a Corning® Costar® 24 well cell culture plate (Merck KGaA, Darmstadt, Germany). A Whatman® 3MM Chromatography paper was cut to the well’s size and then submerged on a freshly prepared 20% (w/v) silver nitrate (AgNO3) solution. The membrane was blotted to remove excess liquid and laid over the microtiter plate wells. After sealing with parafilm and the microplate’s lid, the plates were wrapped in aluminium foil in order to protect the reaction from light and incubated statically at 28 °C. After 24 h of incubation, the strip was photographed using a digital camera.
2.6. Analytical Methods
Analytical methods were based on OIV’s International Methods of Analysis of Wines and Musts: (a) ethanol content: OIV-MA-AS312-01A (distillation + pycnometry), (b) volatile acidity: OIV MA-AS313-02 (steam distillation + titration), (c) total acidity: OIV MA-AS313-01 (titration with bromothymol blue), (d) YAN: internal method enzymatic (ΥAΝ), (e) density: OIV-MA-AS2-01A (pycnometry), (f) pH: OIV –MA-AS313-15-PH (pHmeter), g) free SO2: OIV-MA-AS323-04A (titrimetry), (h) total SO2: OIV-MA-AS323-04A (Titrimetry), (i) glucose/fructose (total sugars): OIV-MA-AS311-02 (enzymatic), k) colour intensity/ colour shade: OIV-MA-AS2-07B (spectroscopy). Dry weight was measured after drying 1 mL of fermentation samples at 65 °C overnight. The samples were then weighted in a Kern Model ALS 120-4 analytical balance (KERN & SOHN GmbH, Balingen, Germany).
Volatile compound determination was performed using solid phase micro-extraction followed by GC-FID detection. A total 10 mL of the wine sample was supplemented with 10 μL of the internal 3-octanol (1 g/L) (Merck KGaA, Darmstadt, Germany)and then placed to a 40 mL vial at 20 °C with 3 g NaCl (Merck KGaA, Darmstadt, Germany) and a magnetic stir bar. The vial was sealed with a screw-top cap with a silicon septa. The mixture was equilibrated by magnetic stirring at 750 rpm for 5 min at 30 °C. Then the SPME needle inserted manually through the vial septum and the fiber (CAR/PDMS 75 μm) was exposed to the headspace of the sample for 30 min at 30 °C. The fibre then was retracted, the SPME device removed from the vial and inserted into the injector of GC for thermal desorption for 10 min. The analysis of the target volatile compounds was performed using a Hewlett-Packard 5890 II GC (Agilent, Santa Clara, CA, USA). The column used was a DB-WAX (30 m × 0.32 mm i.d., 0.25 μm film thickness) (Agilent, Santa Clara, CA, USA) and the gas carrier was helium with a flow rate of 1 mL/min. The injector was maintained at 250 °C and the FID detector at 290 °C, respectively. Oven temperature was held at 30 °C for 3 min and raised to 230 °C at 3 °C/min and remained at this temperature for further 10 min. Calibrations were achieved for each analyte using reference standards and model wine as a media.
2.7. Microvinifications
Microvinifications were performed in sterile concentrated grape must of Savvatiano grape variety. Bé were adjusted at 12, pH = 3.45 and total acidity at 4.95 g/L. Fermentations were performed in sterilized 30 L fermenters each containing 20 L of must. The temperature was adjusted at 28 °C. Yeast pitching was performed by streaking both industrial and indigenous yeast strains in agar YPD plates. Single colonies were first grown in 5 mL YPD medium and after shaking for 2 days at 28 °C the cultures were re-inoculated in 500 mL of grape must. Absorbance at 660 nm was measured and appropriate amounts of cells were inoculated in 20 l of grape must in order to achieve concentrations of 1 × 107 cells/mL. The experiments were performed in triplicates.
2.8. Industrial-Scale Vinifications
Industrial vinifications were carried out in 5000 L variable capacity conical bottom stainless steel wine tanks each containing 4000 L of must. For Agiorgitiko and Moschofilero fermentations, 8 g/hL of metabisulfite was added while for Agiorgitiko variety, cold maceration was performed at 15 °C for 2 days. Moschofilero grapes were pressed directly using a pneumatic press and the juice was decanted down to ~80 NTU prior fermentation. After YAN correction, yeasts were inoculated as described previously. The temperature was kept below 20 °C. All fermentations were sampled daily (or every 2 days) for temperature and sugar content (Bé) measurements. At the end of the fermentations, white wines were adjusted by adding 8 g/hL of metabisulfite while red wines were let to proceed to malolactic fermentations. All experiments were performed in triplicates and data were expressed as mean ±SD of 3 experimental replicates.
2.9. Sensory Evaluation
Sensory evaluations were conducted according the global sensory quality assay [53,54,55]. The wines were evaluated by a panel of 17 wine experts with previous experience in sensory analysis of wines. The subjects were asked to rate the global sensory quality by assigning it a value ranging from 1 to 9. Three independent tests were carried out for each replicate of fermentation (total 9 tests). Wines were offered in clear glass bottles and aliquot samples were served in wine-taster glasses. Evaluations were conducted at room temperature. The final scores were normalized on a scale from 1 to 10. According to the sensory quality system a score of 9–10 is equal to a match (the sample has virtually identical sensory characteristics to the control by appearance, aroma, flavour and texture), a score of 6–8 is equal to acceptable (the sample meets the definition for the product, but has differences that are easily spotted when comparing side by side with the control), a score of 3–5 is equal to unacceptable [52].
2.10. Statistical Analysis
Statistical analyses were performed by Student’s t-test and ANOVA followed by the Tukey’s post hoc test. All analyses were performed using the IBM SPSS software (IBM Analytics). Differences were considered to be significant at the p < 0.05 level.
3. Results
3.1. Molecular Characterization of Agiorgitiko and Moschofilero S. cerevisiae Strains
Since the target of the study was to isolate the most phylogenetically scarce S. cerevisiae yeast strains, from both Agiorgitiko and Moschofilero wines fermented spontaneously, a search for appropriate RAPD-PCR primers was conducted.
A total of 201 yeasts isolates from Agiorgitiko and 229 yeasts isolates from Moschofilero were DNA extracted and analyzed in RAPD-PCR assays using the RAPD12 primer. RAPD12 primer resulted in three most abundant types of RAPD-PCR patterns. In Pattern A, six major DNA bands were observed, a ~1250 bp (strong), a band of ~800 bp, two bands of ~600–750 bp, one band of ~500 bp and finally one band of ~350 bp (Figure 1). In Pattern B, two major bands were observed, a ~1250 bp and a ~350 bp (both strong) (Figure 1). In Pattern C, only one band was observed, this of ~350 bp (Figure 1).
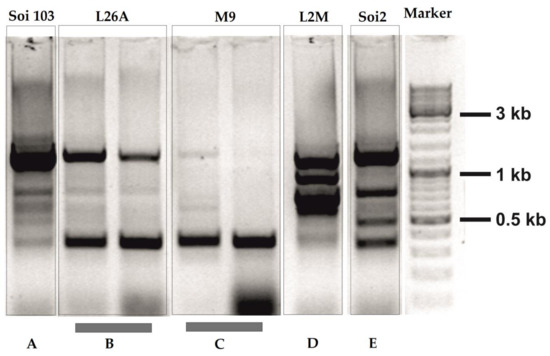
Figure 1.
RAPD-PCR analysis of Agiorgitiko and Moschofilero indigenous yeast strains primed-off with the RAPD-12 primer. (A–E) represent the 5 RAPD-PCR patterns which were detected among a total population of 201 yeasts from Agiorgitiko and 229 yeasts from Moschofilero spontaneously fermented musts. L26A, L2M and M9 represent autochthonous yeast strains isolated from spontaneous fermentation of Moschofilero must, while Soi2 and Soi103 represent autochthonous yeast strains isolated from spontaneous fermentation of Agiorgitiko must. Commercial strains: SC22 (Fermentis), Saint Georges S101 (Fermentis), 58W3 (Martin Vialatte).
Furthermore, the three kinds of patterns (A,B,C) were observed in the isolated yeasts of both Agiorgitiko and Moschofilero varieties respectively (Figure 1). Two strains out of 201 (1%) from Agiorgitiko and two strains out of 229 (0.87%) from Moschofilero spontaneously fermented musts presented unique RAPD-PCR patterns when compared with the dominant ones. In the case of Moschofilero, the unique pattern consisted of 4 (strong) bands of ~1200 bp, ~1000 bp, ~800 bp and ~750 bp respectively (Pattern D), and the strain was named as L2M while in the case of Agiorgitiko the unique pattern consisted of four (strong) bands of ~1250 bp, ~800 bp, ~450 bp and ~350 bp (Pattern E), and the strain was named as Soi2 (Figure 1).
One representative strain from each RAPD-PCR pattern was further analysed in molecular microbiological and biochemical assays. In order to increase RAPD sensitivity, we performed a second round of RAPD-PCR screens, using OPA-12 and OPC-06 primers, which have extensively been used in the past for phylogenetically analysing indigenous yeast strains [56,57,58].
Using the aforementioned primers, we analyzed genomic DNA isolated from the 5 representative indigenous S. cerevisiae yeast strains which were described in the previous paragraph and from three commercial yeast strains that have been used extensively from the two wineries in the past. The commercial yeast strains were (a) the SC22 (Fermentis), which is particularly adapted to cultivars like Cabernet Sauvignon, Cabernet Franc and Merlot, (b) the Saint Georges S101 (Fermentis) which is adapted to cultivars like Pinot Noir & Gamay as well as to all other light-body red varieties intended to produce wines with a rapid turnover (Young Tempranillo, Sangiovese, Cinsault, Pinotage, Grenache), and (c) 58W3 strain (Martin Vialatte).
RAPD-PCR analysis with OPA-12 and OPC-06 primers showed a clear phylogenetic relationship among all strains tested (Figure 2). OPC-06 resulted in bands of low polymorphism so it was discarded from further analysis (Figure 2). The RAPD-PCR polymorphisms that were generated using OPA-12 primer were further analysed with the PyElph 1.4 software. To prepare the dendrogram we used the neighbour-joining method [59]. Reproducibility of banding patterns for the eight strains was also evaluated. Phylogenetic analysis revealed that the two Moschofilero isolates (L2M and M9) grouped together in a separate clade from the other yeast strains (L26A, Soi2, Soi103, 58W3 and Sc22) while the Soi2 strain which was isolated from the Agiorgitiko spontaneous fermentation was grouped in a second clade separated from the L26A, Soi103, 58W3 and Sc22 ones (Figure 3). Further, 58W3, Soi103, S101, Sc22 and L26A seemed to be the most phylogenetically related compared with the other strains (Figure 3).
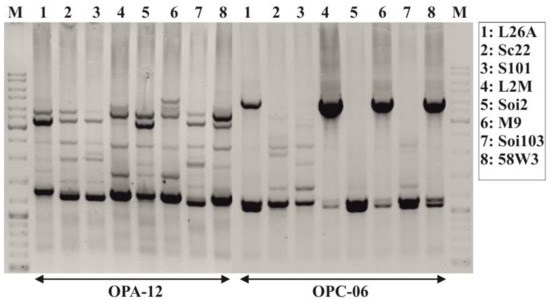
Figure 2.
RAPD-PCR analysis of Agiorgitiko and Moschofilero indigenous yeast strains primed-off with the OPA-12 and OPC-06 primers. M: represents the molecular marker. L26A, L2M and M9 represent autochthonous yeast strains isolated from spontaneous fermentation of Moschofilero must, while Soi2 and Soi103 represent autochthonous yeast strains isolated from spontaneous fermentation of Agiorgitiko must. Commercial strains: SC22 (Fermentis), Saint Georges S101 (Fermentis), 58W3 (Martin Vialatte).
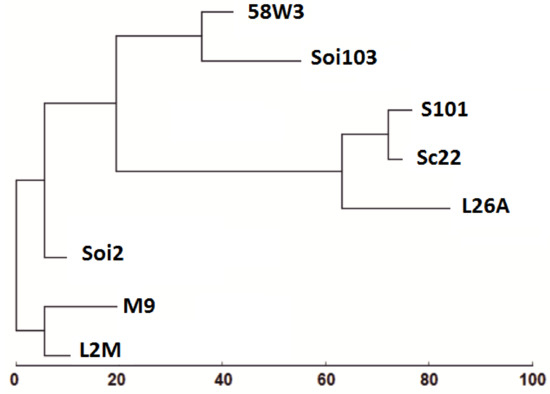
Figure 3.
Phylogenetic analysis of RAPD-PCR patterns from Agiorgitiko and Moschofilero indigenous yeast strains primed-off with the OPA-12 primers. L26A, L2M and M9 represent autochthonous yeast strains isolated from spontaneous fermentation of Moschofilero must, while Soi2 and Soi103 represent autochthonous yeast strains isolated from spontaneous fermentation of Agiorgitiko must. Commercial strains: SC22 (Fermentis), Saint Georges S101 (Fermentis), 58W3 (Martin Vialatte).
In order to validate the results of RAPD-PCR analysis, we sequenced the internal transcribed spacer (ITS) rDNA sequences of the isolated indigenous yeast strains and this of the commercial 58W3 strain. ITS sequences were subjected to BLAST analysis against other sequenced S. cerevisiae strains. The retrieved sequences were first subjected to multiple sequence alignments (Figure S1) and then were analysed using the PyElph 1.4 software. Phylogenetic analyses showed that M9 and Soi2 were grouped together with yeast strains isolated from the Montepulciano d’Abruzzo wines [60] and with the YJM993 model yeast strain (Figure 4). Additionally, L2M and Soi103 were grouped with the AWRI1631 and the reference S288C yeast strains respectively (Figure 4) while the L26A and 58W3 were grouped with a vast variety of yeast strains like JAY291, CHFY0321, T73, TB24 and HA1829 (Figure 4).
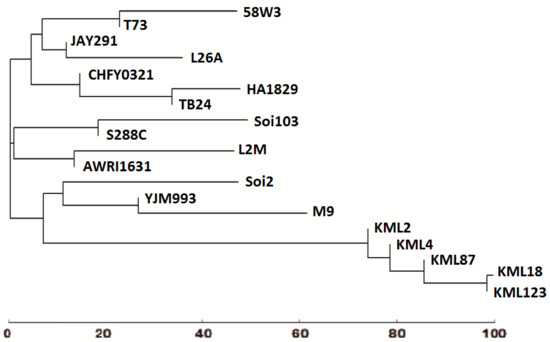
Figure 4.
Phylogenetic analysis of rDNA ITS sequences of Agiorgitiko and Moschofilero indigenous yeast strains compared with BLAST-NCBI retrieved ITS rDNAs of different S. cerevisiae yeast strains. L2M (GenBank KP723678), L26A (GenBank KP723679), M9 (GenBank KP723680), Soi2 (GenBank KP723681), Soi103 (GenBank KP723682). KML2 (GenBank KF486910), KML4 (GenBank KF486909), KML18 (GenBank KF486908), KML87 (GenBank KF486911) and KML123 (GenBank KF486912) were isolated from the ‘terroir’ of the Montepulciano d’Abruzzo ‘Colline Teramane’, [61], S288C strain is the reference genome sequenced S. cerevisiae strain while T73, AWRI1631, YJM993 and JAY291 represent additional genome sequenced yeast strains. TB24 was isolated from tree-saps and flowers at Ise-Shima area (GenBank: AB910265). HA1829 isolated from Austrian vine-growing regions ([62], GenBank: AM262826), while CHFY0321 represents a flocculent Saccharomyces bayanus × Saccharomyces cerevisiae hybrid which was generated by protoplast fusion between Saccharomyces cerevisiae and Saccharomyces bayanus ([63], Genbank: EU719073).
3.2. Microvinifications Trials with Indigenous S. cerevisiae Strains
Microvinifications using sterile concentrated grape must of Savvatiano grape variety were performed in order to evaluate the fermentation kinetics and assess the oenological characteristics of the selected strains. The commercial yeast strain 58W3 was used as a reference strain for comparison reasons. This strain is commonly used for fermentations of white grape musts. Fermentations were completed in the fifth day post inoculation for the L2M, Soi2, Soi103 and 58W3 yeast strains, while for M9 and L26A fermentations were completed in the eighth day post inoculation (Table S1; Figure S2). Residual YAN (initial 131.98 mg/L) was almost similar for L2M, L26A, M9, Soi2 and Soi103, while for the commercial yeast strain was insignificantly higher (ANOVA, p > 0.05) (Table S1; Figure S2). Residual dry mass was significantly higher for Soi2, Soi103 and L26A yeast strains comparing to the M9 and 58W3 ones (ANOVA, p < 0.05) (Table S1; Figure S2).
Moreover, the ethanol content of the completed wines was the same for all yeast strains tested (ANOVA, p > 0.05) (Table S1.). Soi2 and M9 resulted in statistically lower reducing sugar concentrations (ANOVA, p < 0.05), while L26A and M9 strains resulted in the higher volatile acidity levels (ANOVA, p < 0.05) (Table S1). Total acidity and pH levels were almost similar for all strains tested while the L2M strain resulted in the statistically lower free SO2 concentration (ANOVA, p < 0.05) (Table S1). Final density was almost equal for all strains tested (Table S1).
Basic biochemical parameters like SO2 and alcohol resistance as also H2S formation were also evaluated. The strains were separated in 4 categories according to the H2S formation: (a) M9 and 58W3: high H2S formation, (b) Soi2 and Soi103: moderate H2S formation, (c) L26A moderate low: H2S formation, and (d) L2M: low H2S formation (Figure S3). Moreover, alcohol and SO2 resistance assays showed that all strains were tolerant above the concentration of 14% v/v alcohol and in 300 ppm of SO2 (Figure S4). Finally, sensory evaluation of the produced wines was performed in sterile concentrated grape must of Savvatiano by selected indigenous yeast strains. According to average score tasting the significantly higher rates were observed for the wines fermented with the strain L2M and Soi2 (Table 1) and thus these two strains were selected for their further study at industrial level vinifications trials.

Table 1.
Sensory evaluation of microvinificated wines with indigenous yeast strains. Data are expressed as mean ± SD of 9 experimental replicates (three independent tests were carried out for each replicate of fermentation).
3.3. Industrial Fermentations
The combination of phylogenetic, biochemical and oenological analyses revealed that Soi2 (Agiorgitiko) and L2M (Moschofilero) seem to be the best candidates for proceeding into industrial fermentations of Agiorgitiko and Moschofilero musts. As a result, during the 2014 vintage these strains were used in order to perform industrial vinifications. A total of three industrial vinifications per yeast strain were performed in 5000 L capacity stainless steel wine tanks. Both Soi2 and L2M were used for fermenting both Agiorgitiko and Moschofilero musts. For Moschofilero fermentation, it had been used as reference, a commercial yeast strain commonly associated with Moschofilero wine production (SC22) while for Agiorgitiko fermentation a commercial yeast strain commonly associated with Agiorgitiko wine production (STG S101). The behaviour of the fermentation was checked daily by temperature and sugar content measurements. Average sugar content consumption slopes were calculated during fermentations while wine’s analytical parameters were calculated after the completion of the fermentations.
Our results showed similar fermentation behaviour for L2M and Soi2 strains in both Moschofilero and Agiorgitiko fermentations when compared to the SC22 and STG S101 reference yeasts (Figure S5). For Moschofilero fermentations, similar analytical characteristics were observed for both indigenous strains compared to the control SC22 strain (Table S3). Ethanol content, free and total SO2, pH, total acidity and density were similar in all strains tested (Table S3). SC22 resulted in the statistically lowest concentration levels of reducing sugars in Moschofilero fermentations (ANOVA, p < 0.05) (Table S3) while both indigenous yeast strains resulted in higher but not prohibitive volatile acidity levels (ANOVA, p < 0.05) (Table S3). For Agiorgitiko fermentations similar analytical characteristics were observed between all strains tested considering ethanol content, free and total SO2, pH, total acidity and density (Table S4). Further, in this case the L2M resulted in statistically higher volatile acidity and reduced sugar levels (ANOVA, p < 0.05) (Table S4), while Soi2 and STG S101 resulted into wines with similar characteristics regarding these two parameters. Soi2 resulted in statistically higher colour density and similar hue levels (ANOVA, p < 0.05), (Table S4).
3.4. Sensory Evaluation and Volatile Compound Identifications
For Moschofilero and Agiorgitiko wines, sensory evaluation tests resulted in almost similar scores (Figure 5). According to the global sensory quality, including the evaluation of wine appearance, aroma, flavour and texture, the wines produced with the indigenous S. cerevisae strains resulted in similar sensory profiles when compared to the control ones. More precisely, the average tasting score for both fermented must varieties, Moschofilero and Agiorgitiko, were similar for L2M and Soi2 as well as with the two commercial (SC22 and STG S101) strains. Nevertheless, the two commercial reference strains exhibited greater variability to the tasting score between the two grape varieties compared to the indigenous strains.
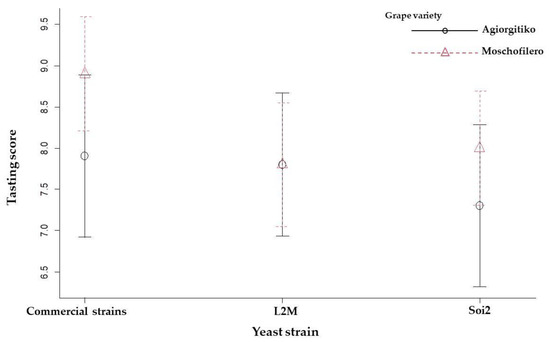
Figure 5.
Plot for max, min and mean values of wine tasting scores by two-factor interaction: the grape variety (Agiorgitiko and Moschofilero) and the yeast strain (Commercial strains, L2M and Soi2). The commercial strains, SC22 and STG S101, were used as references strains.
GC-FID analysis showed similar volatile compounds profile for all strains tested in all industrial fermentations for most of the analysed volatiles. Significant variations were observed for ethyl-butyrate, ethyl-isovalerate, isoamyl-alcohol, 2-phenyl-ethyl acetate and phenethyl alcohol in which concentrations of 0.206 ± 0.007, 0.135 ± 0.009, 608.241 ± 11.514, 0.338 ± 0.225, 109.168 ± 4.732 and 0.189 ± 0.043, 0.117 ± 0.001, 371.814 ± 10.033, 0.046 ± 0.079, 35.920 ± 4.701 mg/L were detected for Soi2 and L2M yeasts respectively (Table 2 and Table 3).

Table 2.
Concentration of volatile fermentation products for industrial fermented Moschofilero wines with Soi2 and L2M indigenous yeast strains. SC22 was used as a reference strain. Data are expressed as mean ± SD of 3 experimental replicates.

Table 3.
Concentration of volatile fermentation products for industrial fermented Agiorgitiko wines with Soi2 and L2M indigenous yeast strains. STG S101 was used as a reference strain. Data are expressed as mean ± SD of 3 experimental replicates.
Additionally, for Agiortgitiko fermentations, ethyl-isovalerate, isoamyl-alcohol and phenethyl-alcohol varied significantly for Soi2 and L2M indigenous yeast strains (Table 3). Ethyl-isovalerate was detected at concentrations of 0.069 ± 0.062 and 0.137 ± 0.057 mg/L for Soi2 and L2M yeast strains comparing with the reference STG S101 yeast in which was not detectable while for phenethyl alcohol the significant higher concentration of 98.756 ± 36.505 mg/L was observed in the commercial STG S101 yeast. Isoamyl alcohol levels were significantly higher in Soi2 and L2M indigenous S. cerevisiae strains (Table 3).
4. Discussion
The increasing need of wine industry for new Saccharomyces cerevisiae yeast strains with even more promising oenological features as potential must fermentation starters for industrial applications led to discoveries of new molecular, biochemical and technical approaches which could be directly applied. These approaches involve the isolation of yeast strains from their natural habitats followed by an extended discrimination using phylogenetic and qualitative markers [9,10,62,63,64,65,66,67]. The qualitative traits of S. cerevisiae to be evaluated in the selection of wine yeast strains consider, (a) fermentation by-products, (e.g., glycerol, acetic acid, H2S, SO2 etc.), (b) aroma and flavour compounds (higher alcohols: 2-phenylethanol, isoamyl-alcohol, ethyl-esters of fatty acids: ethyl-butanoate, ethyl-hexanoate and acetates: isoamyl-acetate, hexyl-acetate), and (c) phenolic compounds (e.g., anthocyanins). The concentration of these compounds varies based upon yeast species and among yeast strains [9,60,68,69].
In the present study, the use of molecular markers contributed to the isolation of indigenous S. cerevisiae yeast strains from spontaneous fermentations of Agiorgitiko and Moschofilero grape musts and were used as must fermentation starters for industrial fermentations. Phylogenetic analyses showed RAPD-PCR profiles, distributed among different populations of S. cerevisiae, which were isolated from two different wineries of Greece. A low percentage of the analyzed yeasts were phylogenetically unique and specific between the two regions. Geographically common yeasts one from each phylogenetic profile (named as Soi103, M9 and L26A) and two region-specific yeasts (named as L2M for Moschofilero and Soi2 for Agiorgitiko), were used for further technological assessment while compared to reference commercial yeast strains.
Our isolated strains seem to belong to the S. cerevisiae species using carbon assimilation, RAPD-PCR and rDNA ITS sequencing assays. However, we cannot exclude the possibility that these isolates belong to the Saccharomyces bayanus and/or the Saccharomyces kudriavzevii species or to their potential hybrids as it was proposed by König and Claus, 2018 [70]. Therefore, our ethanol tolerance assays showed a range of restricted tolerance fluctuating from 14% to 15% ethanol content. This seems to be sufficient enough to minimize the possibility to have isolated S. bayanus. Additionally rDNA ITS BLAST analysis best matched S. cerevisiae strains. To further elucidate this possibility we performed an extended genomic sequencing to our selected strains. Our work showed that the isolates have produced high quality wines.
According to our results, five strains were identified from the two viticulture regions. Previous studies showed that commonly used agricultural practices have a great influence to the microbial diversity of the local farm ecosystem [71,72,73,74,75]. Depending on the agronomic practices, differences in the composition and distribution of yeast populations in the vineyard have been described [74], but certain oenological practices applied in the winery may also affect the surrounding vineyard yeast diversity [73,75]. To our knowledge, the grapes of which we used to perform the spontaneous fermentations from both vineyards have been extensively using conventional agricultural practices and a wide spectrum of commercial yeast strains for inoculating their fermentations.
S. cerevisiae strains generally possess the technological characteristics required to perform an efficient fermentation process [9,10,62,63,64,65,66,67]. The determination of these traits is, however necessary, since most of these characteristics can vary among the strains: ethanol tolerance, fermentation vigour, sugar depletion, resistance to SO2, type of growth in liquid media, dispersed cells, aggregates cells, flocculence, foam formation, film formation, sedimentation speed growth at high and low temperatures, and the presence of killer factors, etc. [9,64,69]. For Soi103, M9 and L26A, yeasts undesirable oenological characteristics were observed; L26A and M9 strains resulted in wines with relatively high volatile acidity levels, while M9 was produced high amounts of H2S. Even if fermentation kinetics, ethanol tolerance and SO2 resistance were similar for all indigenous yeast strains tested, comparing with the commercial reference yeast (with the exception in dry mass production which was lower in the commercial strain), sensory evaluation results for the wines produced by the application of microvinification led us to discard L26A, M9 and Soi103 from further evaluation. Our biomass production assays were consistent with previous observations in which limited biomass formation was exhibited by commercial strains comparing to the indigenous ones [17].
L2M and Soi2 indigenous yeasts were further evaluated in industrial vinifications using as reference commercial yeasts, which were extensively used in industrial production of Moschofilero and Agiorgitiko wines. The fermentative behaviour was appropriate in both vinifications for indigenous yeast strains tested. L2M resulted in higher volatile acidity levels in both fermentations (Agiorgitiko and Moschofilero) while for Moschofilero wine the two indigenous strains resulted in higher reducing sugar levels. For Agiorgitiko wine, only L2M resulted in higher reducing sugar levels. All the other characteristics were similar for all analysed strains with the exception of colour intensity which was higher for the Soi2 produced Agiorgitiko wine. Wine yeasts produce acetic acid by the oxidation of acetaldehyde to acetate by NAD(P)+- dependent (acet)aldehyde dehydrogenases [76] and vary greatly in their ability to form acetic acid [77]. The overproduction of glycerol during wine fermentations for its positive sensory attributes leads to an increase in acetic acid concentration [78,79]. However, the degree of acetic acid formation is yeast-strain dependent [78]. Additionally, it has been observed, that the glucose consumption rate, is genetically variable among S. cerevisiae yeast strains [80,81]. Consequently, variations in acetic acid formation and reducing sugar levels among indigenous and commercial yeast strains, observed in this study, could explain the genetic basis of the strain-specific differences.
It has been proposed that, specific traits characterize strains originate from the same ecological niche [3,5,17,28,81,82]. Examples include the low-level production of fermentation by-products (e.g., glycerol, acetate and ethyl succinate) by baker’s yeasts or the very high production levels of ethyl-butyrate and limited biomass formation exhibited by commercial strains [17]. It has been also identified several variables relevant to discriminating the strains on the basis of their origin like dry weight, population size, glycerol, acetate and ethyl butyrate [17]. Τhe results found during the volatile compound analysis in the present study showed significant variations for ethyl-butyrate, ethyl-isovalerate, 2-phenyl-ethyl-acetate and phenethyl-alcohol among indigenous and commercial S. cerevisiae yeast strains. Soi2 resulted in significantly higher ethyl-butyrate levels in Moschofilero vinifications comparing with the other indigenous and commercial yeast strains. For instance, the influence of the matrix is of great importance and directly correlated to the used yeast strain.
Selected strains can be used as starters in wine fermentation only if the major characteristics of wine flavour remain essentially unchanged and when the oenological parameters remain at appropriate level [67]. According to the results found in this study, L2M and Soi2 strains resulted in quality wines with equal organoleptic and chemical characteristics with those of the commercial yeast strains. Fermentation kinetics was similar for all strains tested. Moreover, Soi2 resulted in an improved aromatic profile for Moschofilero and partially for Agiorgitiko fermentations, at least based on the analysis of the selected volatile compounds that have been analysed in this study. We propose that both strains could be easily used as wine starters for alcoholic fermentations of Moschofilero and Agiorgitiko musts; surprisingly Soi2, even if it was isolated from Agiorgitiko spontaneous fermentations, it could be also exploited for the fermentation of Moschofilero or more generally for the industrial fermentations of white grape musts.
5. Conclusions
Genetic analysis of L2M and Soi2 S. cerevisiae strains, isolated from spontaneous fermentations of Moschofilero and Agiorgitiko grape musts indicate a Geographic origin specific for Mantinia and Nemea regions. To our knowledge, this the first time ever reported a Greek vineyard-specific isolation of wine-making yeasts with oenological interest. Our yeasts presented particular genetic and aromatic profiles, which could be able to strengthen the quality and the regional characteristics of the Greek wines. Further research should be performed in order to exploit the particular genetic diversity of Greek S. cerevisiae strains and analyze their importance in the local and global wine industry.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/beverages7020027/s1, Figure S1: Multiple sequence alignment of ITS rDNAs of Agiorgitiko and Moschofilero yeast strains with BLAST-NCBI retrieved ITS rDNAs of different S. cerevisiae yeast strains, Figure S2: Fermentation kinetics and characteristics of experimental wines using several indigenous Agiorgitiko and Moschofilero S. cerevisiae strains, Figure S3: H2S production assays of several indigenous Agiorgitiko and Moschofilero S. cerevisiae strains, Figure S4: Ability to grow of several indigenous Agiorgitiko and Moschofilero S. cerevisiae strains in the presence of 0, 150 and 300 ppm of SO2, Figure S5: Sugar consumption slopes (oBé at 20 °C) of industrial vinifications of Moschofilero (A) and Agiorgitiko (B) grape musts which were fermented with the L2M and Soi2 S. cerevisiae strains, Table S1: Oenological characteristics of experimental wines using several indigenous Agiorgitiko and Moschofilero S. cerevisiae strains, Table S2: Ethanol tolerance assays of several indigenous Agiorgitiko and Moschofilero S. cerevisiae strains with the maximum ethanol production method, Table S3: Oenological characteristics of industrial vinification of Moschofilero grape must which was fermented with L2M and Soi2 S. cerevisiae strains, Table S4: Oenological characteristics of industrial vinification of Agiorgitiko grape must which was fermented with L2M and Soi2 S. cerevisiae strains.
Author Contributions
Conceptualization, Y.K. and V.T.; methodology, D.K.; software, D.K.; validation, Y.K., P.H. and V.T.; formal analysis, M.D.; investigation, D.K.; resources, Y.K. and P.H.; data curation, M.D.; writing—original draft preparation, D.K.; writing—review and editing, Y.K. and M.D.; visualization, D.K.; supervision, Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
This program is funded by the operational programme, “Human Resources Development” of the National Strategic Framework (NSRF) 2007–2013.
Acknowledgments
We would like to thank K. Zafeiri (Domaine Zafeiri, Nemea, Greece) and G. Papadopoulos (Domaine Papadopoulos, Mantinia, Greece) for providing us with the Agiorgitiko and Moschofilero grape musts, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bisson, L.F. The biotechnology of wine yeast. Food Biotechnol. 2004, 18, 63–96. [Google Scholar] [CrossRef]
- Bisson, L.F.; Kunkee, R.E. Microbial interactions during wine production. In Mixed Cultures in Biotechnology; Zeikus, G., Johnson, E.A., Eds.; McGraw-Hill, Inc.: New York, NY, USA, 1991; pp. 37–68. [Google Scholar]
- Lachance, M.A.; Stramer, W.T. Ecology and yeasts. In The Yeasts: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Eds.; Elsevier Sciences: Amsterdam, The Netherlands, 1998; pp. 21–30. [Google Scholar]
- Mortimer, R.K.; Polsinelli, M. On the origins of wine yeast. Res. Microbiol. 1999, 150, 199–204. [Google Scholar] [CrossRef]
- Pretorius, I.S.; van der Westhuizen, T.J.; Augustyn, O.P.H. Yeast biodiversity in vineyards and wineries and its importance to the South African wine industry. A review. S. Afr. J. Enol. Vitic. 1999, 20, 61–74. [Google Scholar] [CrossRef]
- Stefanini, I.; Dapporto, L.; Legras, J.L.; Calabretta, A.; Di Paola, M.; De Filippo, C.; Viola, R.; Capretti, P.; Polsinelli, M.; Turillazzi, S.; et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.L.; Galeote, V.; Bigey, F.; Camarasa, C.; Marsit, S.; Nidelet, T.; Sanchez, I.; Couloux, A.; Guy, J.; Franco-Duarte, R.; et al. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 2018, 35, 1712–1727. [Google Scholar] [PubMed]
- Vaughan, A.; Martini, A. Facts, myths and legends on the prime industrial microorganism. J. Indust. Microbiol. 1995, 14, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef]
- Tempère, S.; Marchal, A.; Barbe, J.C.; Bely, M.; Masneuf-Pomarède, I.; Marullo, P.; Albertin, W. The complexity of wine: Clarifying the role of microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 3995–4007. [Google Scholar] [CrossRef]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and its importance in wine aroma-a review. S. Afr. J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavor. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial Contribution to Wine Aroma and Its Intended Use for Wine Quality Improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef]
- Fia, G.; Giovani, G.; Rosi, I. Study of B-glucosidase production by wine-related yeasts during alcoholic fermentation. A new rapid fluorimetric method to determine enzyme activity. J. Appl. Microbiol. 2005, 99, 509–517. [Google Scholar] [CrossRef]
- Maicas, S.; Mateo, J.J. Hydrolysis of terpenyl glycosides in grape juice and other fruit juices: A review. Appl. Microbiol. Biotechnol. 2005, 67, 322–335. [Google Scholar] [CrossRef]
- Villena, M.A.; Iranzo, J.F.U.; Perez, A.I.B. B-Glucosidase activity in wine yeasts: Application in enology. Enzym. Microb. Technol. 2007, 40, 420–425. [Google Scholar] [CrossRef]
- Camarasa, C.; Sanchez, I.; Brial, P.; Bigey, F.; Dequin, S. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: Evidence for origin-dependent metabolic traits. PLoS ONE 2011, 6, e25147. [Google Scholar] [CrossRef] [PubMed]
- Dubourdieu, D.; Tominaga, T.; Masneuf, I.; Peyrot des Gachons, C.; Murat, M.L. The role of yeasts in grape flavour development during fermentation: The example of Sauvignon blanc. Am. J. Enol. Vitic. 2006, 57, 81–88. [Google Scholar]
- Klis, F.M.; Boorsma, A.; de Groot, P.W. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef]
- Caridi, A. New perspectives in safety and quality enhancement of wine through selection of yeasts based on the parietal adsorption capacity. Int. J. Food Microbiol. 2007, 120, 167–172. [Google Scholar] [CrossRef]
- Hayasaka, Y.; Birse, M.; Eglinton, J.; Herderich, M. The effect of Saccharomyces bayanus yeast on colour properties and pigment profiles of a Cabernet Sauvignon red wine. Aust. J. Grape Wine Res. 2007, 13, 176–185. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Yeast interactions with anthocyanins during red wine fermentation. Am. J. Enol. Vitic. 2005, 56, 104–108. [Google Scholar]
- Martínez, C.; Cosgaya, P.; Vásquez, C.; Gac, S.; Ganga, A. High degree of polymorphism and geographic origin of wine yeast strains. J. Appl. Microbiol. 2007, 103, 2185–2195. [Google Scholar] [CrossRef]
- Raspor, P.; Milek, D.M.; Polanc, J.; Mozina, S.S.; Nadez, N. Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int. J. Food Microbiol. 2006, 109, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Valero, E.; Cambon, B.; Schuller, D.; Casal, M.; Dequin, S. Biodiversity of Saccharomyces yeast strains from grape berries from wine producing areas using starter commercial yeasts. FEMS Yeast Res. 2007, 7, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Vezinhet, F.; Hallet, J.; Valade, M.; Poulard, A. Ecological survey of wine yeast strains by molecular methods of identification. Am. J. Enol. Vitic. 1992, 43, 83–86. [Google Scholar]
- Mercado, L.; Combina, M. Exploring the biodiversity of a wine region: Saccharomyces yeasts associated with wineries and vineyards. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex Research Center SL: Badajos, Spain, 2010; pp. 1042–1053. [Google Scholar]
- Van Leeuwen, C.; Seguin, G. The concept of terroir in viticulture. J. Wine Res. 2006, 17, 1–10. [Google Scholar] [CrossRef]
- Gilbert, J.A.; van der Lelie, D.; Zarraonaindia, I. Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Aa, E.; Townsend, J.P.; Adams, R.I.; Nielsen, K.M.; Taylor, J.W. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006, 6, 702–715. [Google Scholar] [CrossRef]
- Fay, J.C.; Benavides, J.A. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005, 1, e5. [Google Scholar] [CrossRef]
- Schacherer, J.; Ruderfer, D.M.; Gresham, D.; Dolinski, K.; Botstein, D.; Kruglyak, L. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE 2007, 2, e322. [Google Scholar] [CrossRef]
- Schacherer, J.; Shapiro, J.A.; Ruderfer, D.M.; Kruglyak, L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 2009, 458, 342–345. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Castillo-Davis, C.I.; Oshiro, G.; Liang, D.; Richards, D.R.; Zhou, Y.; Hartl, D.L. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 2003, 163, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Fernàndez-Espinar, T.M.; Barrio, E.; Querol, A. Analysis of the genetic variability in the species of the Saccharomyces sensu stricto complex. Yeast 2003, 20, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, J.; Barrio, E.; Querol, A. Characterization of wine yeast strains of the Saccharomyces genus on the basis of molecular markers: Relationships between genetic distance and geographic or ecological origin. Syst. Appl. Microbiol. 1996, 19, 122–132. [Google Scholar] [CrossRef]
- Nadal, D.; Colomer, B.; Pinã, B. Molecular polymorphism distribution in phenotypically distinct populations of wine yeast strains. Appl. Environ. Microbiol. 1996, 62, 1944–1950. [Google Scholar] [CrossRef]
- Sabate, J.; Cano, J.; Querol, A.; Guillamón, J. Diversity of Saccharomyces strains in wine fermentations: Analysis for two consecutive years. Lett. Appl. Microbiol. 1998, 26, 452–455. [Google Scholar] [CrossRef]
- Schuller, D.; Valero, E.; Dequin, S.; Casal, M. Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol. Lett. 2004, 231, 19–26. [Google Scholar] [CrossRef]
- Versavaud, A.; Courcoux, P.; Rouland, L.; Hallet, J. Genetic diversity and geographical distribution of wild Saccharomyces cerevisiae strains from the wine-producing area of Charentes, France. Appl. Environ. Microbiol. 1995, 61, 3521–3529. [Google Scholar] [CrossRef] [PubMed]
- APIWEBTM. Available online: https://apiweb.biomerieux.com/ (accessed on 23 December 2013).
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR based applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Pavel, A.B.; Vasile, C.I. PyElph—A software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012, 13, 9. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Inns, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- GenBank Nucleotide Database. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 1 April 2013).
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- GeneDoc. Available online: http://www.nrbsc.org/gfx/genedoc/ (accessed on 1 April 2013).
- Page, R.D. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef]
- Hayashida, S.; Feng, D.D.; Hungo, M. Physiological properties of yeast cells grown in the proteolipid-supplementation media. Agric. Biol. Chem. 1975, 39, 1025–1031. [Google Scholar]
- Duan, W.; Roddick, F.A.; Higgins, V.J.; Rogers, P.J. A parallel analysis of H2S and SO2 formation by brewing yeast in response to sulfur-containing amino acids and ammonium ions. J. Am. Soc. Brew. Chem. 2004, 62, 35–41. [Google Scholar] [CrossRef]
- Jesús, M.; Palero, R.; Fierro-Risco, J.; Codón, A.C.; Benítez, T.; Valcárcel, M.J. Selection of an autochthonous Saccharomyces strain starter for alcoholic fermentation of Sherry base wines. J. Ind. Microbiol. Biotechnol. 2013, 40, 613–623. [Google Scholar]
- Kinga, S.; Gillette, M.; Titman, D.; Adams, J.; Ridgely, M. The Sensory Quality System: A global quality control solution. Food Qual. Prefer. 2002, 13, 385–395. [Google Scholar] [CrossRef]
- Torrens, J.; Urpi, P.; Riu-Aumatell, M.; Vichi, S.; López-Tamames, E.; Buxaderas, S. Different commercial yeast strains affecting the volatile and sensory profile of cava base wine. Int. J. Food Microbiol. 2008, 124, 48–55. [Google Scholar] [CrossRef]
- EL-Fiky, Z.A.; Hassan, G.M.; Emam, M. Quality parameters and RAPD-PCR differentiation of commercial baker’s yeast and hybrid strains. J. Food Sci. 2012, 77, M312–M317. [Google Scholar] [CrossRef]
- Gallego, F.J.; Pérez, M.A.; Núñez, Y.; Hidalgo, P. Comparison of RAPDs, AFLPs and SSR markers for the genetic analysis of yeast strains of Saccharomyces cerevisiae. Food Microbiol. 2005, 22, 561–568. [Google Scholar] [CrossRef]
- Lathar, P.K.; Sharma, A.; Thakur, I. Isolation and random amplified polymorphic DNA (RAPD) analysis of wild yeast species from 17 different fruits. J. Yeast Fungal Res. 2010, 1, 146–151. [Google Scholar]
- Saitou, N.; Masatoshi, N. The Neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Monagas, M.; Gómez-Cordovés, C.; Bartolomé, B. Evaluation of different Saccharomyces cerevisiae strains for red winemaking. Influence on the anthocyanin, pyranoanthocyanin and non-anthocyanin phenolic content and colour characteristics of wines. Food Chem. 2007, 104, 814–823. [Google Scholar] [CrossRef]
- Tofalo, R.; Perpetuini, G.; Fasoli, G.; Schirone, M.; Corsetti, A.; Suzzi, G. Biodiversity study of wine yeasts belonging to the ‘terroir’ of Montepulciano d’Abruzzo ‘Colline Teramane’ revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8S-ITS restriction patterns. Food Microbiol. 2014, 39, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Lopandic, K.; Gangl, H.; Wallner, E.; Tscheik, G.; Leitner, G.; Querol, A.; Borth, N.; Breitenbach, M.; Prillinger, H.; Tiefenbrunner, W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007, 7, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.W.; Um, H.J.; Kang, H.W.; Kim, Y.; Kim, M.; Kim, Y.H. Bioethanol production by a flocculent hybrid, CHFY0321 obtained by protoplast fusion between Saccharomyces cerevisiae and Saccharomyces bayanus. Biomass Bioenergy 2010, 34, 1232–1242. [Google Scholar] [CrossRef]
- Albertin, W.; Masneuf-Pomarède, I.; Galeote, V.; Legras, J.L. New Insights into Wine Yeast Diversities. In Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer: New York, NY, USA, 2019; pp. 117–163. [Google Scholar]
- Pretorius, I.S. Tasting the terroir of wine yeast innovation. FEMS Yeast. Res. 2020, 20, foz084. [Google Scholar] [CrossRef]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Dimopoulou, M.; Troianou, V.; Toumpeki, C.; Gosselin, Y.; Dorignac, É.; Kotseridis, Y. Effect of strains from different Saccharomyces species used in different inoculation schemes on chemical composition and sensory characteristics of Sauvignon blanc wine. OENO ONE 2020, 54, 4. [Google Scholar] [CrossRef]
- Rainieri, S.; Pretorius, I.S. Selection and improvement of wine yeasts. Ann. Microbiol. 2000, 50, 15–31. [Google Scholar]
- König, H.; Claus, H. A future place for Saccharomyces mixtures and hybrids in wine making. Fermentation 2018, 4, 67. [Google Scholar] [CrossRef]
- Ashworth, A.J.; DeBruyn, J.M.; Allen, F.L.; Radosevich, M.; Owens, P.R. Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol. Biochem. 2017, 114, 210–219. [Google Scholar] [CrossRef]
- De Celis, M.; Ruiz, J.; Martín-Santamaría, M.; Alonso, A.; Marquina, D.; Navascués, E.; Gómez-Flechoso, M.Á.; Belda, I.; Santos, A. Diversity of Saccharomyces cerevisiae yeasts associated to spontaneous and inoculated fermenting grapes from Spanish vineyards. Lett. Appl. Microbiol. 2019, 68, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.; Picot, A.; Cobo-Diaz, J.F.; Carof, M.; Chen, W.; Le Floch, G. Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 2018, 132, 135–145. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Arroyo, T.; Serrano, A.; Valero, E. Remanence and survival of commercial yeast in different ecological niches of the vineyard. FEMS Microbiol. Ecol. 2011, 77, 429–437. [Google Scholar] [CrossRef]
- Grangeteau, C.; Gerhards, D.; von Wallbrunn, C.; Alexandre, H.; Guilloux-Benatier, M. Persistence of two non-Saccharomyces yeasts (Hanseniaspora and Starmerella) in the cellar. Front. Microbiol. 2016, 7, 268. [Google Scholar] [CrossRef]
- Remize, F.; Roustan, J.L.; Sablayrolles, J.M.; Barre, P.; Dequin, S. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in byproduct formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 1999, 65, 143–149. [Google Scholar] [CrossRef]
- Delfini, C.; Cervetti, F. Metabolic and technological factors affecting acetic acid production by yeasts during alcoholic fermentation. Vitic Enol. Sci. 1991, 46, 142–150. [Google Scholar]
- Erasmus, D.J.; Cliff, M.; van Vuuren, H.J.J. Impact of Yeast Strain on the Production of Acetic Acid, Glycerol, and the Sensory Attributes of Icewine. Am. J. Enol. Vitic. 2004, 55, 4. [Google Scholar]
- Zhao, X.; Procopio, S.; Becker, T. Flavor impacts of glycerol in the processing of yeast fermented beverages: A review. J. Food Sci. Technol. 2015, 52, 7588–7598. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Wang, S.; Dillmann, C.; de Vienne, D.; Sicard, D. “Ant” and “grasshopper” life-history strategies in Saccharomyces cerevisiae. PLoS ONE 2008, 3, e1579. [Google Scholar] [CrossRef] [PubMed]
- Guzzon, R.; Roman, T.; Larcher, R.; Francesca, N.; Guarcello, R.; Moschetti, G. Biodiversity and oenological attitude of Saccharomyces cerevisiae strains isolated in the Montalcino district: Biodiversity of S. cerevisiae strains of Montalcino wines. FEMS Microbiol. Lett. 2021, 368. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Tu, C.; Hoyt, G.D.; DeForest, J.L.; Hu, S. Long-term no-tillage and organic input management enhanced the diversity and stability of soil microbial community. Sci. Total Environ. 2017, 609, 341–347. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).