Development and Validation of a Fast UHPLC–HRMS Method for the Analysis of Amino Acids and Biogenic Amines in Fermented Beverages
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Standard and Sample Solutions Preparation
2.3. LC–MS Parameters
2.4. Method Validation
2.5. Data Processing, Statistical Analysis and Modelling
3. Results and Discussion
3.1. Gradient Elution Study
3.2. Optimization of the MS/MS Conditions
3.3. Matrix Effect and Sample Preparation
3.4. Precision
3.5. Application of the Validated Method to FBs and Must Samples
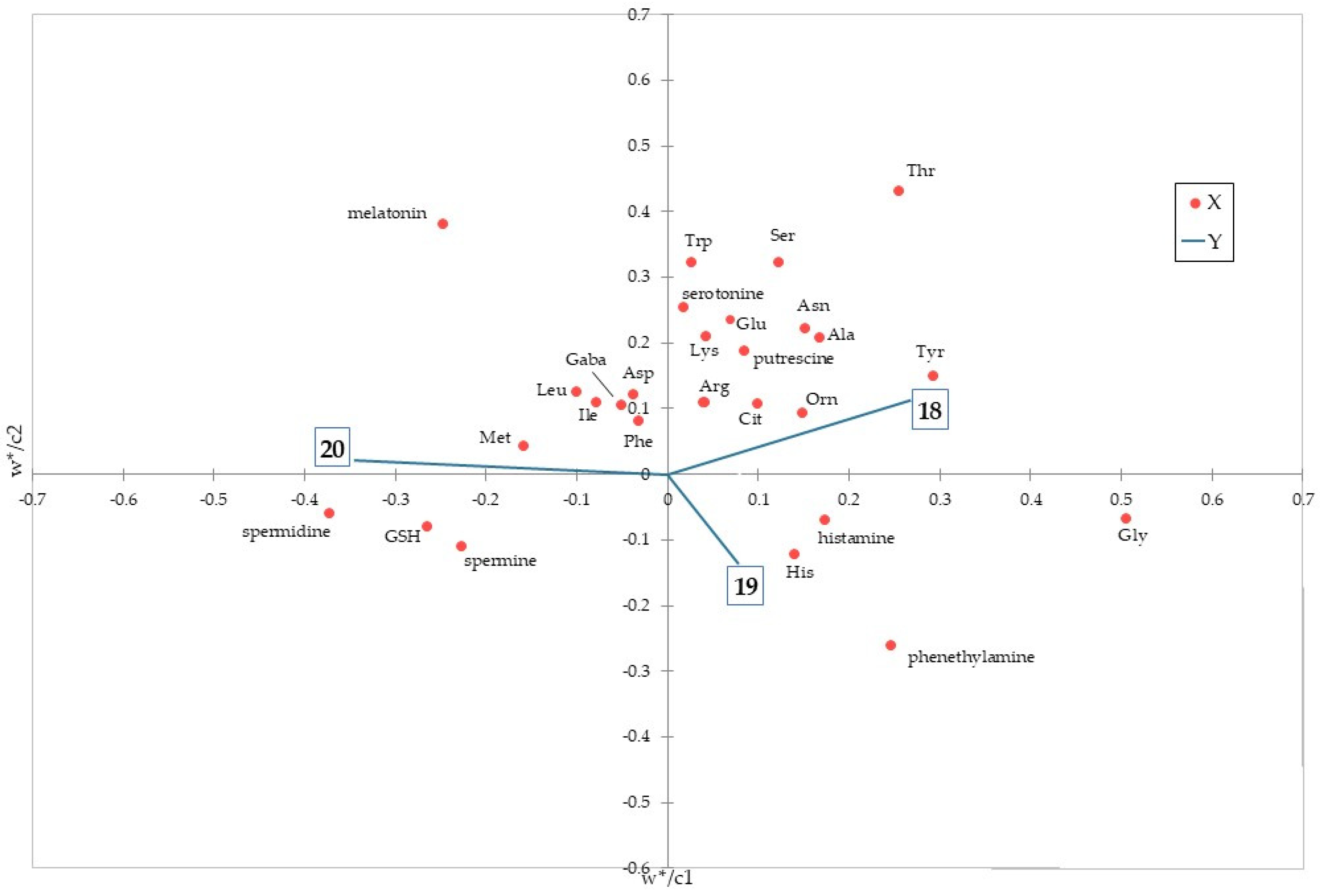
3.6. Modelling Based on Data Obtained from the Validated Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FAAs | Free amino acids |
| BAs | Biogenic amines |
| FB | Fermented beverages |
| UHPLC | Ultra-high-performance liquid chromatography |
| HRMS | High-resolution mass spectrometry |
| N | Nitrogen |
| AA | Amino acid |
| HPLC | High-performance liquid chromatography |
| MS | Mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| GC | Gas chromatography |
| FMOC | Fluorenylmethyloxycarbonyl chloride |
| DEMM | Diethyl ethoxymethylene malonate |
| LC–MS/MS | Liquid chromatography coupled to mass spectrometry |
| Leu | Leucine |
| Ile | Isoleucine |
| GSH | Glutathione |
| Cys | Cysteine |
| Gly | Glycine |
| MeOH | Methanol |
| HCl | Hydrochloric acid |
| PTFE | Polytetrafluoroethylene |
| CAN | Acetonitrile |
| FA | Formic acid |
| FDA | Food and Drug Administration |
| LOD | Limit of detection |
| LOQ | Limit of quantification |
| RSD | Relative standard deviation |
| HILIC | Hydrophilic interaction liquid chromatography |
| Gaba | γ-aminobutyric acid |
| Asp | Aspartic acid |
| Glu | Glutamic acid |
| Ala | Alanine |
| Arg | Arginine |
| Asn | Asparagine |
| Cit | Citrulline |
| Phe | Phenylalanine |
| Hyp | Hydroxyproline |
| His | Histidine |
| Lys | Lysine |
| Met | Methionine |
| Orn | Ornithine |
| Ser | Serine |
| Tyr | Tyrosine |
| Thr | Threonine |
| Trp | Tryptophan |
| NCE | Normalized collision energy |
| FWHM | Full-width at half-maximum |
| AGC | Automatic gain control |
| EIC | Extracted ion chromatogram |
| RT | Retention time |
| SD | Standard deviation |
| PLS-DA | Partial least squares discriminant analysis |
References
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nuñez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.-S.; et al. Fermented Beverages of Pre- and Proto-Historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Garg, P.; Kumar, P.; Bhatia, S.K.; Kulshrestha, S. Microbial Fermentation and Its Role in Quality Improvement of Fermented Foods. Fermentation 2020, 6, 106. [Google Scholar] [CrossRef]
- Marsh, A.J.; Hill, C.; Ross, R.P.; Cotter, P.D. Fermented Beverages with Health-Promoting Potential: Past and Future Perspectives. Trends Food Sci. Technol. 2014, 38, 113–124. [Google Scholar] [CrossRef]
- Blandino, A.; Al-Aseeri, M.E.; Pandiella, S.S.; Cantero, D.; Webb, C. Cereal-Based Fermented Foods and Beverages. Food Res. Int. 2003, 36, 527–543. [Google Scholar] [CrossRef]
- Sablayrolles, J.-M.; Mouret, J.-R. Chapter 12—Nitrogen Management during Fermentation. In White Wine Technology; Morata, A., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 143–154. ISBN 9780128234976. [Google Scholar]
- Ferreira, I.M.; Guido, L.F. Impact of Wort Amino Acids on Beer Flavour: A Review. Fermentation 2018, 4, 23. [Google Scholar] [CrossRef]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and Bacterial Modulation of Wine Aroma and Flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Hernández-Orte, P.; Cacho, J.F.; Ferreira, V. Relationship between Varietal Amino Acid Profile of Grapes and Wine Aromatic Composition. Experiments with Model Solutions and Chemometric Study. J. Agric. Food Chem. 2002, 50, 2891–2899. [Google Scholar] [CrossRef]
- Chung, C.; Rojanasasithara, T.; Mutilangi, W.; McClements, D.J. Stability Improvement of Natural Food Colors: Impact of Amino Acid and Peptide Addition on Anthocyanin Stability in Model Beverages. Food Chem. 2017, 218, 277–284. [Google Scholar] [CrossRef]
- Procopio, S.; Sprung, P.; Becker, T. Effect of Amino Acid Supply on the Transcription of Flavour-Related Genes and Aroma Compound Production during Lager Yeast Fermentation. LWT-Food Sci. Technol. 2015, 63, 289–297. [Google Scholar] [CrossRef]
- Qiu, L.; Zhang, M.; Chang, L. Effects of Lactic Acid Bacteria Fermentation on the Phytochemicals Content, Taste and Aroma of Blended Edible Rose and Shiitake Beverage. Food Chem. 2023, 405, 134722. [Google Scholar] [CrossRef]
- Kaspar, H.; Dettmer, K.; Gronwald, W.; Oefner, P.J. Advances in Amino Acid Analysis. Anal. Bioanal. Chem. 2009, 393, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, A.; Dosi, R.; Ferrara, L.; Rocco, M. Free Amino Acid Profile of Malus Domestica Borkh Cv. Annurca from the Campania Region and Other Italian Vegetables. Aust. J. Crop Sci. 2011, 5, 154–161. [Google Scholar]
- González-Domínguez, R. Food Authentication: Techniques, Trends and Emerging Approaches. Foods 2020, 9, 346. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, A.R. Significance of Biogenic Amines to Food Safety and Human Health. Food Res. Int. 1996, 29, 675–690. [Google Scholar] [CrossRef]
- Silla Santos, M.H. Biogenic Amines: Their Importance in Foods. Int. J. Food Microbiol. 1996, 29, 213–231. [Google Scholar] [CrossRef]
- Visciano, P.; Schirone, M. Update on Biogenic Amines in Fermented and Non-Fermented Beverages. Foods 2022, 11, 353. [Google Scholar] [CrossRef]
- Aung, T.; Lee, W.-H.; Eun, J.-B. Metabolite Profiling and Pathway Prediction of Laver (Porphyra Dentata) Kombucha during Fermentation at Different Temperatures. Food Chem. 2022, 397, 133636. [Google Scholar] [CrossRef]
- Callejón, R.M.; Troncoso, A.M.; Morales, M.L. Determination of Amino Acids in Grape-Derived Products: A Review. Talanta 2010, 81, 1143–1152. [Google Scholar] [CrossRef]
- Tırıs, G.; Sare Yanıkoğlu, R.; Ceylan, B.; Egeli, D.; Kepekci Tekkeli, E.; Önal, A. A Review of the Currently Developed Analytical Methods for the Determination of Biogenic Amines in Food Products. Food Chem. 2023, 398, 133919. [Google Scholar] [CrossRef]
- Gong, X.; Wang, X.; Qi, N.; Li, J.; Lin, L.; Han, Z. Determination of Biogenic Amines in Traditional Chinese Fermented Foods by Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC). Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2014, 31, 1431–1437. [Google Scholar] [CrossRef]
- Izco, J.M.; Torre, P.; Barcina, Y. Ripening of Ossau-Iraty Cheese: Determination of Free Amino Acids by RP-HPLC and of Total Free Amino Acids by the TNBS Method. Food Control 2000, 11, 7–11. [Google Scholar] [CrossRef]
- Liu, P.-H.; Vrigneau, C.; Salmon, T.; Hoang, D.A.; Boulet, J.-C.; Jégou, S.; Marchal, R. Influence of Grape Berry Maturity on Juice and Base Wine Composition and Foaming Properties of Sparkling Wines from the Champagne Region. Molecules 2018, 23, 1372. [Google Scholar] [CrossRef]
- Schwarz, E.L.; Roberts, W.L.; Pasquali, M. Analysis of Plasma Amino Acids by HPLC with Photodiode Array and Fluorescence Detection. Clin. Chim. Acta 2005, 354, 83–90. [Google Scholar] [CrossRef]
- Ohtake, N.; Takano, A.; Ito, S.; Yamazaki, A.; Fujikake, H.; Sueyoshi, K.; Ohyama, T. Quantitative and Isotopic Analysis of Amino Acids, Allantoin, and Allantoic Acid in Soybeans by LC-MS Using the Atmospheric Pressure Chemical Ionization Method. Soil Sci. Plant Nutr. 2004, 50, 241–248. [Google Scholar] [CrossRef]
- Rebane, R.; Oldekop, M.-L.; Herodes, K. Comparison of Amino Acid Derivatization Reagents for LC–ESI-MS Analysis. Introducing a Novel Phosphazene-Based Derivatization Reagent. J. Chromatogr. B 2012, 904, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Nimbalkar, M.S.; Pai, S.R.; Pawar, N.V.; Oulkar, D.; Dixit, G.B. Free Amino Acid Profiling in Grain Amaranth Using LC–MS/MS. Food Chem. 2012, 134, 2565–2569. [Google Scholar] [CrossRef] [PubMed]
- Thiele, B.; Füllner, K.; Stein, N.; Oldiges, M.; Kuhn, A.J.; Hofmann, D. Analysis of Amino Acids without Derivatization in Barley Extracts by LC-MS-MS. Anal. Bioanal. Chem. 2008, 391, 2663–2672. [Google Scholar] [CrossRef] [PubMed]
- Mudiam, M.K.R.; Ch, R.; Jain, R.; Saxena, P.N.; Chauhan, A.; Murthy, R.C. Rapid and Simultaneous Determination of Twenty Amino Acids in Complex Biological and Food Samples by Solid-Phase Microextraction and Gas Chromatography--Mass Spectrometry with the Aid of Experimental Design after Ethyl Chloroformate Derivatization. J. Chromatogr. B 2012, 907, 56–64. [Google Scholar] [CrossRef]
- Nozal, M.J.; Bernal, J.L.; Toribio, M.L.; Diego, J.C.; Ruiz, A. Rapid and Sensitive Method for Determining Free Amino Acids in Honey by Gas Chromatography with Flame Ionization or Mass Spectrometric Detection. J. Chromatogr. A 2004, 1047, 137–146. [Google Scholar] [CrossRef]
- Bobbitt, D.R.; Jackson, W.A.; Hendrickson, H.P. Chemiluminescent Detection of Amines and Amino Acids Using in Situ Generated Ru(Bpy)33+ Following Separation by Capillary Electrophoresis. Talanta 1998, 46, 565–572. [Google Scholar] [CrossRef]
- Zhan, W.; Wang, T.; Li, S.F. Derivatization, Extraction and Concentration of Amino Acids and Peptides by Using Organic/Aqueous Phases in Capillary Electrophoresis with Fluorescence Detection. Electrophoresis 2000, 21, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- Hogenboom, J.A.; D’Incecco, P.; Fuselli, F.; Pellegrino, L. Ion-Exchange Chromatographic Method for the Determination of the Free Amino Acid Composition of Cheese and Other Dairy Products: An Inter-Laboratory Validation Study. Food Anal. Methods 2017, 10, 3137–3148. [Google Scholar] [CrossRef]
- Michalski, R.; Pecyna-Utylska, P.; Kernert, J. Determination of Ammonium and Biogenic Amines by Ion Chromatography. A Review. J. Chromatogr. A 2021, 1651, 462319. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.P.; Prats-Moya, M.S. Free Amino Acids and Biogenic Amines in Alicante Monastrell Wines. Food Chem. 2012, 135, 1511–1519. [Google Scholar] [CrossRef]
- Kalkan Yıldırım, H.; Üren, A.; Yücel, U. Evaluation of Biogenic Amines in Organic and Non-Organic Wines by HPLC OPA Derivatization. Food Technol. 2007, 45, 62–68. [Google Scholar]
- Bank, R.A.; Jansen, E.J.; Beekman, B.; te Koppele, J.M. Amino Acid Analysis by Reverse-Phase High-Performance Liquid Chromatography: Improved Derivatization and Detection Conditions with 9-Fluorenylmethyl Chloroformate. Anal. Biochem. 1996, 240, 167–176. [Google Scholar] [CrossRef]
- Bernal, J.L.; Nozal, M.J.; Toribio, L.; Diego, J.C.; Ruiz, A. A Comparative Study of Several HPLC Methods for Determining Free Amino Acid Profiles in Honey. J. Sep. Sci. 2005, 28, 1039–1047. [Google Scholar] [CrossRef]
- Iwanicka-Grzegorek, K.; Lipkowska, E.; Kepa, J.; Michalik, J.; Wierzbicka, M. Comparison of Ninhydrin Method of Detecting Amine Compounds with Other Methods of Halitosis Detection. Oral Dis. 2005, 11 (Suppl. S1), 37–39. [Google Scholar] [CrossRef]
- Zhang, H. Determination of Twenty Amino Acids by Ninhydrin Reaction with FIA. In Proceedings of the 2011 International Conference on Remote Sensing, Environment and Transportation Engineering, Nanjing, China, 24–26 June 2011; pp. 6735–6737. [Google Scholar]
- Jain, A.; Verma, K.K. Strategies in Liquid Chromatographic Methods for the Analysis of Biogenic Amines without and with Derivatization. Trends Analyt. Chem. 2018, 109, 62–82. [Google Scholar] [CrossRef]
- Milheiro, J.; Ferreira, L.C.; Filipe-Ribeiro, L.; Cosme, F.; Nunes, F.M. A Simple Dispersive Solid Phase Extraction Clean-up/Concentration Method for Selective and Sensitive Quantification of Biogenic Amines in Wines Using Benzoyl Chloride Derivatisation. Food Chem. 2019, 274, 110–117. [Google Scholar] [CrossRef]
- Guo, N.; Yang, D.; Yang, X.; Yan, H.; Fan, B. A Rapid, Sensitive, and Widely Applicable Method for Quantitative Analysis of Underivatized Amino Acids in Different Biological Matrices by UHPLC-MS/MS. J. Sep. Sci. 2019, 42, 3173–3181. [Google Scholar] [CrossRef]
- Le, A.; Ng, A.; Kwan, T.; Cusmano-Ozog, K.; Cowan, T.M. A Rapid, Sensitive Method for Quantitative Analysis of Underivatized Amino Acids by Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS). J. Chromatogr. B 2014, 944, 166–174. [Google Scholar] [CrossRef]
- Piri-Moghadam, H.; Miller, A.; Pronger, D.; Vicente, F.; Charrow, J.; Haymond, S.; Lin, D.C. A Rapid LC-MS/MS Assay for Detection and Monitoring of Underivatized Branched-Chain Amino Acids in Maple Syrup Urine Disease. JMSACL 2022, 24, 107–117. [Google Scholar] [CrossRef]
- Plenis, A.; Olędzka, I.; Kowalski, P.; Miękus, N.; Bączek, T. Recent Trends in the Quantification of Biogenic Amines in Biofluids as Biomarkers of Various Disorders: A Review. J. Clin. Med. Res. 2019, 8, 640. [Google Scholar] [CrossRef] [PubMed]
- Thiele, B.; Stein, N.; Oldiges, M.; Hofmann, D. Direct Analysis of Underivatized Amino Acids in Plant Extracts by LC-MS-MS. Methods Mol. Biol. 2012, 828, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Chaimbault, P.; Petritis, K.; Elfakir, C.; Dreux, M. Determination of 20 Underivatized Proteinic Amino Acids by Ion-Pairing Chromatography and Pneumatically Assisted Electrospray Mass Spectrometry. J. Chromatogr. A 1999, 855, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Held, P.K.; White, L.; Pasquali, M. Quantitative Urine Amino Acid Analysis Using Liquid Chromatography Tandem Mass Spectrometry and ATRAQ® Reagents. J. Chromatogr. B 2011, 879, 2695–2703. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Song, F.; Liu, Z.; Liu, S. A Simple Method for the Analysis by MS/MS of Underivatized Amino Acids on Dry Blood Spots from Newborn Screening. Amino Acids 2012, 42, 1889–1895. [Google Scholar] [CrossRef]
- Xia, T.; Gao, S.; Shu, C.; Wen, Y.; Yun, Y.; Tao, X.; Chen, W.; Zhang, F. Analysis of Amino Acids in Human Blood Using UHPLC-MS/MS: Potential Interferences of Storage Time and Vacutainer Tube in Pre-Analytical Procedure. Clin. Biochem. 2016, 49, 1372–1378. [Google Scholar] [CrossRef]
- Liu, Z.; Tu, M.-J.; Zhang, C.; Jilek, J.L.; Zhang, Q.-Y.; Yu, A.-M. A Reliable LC-MS/MS Method for the Quantification of Natural Amino Acids in Mouse Plasma: Method Validation and Application to a Study on Amino Acid Dynamics during Hepatocellular Carcinoma Progression. J. Chromatogr. B 2019, 1124, 72–81. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, X.; Xu, Y.; Liu, X.; Zhang, J.; He, Z. Simultaneous Determination of 49 Amino Acids, B Vitamins, Flavonoids, and Phenolic Acids in Commonly Consumed Vegetables by Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry. Food Chem. 2021, 344, 128712. [Google Scholar] [CrossRef]
- Gökmen, V.; Serpen, A.; Mogol, B.A. Rapid Determination of Amino Acids in Foods by Hydrophilic Interaction Liquid Chromatography Coupled to High-Resolution Mass Spectrometry. Anal. Bioanal. Chem. 2012, 403, 2915–2922. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, Q.; Shi, Y.; Qu, J.; Song, F.; Liu, Z. Direct quantitative analysis of amino acids in fermented beverage of plant extract using high performance liquid chromatography-tandem mass spectrometry. Se Pu 2015, 33, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Taniguchi, M.; Fukusaki, E. High-Sensitive Liquid Chromatography-Tandem Mass Spectrometry-Based Chiral Metabolic Profiling Focusing on Amino Acids and Related Metabolites. J. Biosci. Bioeng. 2019, 127, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Kritzinger, E.C.; Bauer, F.F.; du Toit, W.J. Role of Glutathione in Winemaking: A Review. J. Agric. Food Chem. 2013, 61, 269–277. [Google Scholar] [CrossRef]
- Stephen, D.W.S.; Jamieson, D.J. Glutathione Is an Important Antioxidant Molecule in the YeastSaccharomyces Cerevisiae. FEMS Microbiol. Lett. 1996, 141, 207–212. [Google Scholar] [CrossRef]
- Gijs, L.; Perpète, P.; Timmermans, A.; Guyot-Declerck, C.; Delincé, P.; Collin, S. Assessment of Added Glutathione in Yeast Propagations, Wort Fermentations, and Beer Storage. J. Am. Soc. Brew. Chem. 2004, 62, 97–102. [Google Scholar] [CrossRef]
- Kankolongo Cibaka, M.-L.; Decourrière, L.; Lorenzo-Alonso, C.-J.; Bodart, E.; Robiette, R.; Collin, S. 3-Sulfanyl-4-Methylpentan-1-Ol in Dry-Hopped Beers: First Evidence of Glutathione S-Conjugates in Hop (Humulus lupulus L.). J. Agric. Food Chem. 2016, 64, 8572–8582. [Google Scholar] [CrossRef]
- AOAC Official Methods of Analysis. Appendix F: Guidelines for Standard Method Performance Requirements. In AOAC Official Methods of Analysis; AOAC: Rockville, MD, USA, 2016. [Google Scholar]
- U.S. Food and Drug Administration. Guidelines for the Validation of Chemical Methods for the FDA Foods Program; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2019.
- Magnusson, B. The Fitness for Purpose of Analytical Methods: A Laboratory Guide to Method Validation and Related Topics, 2nd ed.; Eurachem: Middlesex, UK, 2014.
- Liigand, J.; Laaniste, A.; Kruve, A. PH Effects on Electrospray Ionization Efficiency. J. Am. Soc. Mass Spectrom. 2017, 28, 461–469. [Google Scholar] [CrossRef]
- Buszewski, B.; Noga, S. Hydrophilic Interaction Liquid Chromatography (HILIC)--A Powerful Separation Technique. Anal. Bioanal. Chem. 2012, 402, 231–247. [Google Scholar] [CrossRef]
- Procopio, S.; Krause, D.; Hofmann, T.; Becker, T. Significant Amino Acids in Aroma Compound Profiling during Yeast Fermentation Analyzed by PLS Regression. LWT-Food Sci. Technol. 2013, 51, 423–432. [Google Scholar] [CrossRef]
- Chua, J.-Y.; Huang, A.; Liu, S.-Q. Comparing the Effects of Isoleucine and Leucine Supplementation at Different Dosage on the Growth and Metabolism of Torulaspora Delbrueckii Biodiva during Soy Whey Fermentation. Food Biosci. 2022, 50, 101963. [Google Scholar] [CrossRef]
- He, Y.; Dong, J.; Yin, H.; Zhao, Y.; Chen, R.; Wan, X.; Chen, P.; Hou, X.; Liu, J.; Chen, L. Wort Composition and Its Impact on the Flavour-Active Higher Alcohol and Ester Formation of Beer—A Review. J. Inst. Brew. 2014, 120, 157–163. [Google Scholar] [CrossRef]
- SANCO/12571/2013; Guidance Document on Analytical Quality Control and Validation Procedures for Pesticide Residues Analysis in Food and Feed. European Commission: Brussels, Belgium, 2013.
- Furey, A.; Moriarty, M.; Bane, V.; Kinsella, B.; Lehane, M. Ion Suppression; a Critical Review on Causes, Evaluation, Prevention and Applications. Talanta 2013, 115, 104–122. [Google Scholar] [CrossRef] [PubMed]
- Garai-Ibabe, G.; Irastorza, A.; Dueñas, M.T.; Martín-Álvarez, P.J.; Moreno-Arribas, V.M. Evolution of Amino Acids and Biogenic Amines in Natural Ciders as a Function of the Year and the Manufacture Steps. Int. J. Food Sci. Technol. 2013, 48, 375–381. [Google Scholar] [CrossRef]
- Gogami, Y.; Okada, K.; Oikawa, T. High-Performance Liquid Chromatography Analysis of Naturally Occurring D-Amino Acids in Sake. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 3259–3267. [Google Scholar] [CrossRef]
- Kong, Y.; Zhang, L.-L.; Sun, Y.; Zhang, Y.-Y.; Sun, B.-G.; Chen, H.-T. Determination of the Free Amino Acid, Organic Acid, and Nucleotide in Commercial Vinegars. J. Food Sci. 2017, 82, 1116–1123. [Google Scholar] [CrossRef]
- Fordellone, M.; Bellincontro, A.; Mencarelli, F. Partial Least Squares Discriminant Analysis: A Dimensionality Reduction Method to Classify Hyperspectral Data. arXiv 2018, arXiv:1806.09347. [Google Scholar]
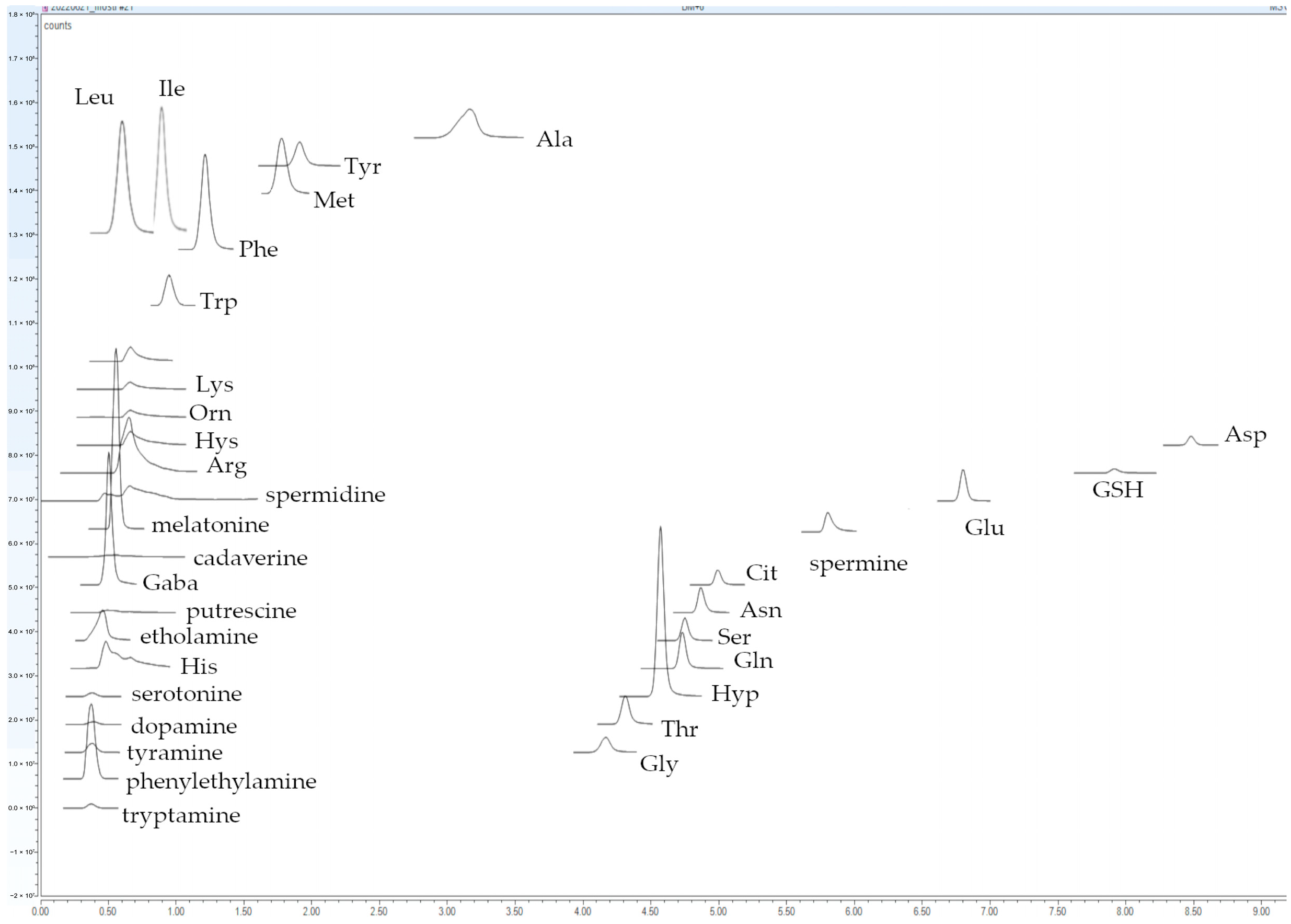
| FAAs/BAs | Chemical Formula | [M + H]+ (m/z) | Δ m/z (ppm) | RT (min) | Fragments (m/z) | LOD (mg/L) | LOQ (mg/L) | Linearity (mg/L) | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gaba | γ-Aminobutyric acid | C4H9NO2 | 104.0706 | 0.5 | 0.53 | 87.0444, 69.0340 | 0.001 | 0.003 | 0.001–2 | 0.99 |
| Asp | L-Aspartic acid | C4H7NO4 | 134.0448 | 0.0 | 8.49 | 88.0397, 74.0241 | 0.0015 | 0.005 | 0.005–5 | 0.99 |
| Glu | L-Glutamic acid | C5H9NO4 | 148.0602 | 1.6 | 6.82 | 130.0498, 84.0448 | 0.0015 | 0.005 | 0.005–5 | 0.99 |
| Ala | L-Alanine | C3H7NO2 | 90.0553 | −4.1 | 3.16 | 72.0444 | 0.003 | 0.01 | 0.01–5 | 0.99 |
| Arg | L-Arginine monohydrochloride | C6H14N4O2 | 175.1186 | 2.2 | 0.69 | 116.0706, 70.0656 | 0.0006 | 0.002 | 0.002–5 | 0.99 |
| Asn | L-Asparagine | C4H8N2O3 | 133.0607 | 0.8 | 4.84 | 99.0063, 74.0242 | 0.003 | 0.01 | 0.01–5 | 0.99 |
| Cit | L-Citrulline | C6H13N3O3 | 176.1031 | −0.7 | 5.00 | 159.0765, 113.0711 | 0.006 | 0.02 | 0.02–5 | 0.99 |
| Phe | L-Phenylalanine | C9H11NO2 | 166.0862 | 0.2 | 1.19 | 120.0808, 95.0494 | 0.0003 | 0.001 | 0.001–1 | 0.99 |
| Gly | Glycine | C2H5NO2 | 76.0394 | −1.6 | 4.18 | - | 0.006 | 0.02 | 0.02–5 | 0.99 |
| Hyp | trans-4-Hydroxy-L-proline | C5H9NO3 | 132.0654 | 1.1 | 4.57 | 100.0139, 86.0604 | 0.0015 | 0.005 | 0.005–5 | 0.99 |
| Ile | L-Isoleucine | C6H13NO2 | 132.1018 | 1.1 | 0.87 | 86.0964, 69.0699 | 0.0003 | 0.001 | 0.001–2 | 0.99 |
| Leu | L-Leucine | C6H13NO2 | 132.1018 | 1.2 | 0.59 | 86.0964 | 0.0001 | 0.001 | 0.001–5 | 0.99 |
| His | L-Histidine monohydrochloride monohydrate | C6H9N3O2 | 156.0766 | 0.1 | 0.61 | 110.0713, 95.0606 | 0.0015 | 0.05 | 0.05–5 | 0.99 |
| Lys | L-Lysine monohydrochloride | C6H14N2O2 | 147.1125 | 2.1 | 0.65 | 130.0862, 84.0811 | 0.015 | 0.05 | 0.05–2 | 0.99 |
| Met | L-Methionine | C5H11NO2S | 150.0582 | 0.6 | 1.79 | 133.0316, 104.0530 | 0.0015 | 0.005 | 0.005–2 | 0.99 |
| Orn | L-Ornitine dihydrochloride | C5H12N2O2 | 133.0971 | 0.5 | 0.67 | 116.0706, 70.0656 | 0.015 | 0.05 | 0.05–5 | 0.99 |
| Ser | L-Serine | C3H7NO3 | 106.0500 | −1.3 | 4.64 | 88.0396, 60.0450 | 0.003 | 0.01 | 0.01–5 | 0.99 |
| Tyr | L-Tyrosine | C9H11NO3 | 182.0809 | 1.4 | 1.87 | 136.0756, 123.0441 | 0.0015 | 0.005 | 0.005–1 | 0.99 |
| Thr | L-Threonine | C4H9NO3 | 120.0654 | 0.9 | 4.29 | 102.0551, 74.0605 | 0.006 | 0.02 | 0.02–5 | 0.99 |
| Trp | L-Tryptophan | C11H12N2O2 | 205.0971 | 0.2 | 0.95 | 188.0702, 146.0599 | 0.0015 | 0.005 | 0.005–5 | 0.99 |
| ethanolamine | Ethanolamine | C2H7NO | 62.0603 | −4.2 | 0.45 | - | 0.0015 | 0.005 | 0.005–1 | 0.99 |
| phenylethylamine | 2-Phenylethylamine hydrochloride | C8H11N | 122.0958 | 4.8 | 0.39 | 105.0701, 79.0546 | 0.0003 | 0.001 | 0.001–0.1 | 0.95 |
| putrescine | 1,4-Diaminobutane dihydrochloride | C4H12N2 | 89.1073 | 0.7 | 0.53 | 72.0813 | 0.03 | 0.1 | 0.1–2 | 0.99 |
| cadaverine | 1,5-Diaminopentane dihydrochloride | C5H14N2 | 103.1233 | −2.9 | 0.56 | 86.0968, 69.0704 | 0.015 | 0.05 | 0.05–1 | 0.99 |
| histamine | Histamine dihydrochloride | C5H9N3 | 112.0869 | 0.3 | 0.45 | 95.0606, 83.0608 | 0.015 | 0.05 | 0.05–2 | 0.99 |
| spermine | Spermine tetrahydrochloride | C10H26N4 | 203.2159 | −3.4 | 5.81 | 112.1122, 84.0811 | 0.006 | 0.02 | 0.02–5 | 0.98 |
| spermidine | Spermidine trihydrochloride | C7H19N3 | 146.1651 | 0.5 | 0.71 | 129.1386, 72.0813 | 0.003 | 0.01 | 0.01–5 | 0.96 |
| tyramine | Tyramine hydrochloride | C8H11NO | 138.0908 | 4.1 | 0.44 | 121.0648, 91.0547 | 0.0003 | 0.001 | 0.001–2 | 0.99 |
| tryptamine | Tryptamine hydrochloride | C10H12N2 | 161.1066 | 4.7 | 0.37 | 144.0805, 117.0697 | 0.0015 | 0.005 | 0.005–1 | 0.99 |
| melatonin | Melatonin | C13H16N2O2 | 233.1283 | −1.1 | 0.63 | 174.0912, 159.0677 | 0.0015 | 0.005 | 0.005–2 | 0.99 |
| dopamine | Dopamine hydrochloride | C8H11NO2 | 154.0864 | −3.0 | 0.39 | 137.0596, 91.0546 | 0.003 | 0.01 | 0.01–1 | 0.99 |
| serotonin | Serotonin hydrochloride | C10H12N2O | 177.1018 | 0.2 | 0.39 | 160.0755, 132.0807 | 0.006 | 0.02 | 0.02–2 | 0.99 |
| GSH | L-Glutathione reduced | C10H17N3O6S | 308.0904 | 2.1 | 7.92 | 162.0217, 76.0220 | 0.0006 | 0.002 | 0.002–5 | 0.99 |
| % Recovery (n = 4) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| White Wine | Red Wine | Must | Cider | Beer | Vinegar | Saké | ||||||||
| Low | High | Low | High | Low | High | Low | High | Low | High | Low | High | Low | High | |
| Gaba | 118 ± 3 | 107 ± 4 | 69 ± 2 | 66 ± 3 | - | 117 ± 5 | 121 ± 5 | 114 ± 1 | 76 ± 6 | 112 ± 5 | 73 ± 3 | 102 ± 5 | 96 ± 4 | 107 ± 8 |
| Asp | 94 ± 5 | 99 ± 7 | 105 ± 2 | 100 ± 2 | 75 ± 7 | 93 ± 4 | 108 ± 3 | 107 ± 2 | 99 ± 2 | 96 ± 4 | 69 ± 2 | 100 ± 7 | 103 ± 4 | 105 ± 6 |
| Glu | 80 ± 3 | 79 ± 3 | 119 ± 2 | 100 ± 2 | 94 ± 5 | 65 ± 6 | 101 ± 6 | 104 ± 7 | 97 ± 4 | 104 ± 5 | 66 ± 7 | 103 ± 8 | 106 ± 6 | 111 ± 7 |
| Ala | 104 ± 6 | 100 ± 3 | 103 ± 4 | 101 ± 3 | 117 ± 7 | - | 83 ± 2 | 112 ± 2 | 84 ± 3 | 63 ± 7 | - | 95 ± 5 | 64 ± 7 | 107 ± 3 |
| Arg | 110 ± 3 | 92 ± 5 | 117 ± 7 | 102 ± 1 | - | - | 98 ± 1 | 105 ± 6 | 81 ± 3 | 110 ± 3 | 60 ± 2 | 107 ± 5 | 97 ± 6 | 116 ± 8 |
| Asn | 90 ± 3 | 103 ± 3 | 103 ± 4 | 98 ± 4 | 95 ± 7 | 99 ± 2 | 98 ± 6 | 105 ± 5 | 90 ± 8 | 99 ± 7 | 83 ± 8 | 98 ± 3 | 99 ± 4 | 110 ± 6 |
| Cit | 78 ± 2 | 94 ± 6 | 93 ± 4 | 98 ± 6 | 83 ± 3 | 97 ± 2 | 87 ± 1 | 96 ± 5 | 103 ± 1 | 94 ± 5 | 79 ± 8 | 96 ± 8 | 75 ± 7 | 99 ± 6 |
| Phe | 82 ± 1 | 94 ± 7 | 116 ± 3 | 113 ± 6 | 115 ± 6 | 109 ± 4 | 114 ± 4 | 111 ± 4 | 73 ± 5 | 64 ± 2 | 67 ± 8 | 98 ± 3 | 79 ± 3 | 112 ± 6 |
| Gly | 89 ± 6 | 95 ± 4 | 106 ± 5 | 108 ± 1 | 103 ± 4 | 101 ± 3 | 90 ± 7 | 107 ± 7 | 96 ± 3 | 92 ± 1 | 73 ± 7 | 97 ± 6 | 122 ± 7 | 100 ± 4 |
| Hyp | 91 ± 3 | 104 ± 3 | 109 ± 3 | 106 ± 2 | 97 ± 1 | 105 ± 1 | 103 ± 4 | 107 ± 3 | 104 ± 1 | 99 ± 1 | 118 ± 1 | 110 ± 2 | 93 ± 1 | 104 ± 1 |
| Ile | 63 ± 3 | 68 ± 7 | 102 ± 4 | 112 ± 4 | 68 ± 7 | 116 ± 4 | 129 ± 1 | 102 ± 8 | 88 ± 8 | 73 ± 6 | 111 ± 3 | 66 ± 4 | 87 ± 8 | 117 ± 4 |
| Leu | 123 ± 2 | 87 ± 8 | 122 ± 2 | 97 ± 6 | 101 ± 8 | 122 ± 2 | 120 ± 5 | 120 ± 8 | 105 ± 3 | 83 ± 6 | 133 ± 5 | 100 ± 6 | 83 ± 4 | 117 ± 1 |
| His | 113 ± 5 | 108 ± 2 | 102 ± 2 | 120 ± 1 | 133 ± 5 | 102 ± 7 | 97 ± 4 | 104 ± 5 | 96 ± 6 | 79 ± 4 | 96 ± 1 | 111 ± 6 | 104 ± 8 | 111 ± 8 |
| Lys | 87 ± 1 | 93 ± 3 | 100 ± 1 | 109 ± 1 | 113 ± 2 | 109 ± 6 | 98 ± 8 | 103 ± 2 | 78 ± 2 | 98 ± 2 | 73 ± 6 | 110 ± 2 | 98 ± 1 | 111 ± 4 |
| Met | 111 ± 1 | 100 ± 3 | 100 ± 6 | 100 ± 8 | 107 ± 7 | 119 ± 5 | 99 ± 2 | 100 ± 6 | 114 ± 7 | 86 ± 8 | 127 ± 5 | 130 ± 4 | 100 ± 6 | 120 ± 1 |
| Orn | 91 ± 5 | 113 ± 8 | 111 ± 3 | 119 ± 6 | 114 ± 8 | 114 ± 3 | 93 ± 8 | 96 ± 1 | 117 ± 5 | 87 ± 4 | 92 ± 5 | 103 ± 6 | 89 ± 5 | 106 ± 5 |
| Ser | 81 ± 8 | 95 ± 2 | 99 ± 4 | 103 ± 5 | 95 ± 5 | 101 ± 7 | 100 ± 1 | 103 ± 2 | 82 ± 7 | 92 ± 2 | 78 ± 3 | 104 ± 2 | 89 ± 6 | 106 ± 5 |
| Tyr | 60 ± 6 | 83 ± 8 | 116 ± 7 | 112 ± 3 | 108 ± 1 | 117 ± 8 | 103 ± 5 | 108 ± 8 | - | 94 ± 8 | 92 ± 7 | 120 ± 1 | 60 ± 3 | 120 ± 2 |
| Thr | 89 ± 4 | 89 ± 6 | 118 ± 6 | 103 ± 1 | 85 ± 6 | 79 ± 1 | 84 ± 6 | 101 ± 3 | 82 ± 3 | 93 ± 7 | 72 ± 7 | 100 ± 2 | 99 ± 5 | 108 ± 6 |
| Trp | 104 ± 7 | 111 ± 5 | 101 ± 3 | 113 ± 1 | 103 ± 8 | 126 ± 5 | 106 ± 8 | 104 ± 7 | 114 ± 3 | 100 ± 1 | 98 ± 1 | 110 ± 4 | 99 ± 1 | 113 ± 8 |
| ethanolamine | 105 ± 1 | 103 ± 8 | 112 ± 5 | 110 ± 5 | 103 ± 1 | 96 ± 4 | 86 ± 8 | 126 ± 2 | 80 ± 3 | 102 ± 4 | 106 ± 1 | 110 ± 1 | 120 ± 7 | 119 ± 1 |
| phenylethylamine | 113 ± 2 | 107 ± 5 | 109 ± 5 | 101 ± 1 | 105 ± 2 | 102 ± 1 | 114 ± 8 | 93 ± 1 | 110 ± 4 | 110 ± 1 | 124 ± 6 | 116 ± 6 | 124 ± 4 | 124 ± 8 |
| putrescine | 97 ± 2 | 104 ± 3 | 137 ± 4 | 121 ± 8 | 108 ± 5 | 114 ± 7 | 92 ± 5 | 122 ± 4 | 69 ± 2 | 98 ± 7 | 89 ± 3 | 116 ± 2 | 103 ± 5 | 110 ± 8 |
| cadaverine | 86 ± 2 | 98 ± 4 | 67 ± 7 | 103 ± 6 | 89 ± 3 | 101 ± 5 | 113 ± 5 | 110 ± 5 | 110 ± 4 | 95 ± 3 | 65 ± 2 | 97 ± 4 | 94 ± 7 | 111 ± 7 |
| histamine | 93 ± 8 | 103 ± 7 | 113 ± 6 | 120 ± 8 | 96 ± 8 | 110 ± 7 | 111 ± 1 | 97 ± 3 | 91 ± 5 | 97 ± 7 | 104 ± 2 | 109 ± 2 | 104 ± 7 | 115 ± 2 |
| spermine | 60 ± 6 | 95 ± 2 | 113 ± 4 | 91 ± 4 | 124 ± 5 | 98 ± 2 | 61 ± 3 | 69 ± 2 | 85 ± 2 | 121 ± 6 | 60 ± 2 | 83 ± 4 | 55 ± 5 | 79 ± 8 |
| spermidine | 142 ± 4 | 125 ± 2 | - | 61 ± 6 | 114 ± 8 | 116 ± 8 | 122 ± 2 | 117 ± 2 | 110 ± 2 | 132 ± 4 | 136 ± 2 | 120 ± 2 | 153 ± 3 | 132 ± 7 |
| tyramine | 92 ± 4 | 98 ± 2 | 122 ± 6 | 117 ± 4 | 115 ± 5 | 117 ± 3 | 131 ± 4 | 120 ± 2 | 88 ± 8 | 99 ± 5 | 79 ± 1 | 103 ± 3 | 101 ± 3 | 115 ± 8 |
| tryptamine | 80 ± 7 | 92 ± 8 | 103 ± 5 | 111 ± 3 | 93 ± 2 | 108 ± 1 | 129 ± 2 | 117 ± 7 | 89 ± 1 | 97 ± 1 | 85 ± 2 | 99 ± 1 | 86 ± 6 | 113 ± 2 |
| melatonin | 105 ± 4 | 102 ± 5 | 104 ± 2 | 131 ± 7 | 118 ± 8 | 118 ± 1 | 127 ± 3 | 96 ± 4 | 90 ± 4 | 93 ± 4 | 127 ± 7 | 130 ± 8 | 126 ± 7 | 116 ± 7 |
| dopamine | 81 ± 1 | 89 ± 7 | 117 ± 7 | 119 ± 2 | 100 ± 1 | 103 ± 8 | 122 ± 3 | 117 ± 7 | 87 ± 2 | 97 ± 6 | 110 ± 2 | 108 ± 6 | 89 ± 1 | 102 ± 5 |
| serotonin | 80 ± 5 | 92 ± 6 | 90 ± 2 | 109 ± 3 | 99 ± 1 | 104 ± 7 | 113 ± 2 | 108 ± 5 | 75 ± 2 | 102 ± 4 | 95 ± 8 | 94 ± 2 | 93 ± 8 | 107 ± 5 |
| GSH | 75 ± 7 | 79 ± 6 | 81 ± 1 | 87 ± 7 | 81 ± 8 | 96 ± 1 | 80 ± 6 | 90 ± 4 | 77 ± 1 | 72 ± 1 | 66 ± 4 | 82 ± 1 | 93 ± 3 | 102 ± 5 |
| White Wine | Red Wine | Must | Cider | Beer | Vinegar | Saké | LOD | LOQ | |
|---|---|---|---|---|---|---|---|---|---|
| Gaba | 78.6 ± 3.8 | 89.2 ± 8.4 | 54.3 ± 9.7 | <LOD | 302 ± 13 | 138 ± 16 | 99.6 ± 22 | 0.05 | 0.15 |
| Asp | 15.3 ± 1.9 | 18.5 ± 3.6 | 16.5 ± 6.5 | 1.46 ± 0.93 | 9.78 ± 2.1 | 17.6 ± 3.4 | 7.69 ± 1.5 | 0.075 | 0.25 |
| Glu | 27.1 ± 4.6 | 34.3 ± 1.9 | 47.5 ± 4.5 | 2.53 ± 0.23 | 14.2 ± 3.7 | 15.4 ± 4.9 | 13.8 ± 2.8 | 0.075 | 0.25 |
| Ala | 95.4 ± 7.8 | 86.7 ± 4.8 | 29.6 ± 3.8 | 1.46 ± 0.34 | 93.6 ± 5.2 | 53.6 ± 12 | 55.1 ± 3.6 | 0.15 | 0.5 |
| Arg | 269 ± 25 | 326 ± 33 | 35.1 ± 8.2 | <LOQ | 57.6 ± 4.6 | 56.3 ± 3.7 | 125 ± 15 | 0.03 | 0.1 |
| Asn | 24.1 ± 3.4 | 15.6 ± 4.8 | 11.3 ± 2.6 | 0.862 ± 0.07 | 5.94 ± 1.9 | 4.03 ± 1.8 | 21.4 ± 3.2 | 0.15 | 0.5 |
| Cit | 1.29 ± 0.3 | 1.97 ± 0.53 | 4.28 ± 1.3 | <LOD | 7.12 ± 2.7 | <LOD | <LOD | 0.3 | 1 |
| Phe | 15.3 ± 2.1 | 17.3 ± 3.5 | 33.2 ± 3.8 | 2.36 ± 0.79 | 51.8 ± 3.2 | 21.8 ± 3.2 | 19.8 ± 3.7 | 0.015 | 0.05 |
| Gly | 33.9 ± 5.8 | 94.1 ± 6.3 | 23.8 ± 1.7 | <LOD | 66.8 ± 2.7 | 39.4 ± 7.2 | 77.5 ± 6.8 | 0.3 | 1 |
| Hyp | 6.18 ± 4.3 | 25.8 ± 1.5 | 16.5 ± 3.4 | 1.58 ± 0.42 | 1.67 ± 0.2 | 5.01 ± 1.6 | <LOD | 0.075 | 0.25 |
| Ile | 14.8 ± 1.6 | 9.31 ± 2.7 | 25.8 ± 6.2 | 0.561 ± 0.21 | 15.2 ± 3.7 | 32.6 ± 4.2 | 12.7 ± 1.7 | 0.015 | 0.05 |
| Leu | 18.6 ± 2.4 | 10.1 ± 2.8 | 23.4 ± 3.9 | <LOQ | 7.96 ± 1.8 | 23.4 ± 2.8 | 13.4 ± 4.1 | 0.005 | 0.05 |
| His | 15.7 ± 5.8 | 18.6 ± 1.8 | 115 ± 14 | <LOQ | 53.6 ± 5.7 | 15.3 ± 1.9 | 22.7 ± 3.9 | 0.075 | 2.5 |
| Lys | 56.1 ± 7.8 | 34.9 ± 3.4 | <LOD | 0.962 ± 0.15 | 6.73 ± 1.47 | 49.2 ± 5.1 | 26.9 ± 2.6 | 0.75 | 2.5 |
| Met | 4.74 ± 1.7 | 3.44 ± 1.7 | 7.62 ± 2.9 | <LOD | 16.9 ± 3.1 | 7.21 ± 1.5 | <LOQ | 0.075 | 0.25 |
| Orn | 11.9 ± 3.5 | 23.6 ± 2.6 | 6.86 ± 1.5 | <LOD | <LOD | <LOQ | <LOD | 0.75 | 2.5 |
| Ser | 19.8 ± 4.1 | 18.2 ± 2.5 | 148 ± 39 | 1.25 ± 0.68 | 7.52 ± 1.5 | 15.9 ± 3.2 | 16.8 ± 3.7 | 0.15 | 0.5 |
| Tyr | 8.56 ± 3.5 | 15.4 ± 1.7 | 22.4 ± 3.5 | 0.843 ± 0.32 | 42.3 ± 6.9 | 5.6 3 ± 2.7 | 11.8 ± 2.6 | 0.075 | 0.25 |
| Thr | 15.2 ± 1.9 | 17.9 ± 5.8 | 128 ± 32 | 2.5 ± 0.97 | 6.02 ± 1.08 | 14.7 ± 4.2 | 7.54 ± 2.6 | 0.3 | 1 |
| Trp | 6.72 ± 2.8 | 6.96 ± 5.2 | 33.9 ± 5.8 | <LOD | 7.39 ± 0.47 | <LOD | 0.984 ± 0.06 | 0.075 | 0.25 |
| ethanolamine | 40.8 ± 4.9 | 47.3 ± 4.7 | 35.6 ± 6.2 | 8.56 ± 1.6 | 26.9 ± 2.6 | 13.7 ± 1.7 | 33.1 ± 1.4 | 0.075 | 0.25 |
| phenylethylamine | 0.468 ± 0.15 | <LOQ | <LOD | <LOD | <LOQ | 1.47 ± 0.4 | 0.864 ± 0.29 | 0.015 | 0.05 |
| putrescine | <LOQ | 16.7 ± 5.2 | 11.6 ± 3.8 | <LOD | <LOD | 11.7 ± 2.6 | <LOD | 1.5 | 5 |
| cadaverine | <LOD | <LOD | <LOD | <LOD | <LOD | 12.5 ± 1.5 | <LOD | 0.75 | 2.5 |
| histamine | <LOD | 9.51 ± 4.7 | <LOQ | <LOD | <LOD | <LOQ | <LOD | 0.75 | 2.5 |
| spermine | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0.3 | 1 |
| spermidine | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0.15 | 0.5 |
| tyramine | <LOQ | 13.9 ± 4.1 | <LOD | 0.487 ± 0.12 | 1.45 ± 0.79 | 19.7 ± 1.8 | 1.71 ± 0.69 | 0.015 | 0.05 |
| tryptamine | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0.075 | 0.25 |
| melatonin | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0.075 | 0.25 |
| dopamine | <LOD | <LOD | <LOD | <LOD | <LOQ | <LOQ | <LOD | 0.15 | 0.5 |
| serotonin | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | <LOD | 0.3 | 1 |
| GSH | 2.85 ± 1.6 | 6.16 ± 3.5 | 3.92 ± 1.4 | <LOD | <LOD | <LOD | <LOD | 0.03 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delaiti, S.; Larcher, R.; Pedò, S.; Nardin, T. Development and Validation of a Fast UHPLC–HRMS Method for the Analysis of Amino Acids and Biogenic Amines in Fermented Beverages. Beverages 2025, 11, 124. https://doi.org/10.3390/beverages11050124
Delaiti S, Larcher R, Pedò S, Nardin T. Development and Validation of a Fast UHPLC–HRMS Method for the Analysis of Amino Acids and Biogenic Amines in Fermented Beverages. Beverages. 2025; 11(5):124. https://doi.org/10.3390/beverages11050124
Chicago/Turabian StyleDelaiti, Simone, Roberto Larcher, Stefano Pedò, and Tiziana Nardin. 2025. "Development and Validation of a Fast UHPLC–HRMS Method for the Analysis of Amino Acids and Biogenic Amines in Fermented Beverages" Beverages 11, no. 5: 124. https://doi.org/10.3390/beverages11050124
APA StyleDelaiti, S., Larcher, R., Pedò, S., & Nardin, T. (2025). Development and Validation of a Fast UHPLC–HRMS Method for the Analysis of Amino Acids and Biogenic Amines in Fermented Beverages. Beverages, 11(5), 124. https://doi.org/10.3390/beverages11050124






