Combinatorial Metabolic Engineering in Saccharomyces cerevisiae for the Enhanced Production of the FPP-Derived Sesquiterpene Germacrene
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning of Constructs
2.2. Strain Construction and Culture Conditions
2.3. GC-MS Analysis
3. Results
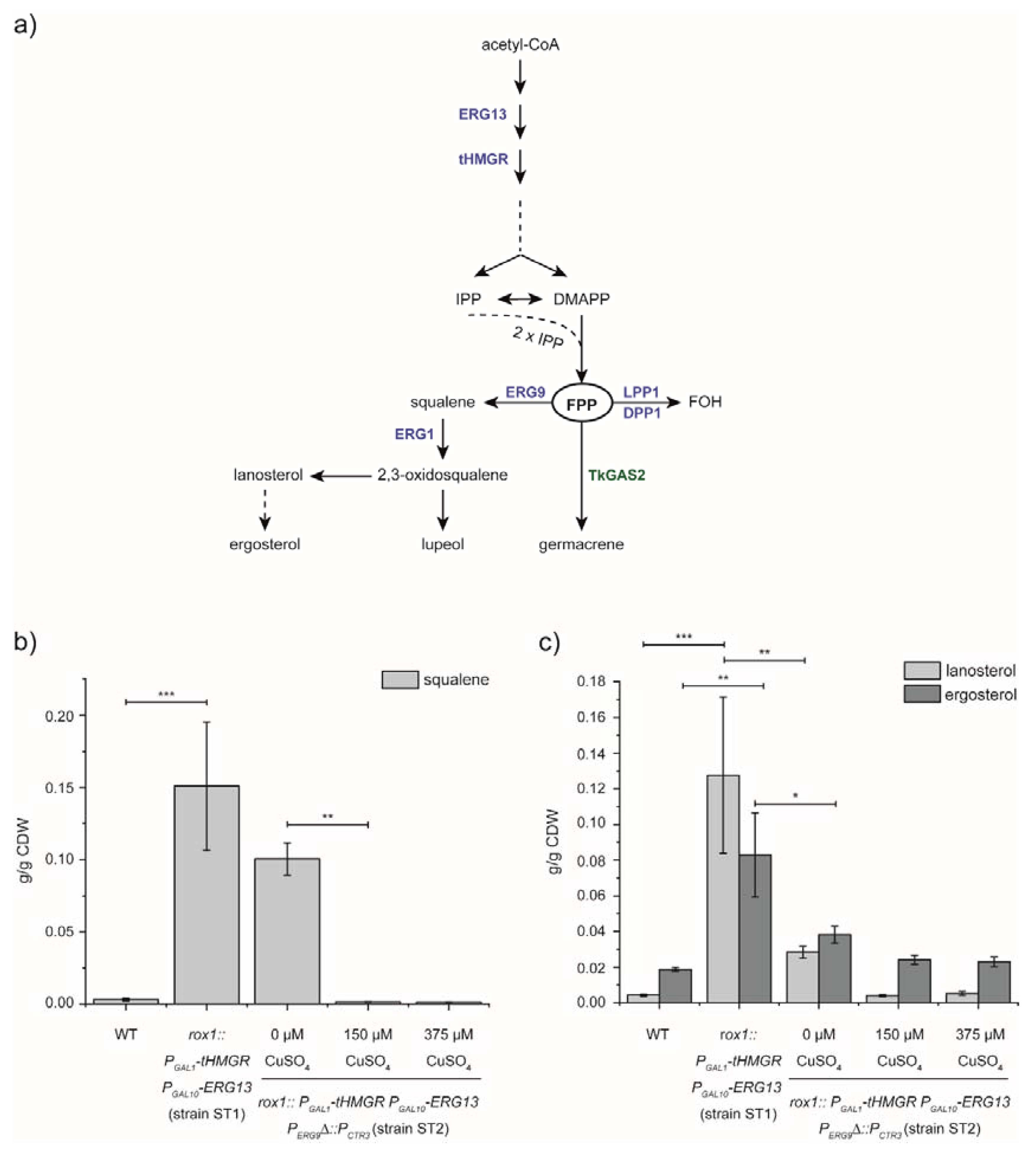
3.1. Manipulation of the MVA-Pathway by Overexpression, Deletion and Repression of Pathway Genes and Regulators
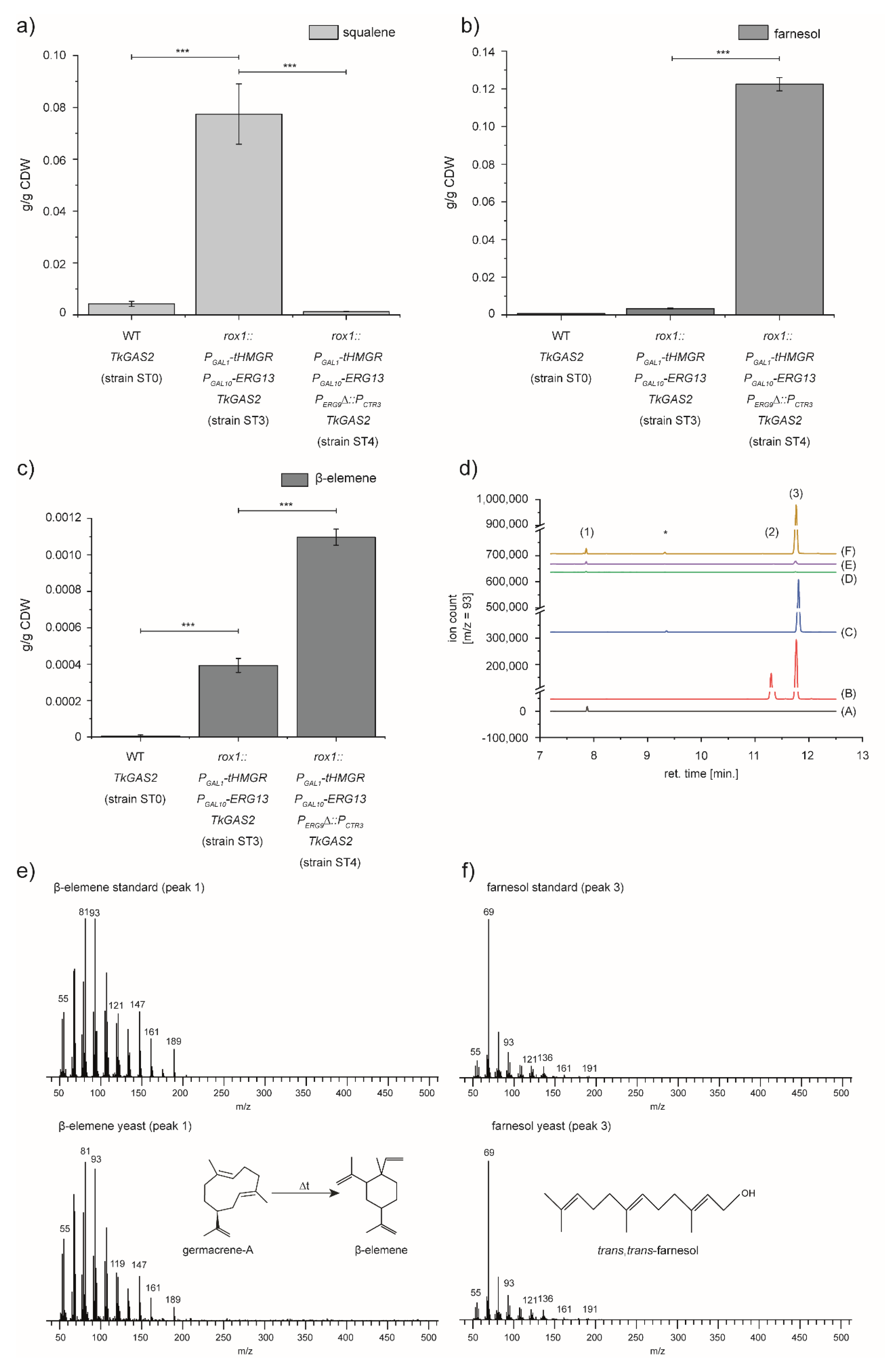
3.2. Expression of TkGAS2 to Validate the Potential of Sesquiterpenoid Production of the Engineered Yeast Strain
3.3. Enhanced Germacrene-A Synthesis by Preventing FPP Hydrolysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d´être for two distinct pathways in the early steps of plant isoprenoid biosynthesis. Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Daviet, L.; Schalk, M. Biotechnology in plant essential oil production: Progress and perspective in metabolic engineering of the terpene pathway. Flavour Fragr. J. 2010, 25, 123–127. [Google Scholar] [CrossRef]
- Schulze Gronover, C.; Wahler, D.; Prüfer, D. Natural Rubber Biosynthesis and Physic-Chemical Studies on Plant Derived Latex. Biotechnol. Biopolym. 2011, 75, 88. [Google Scholar]
- Grabińska, K.A.; Park, E.J.; Sessa, W.C. Cis-Prenyltransferase: New Insights into Protein Glycosylation, Rubber Synthesis, and Human Diseases. J. Biol. Chem. 2016, 291, 18582–18590. [Google Scholar] [CrossRef] [PubMed]
- Post, J.; van Deenen, N.; Fricke, J.; Kowalski, N.; Wurbs, D.; Schaller, H.; Eisenreich, W.; Huber, C.; Twymann, R.M.; Prüfer, D.; et al. Laticifer-specific cis-prenyltransferase silencing affects the rubber, triterpene, and inulin content of Taraxacum brevicorniculatum. Plant Physiol. 2012, 158, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Chakrabarty, R.; Tran, H.T.; Kwon, E.J.; Kwon, M.; Nguyen, T.D.; Ro, D.K. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 2015, 290, 1898–1914. [Google Scholar] [CrossRef]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.; Wu, S.; Shao, X.; Wang, X.; Gan, L.; Qin, B.; Yang, Y.; et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Yamashita, S.; Yamaguchi, H.; Waki, T.; Aoki, Y.; Mizuno, M.; Yanbe, F.; Ishii, T.; Funaki, A.; Tozawa, Y.; Miyagi-Inoue, Y.; et al. Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. Elife 2016, 5, e19022. [Google Scholar] [CrossRef]
- Epping, J.; van Deenen, N.; Niephaus, E.; Stolze, A.; Fricke, J.; Huber, C.; Eisenreich, W.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 2015, 1, 15048. [Google Scholar] [CrossRef]
- Donald, K.A.; Hampton, R.Y.; Fritz, I.B. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 3341–3344. [Google Scholar] [CrossRef]
- Yuan, J.; Ching, C.B. Combinatorial engineering of mevalonate pathway for improved amorpha-4,11-diene production in budding yeast. Biotechnol. Bioeng. 2014, 111, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Jakočiūnas, T.; Bonde, I.; Herrgård, M.; Harrison, S.J.; Kristensen, M.; Pedersen, L.E.; Jensen, M.K.; Keasling, J.D. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 2015, 28, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Bröker, J.N.; Müller, B.; van Deenen, N.; Prüfer, D.; Schulze Gronover, C. Upregulating the mevalonate pathway and repressing sterol synthesis in Saccharomyces cerevisiae enhances the production of triterpenes. Appl. Microbiol. Biotechnol. 2018, 102, 6923–6934. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, A.; Chen, X.; Rush, J.; Horazdovsky, B.; Waechter, C.J.; Carman, G.M.; Sternweis, P.C. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 14831–14837. [Google Scholar] [CrossRef]
- Takahashi, S.; Yeo, Y.; Greenhagen, B.T.; McMullin, T.; Song, L.; Maurina-Brunker, J.; Rosson, R.; Noel, J.P.; Chappell, J. Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol. Bioeng. 2007, 97, 170–181. [Google Scholar] [CrossRef]
- Albertsen, L.; Chen, Y.; Bach, L.S.; Rattleff, S.; Maury, J.; Brix, S.; Nielsen, J.; Mortensen, U.H. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl. Environ. Microbiol. 2011, 77, 1033–1040. [Google Scholar] [CrossRef]
- Scalcinati, G.; Knuf, C.; Partow, S.; Chen, Y.; Maury, J.; Schalk, M.; Daviet, L.; Nielsen, J.; Siewers, V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquiterpene α-santalene in a fed-batch mode. Metab. Eng. 2012, 14, 91–103. [Google Scholar] [CrossRef]
- Scalcinati, G.; Partow, S.; Siewers, V.; Schalk, M.; Daviet, L.; Nielsen, J. Combined metabolic engineering of precursor and co-factor supply to increase α-santalene production by Saccharomyces cerevisiae. Microb. Cell Fact. 2012, 11, 117. [Google Scholar] [CrossRef]
- Muramatsu, M.; Ohto, C.; Obata, S.; Sakuradani, E.; Shimizu, S. Alkaline pH enhances farnesol production by Saccharomyces cerevisiae. J. Biosci. Bioeng. 2009, 108, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Laughery, M.F.; Hunter, T.; Brown, A.; Hoopes, J.; Ostbye, T.; Shumaker, T.; Wyrick, J.J. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 2015, 32, 711–720. [Google Scholar] [CrossRef]
- Alberti, S.; Gitler, A.D.; Lindquist, S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 2007, 24, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAC/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Kirby, J.; Denby, C.M.; Keasling, J.D. Production and quantification of sesquiterpenes in Saccharomyces cerevisiae, including extraction, detection and quantification of terpene products and key metabolites. Nat. Protoc. 2014, 9, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, Y.J.; Bao, J.; Huang, L.; Nielsen, J.; Krivoruchko, A. Metabolic engineering of Saccharomyces cerevisiae for production of germacrene A, a precursor of beta-elemene. J. Ind. Microbiol. Biotechnol. 2017, 44, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Pazouki, L.; Memari, H.R.; Kännaste, A.; Bichele, R.; Niinemets, Ü. Germacrene A synthase in yarrow (Achillea millefolium) is an enzyme with mixed substrate specificity: Gene cloning, functional characterization and expression analysis. Front. Plant Sci. 2015, 6, 111. [Google Scholar] [CrossRef]
- Baidoo, E.E.K.; Wang, G.; Joshua, C.J.; Bentis, V.T.; Keasling, J.D. Liquid Chromatography and Mass Spectrometry Analysis of Isoprenoid Intermediates in Escherichia coli. Methods Mol. Biol. 2019, 1859, 209–224. [Google Scholar]
- Huber, M.; Epping, J.; Schulze Gronover, C.; Fricke, J.; Aziz, Z.; Brillatz, T.; Swyers, M.; Köllner, T.G.; Vogel, H.; Hammerbacher, A.; et al. A Latex Metabolite Benefits Plant Fitness under Root Herbivore Attack. PLoS Biol. 2016, 14, e1002332. [Google Scholar] [CrossRef]
- De Kraker, J.W.; Franssen, M.C.; de Groot, A.; Konig, W.A.; Bouwmeester, H.J. (+)-Germacrene A biosynthesis. The committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol. 1998, 117, 1381–1392. [Google Scholar] [CrossRef]
- Song, L. Detection of farnesyl diphosphate accumulation in yeast ERG9 mutants. Anal. Biochem. 2003, 317, 180–185. [Google Scholar] [CrossRef]
- Asadollahi, M.A.; Maury, J.; Møller, K.; Nielsen, K.F.; Schalk, M.; Clark, A.; Nielsen, J. Production of plant sesquiterpenes in Saccharomyces cerevisiae: Effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol. Bioeng. 2008, 99, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Tippmann, S.; Scalcinati, G.; Siewers, V.; Nielsen, J. Production of farnesene and santalene by Saccharomyces cerevisiae using fed-batch cultivations with RQ-controlled feed. Biotechnol. Bioeng. 2016, 113, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Park, S.H.; Kim, S.; Kim, S.W.; Hahn, J.S. Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. Appl. Microbiol. Biotechnol. 2019, 103, 211–223. [Google Scholar] [CrossRef]
- Garza, R.M.; Tran, P.N.; Hampton, R.Y. Geranylgeranyl pyrophosphate is a potent regulator of HRD-dependent 3-Hydroxy-3-methylglutaryl-CoA reductase degradation in yeast. J. Biol. Chem. 2009, 284, 35368–35380. [Google Scholar] [CrossRef]
- Morsomme, P.; Boutry, M. The plant plasma membrane H(+)-ATPase: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 1–16. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Zhang, Y.; Siewers, V.; Chen, Y.; Nielsen, J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab. Eng. 2015, 28, 24–42. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bröker, J.N.; Müller, B.; Prüfer, D.; Schulze Gronover, C. Combinatorial Metabolic Engineering in Saccharomyces cerevisiae for the Enhanced Production of the FPP-Derived Sesquiterpene Germacrene. Bioengineering 2020, 7, 135. https://doi.org/10.3390/bioengineering7040135
Bröker JN, Müller B, Prüfer D, Schulze Gronover C. Combinatorial Metabolic Engineering in Saccharomyces cerevisiae for the Enhanced Production of the FPP-Derived Sesquiterpene Germacrene. Bioengineering. 2020; 7(4):135. https://doi.org/10.3390/bioengineering7040135
Chicago/Turabian StyleBröker, Jan Niklas, Boje Müller, Dirk Prüfer, and Christian Schulze Gronover. 2020. "Combinatorial Metabolic Engineering in Saccharomyces cerevisiae for the Enhanced Production of the FPP-Derived Sesquiterpene Germacrene" Bioengineering 7, no. 4: 135. https://doi.org/10.3390/bioengineering7040135
APA StyleBröker, J. N., Müller, B., Prüfer, D., & Schulze Gronover, C. (2020). Combinatorial Metabolic Engineering in Saccharomyces cerevisiae for the Enhanced Production of the FPP-Derived Sesquiterpene Germacrene. Bioengineering, 7(4), 135. https://doi.org/10.3390/bioengineering7040135




