A Small Study of Bacterial Contamination of Anaerobic Digestion Materials and Survival in Different Feed Stocks
Abstract
1. Introduction
2. Materials and Methods
2.1. Pathogen Evaluation/Survey
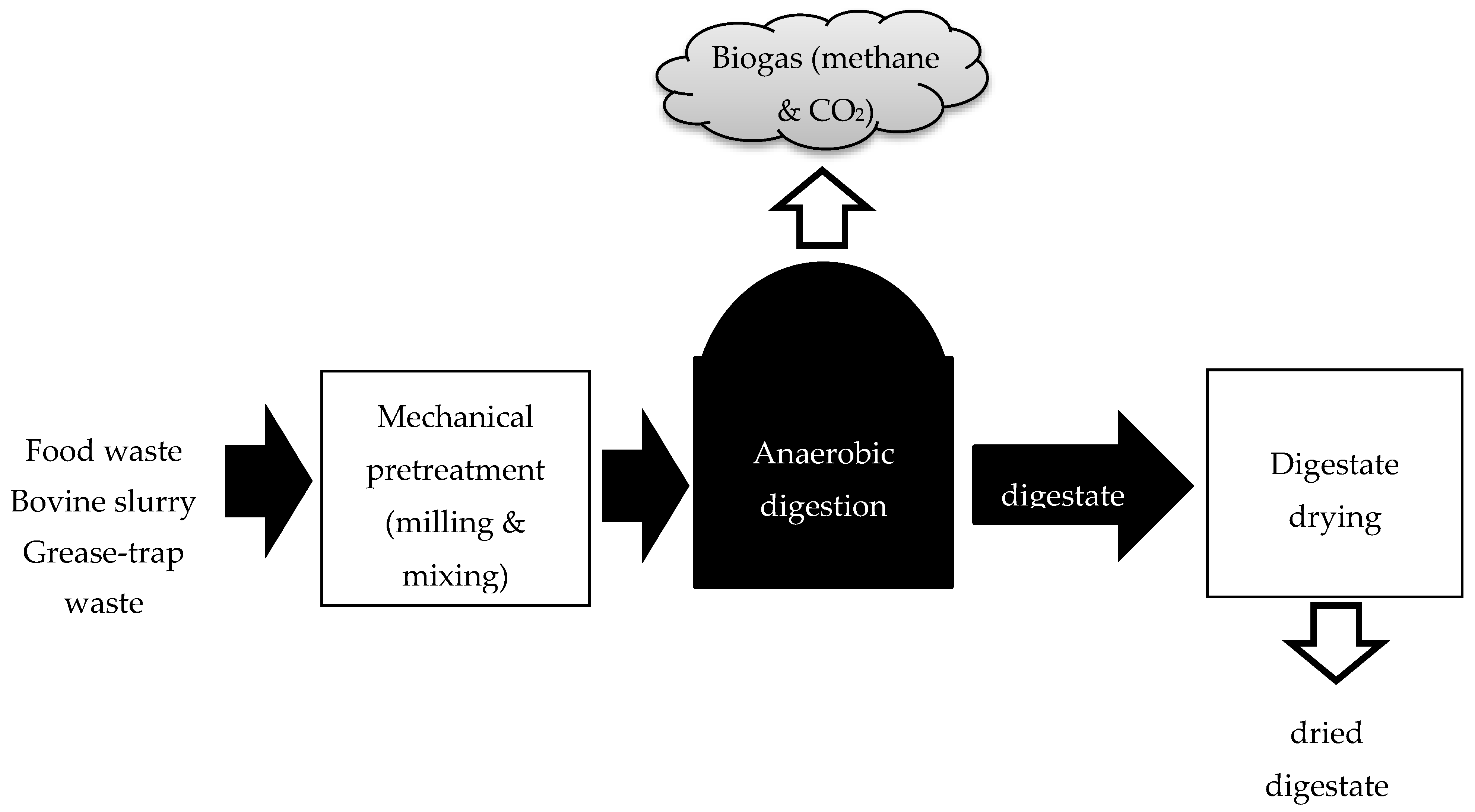
2.1.1. AD Samples
2.1.2. Microbiological Analysis
2.2. Survival Studies
2.2.1. Inoculum Preparation
2.2.2. AD Commercial Formulation Preparation
2.2.3. The Laboratory Model System
2.2.4. Enumeration of Surviving Cells
2.3. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alkanok, G.; Demirel, B.; Onay, T.T. Determination of biogas generation potential as a renewable energy source from supermarket wastes. Waste Manag. 2014, 34, 134–140. [Google Scholar] [CrossRef]
- Johansson, M.; Emmoth, E.; Salomonsson, A.C.; Albihn, A. Potential risks when spreading anaerobic digestion residues on grass silage crops—Survival of bacteria, moulds and viruses. Grass Forage Sci. 2005, 60, 175–185. [Google Scholar] [CrossRef]
- Ramos-Suárez, J.; Arroyo, N.C.; González-Fernández, C. The role of anaerobic digestion in algal biorefineries: Clean energy production, organic waste treatment, and nutrient loop closure. In Algae and Environmental Sustainability; Singh, B., Kuldeep, B., Faizal, B., Eds.; Springer: New Delhi, India, 2015; pp. 53–76. [Google Scholar]
- Anukam, A.; Mohammadi, A.; Naqvi, M.; Granström, K. A review of the chemistry of anaerobic digestion: Methods of accelerating and optimizing process efficiency. Processes 2019, 7, 504. [Google Scholar] [CrossRef]
- Sidhu, J.P.S.; Toze, S.G. Human pathogens and their indicators in biosolids: A literature review. Environ. Int. 2009, 35, 187–201. [Google Scholar] [CrossRef]
- Orzi, V.; Scaglia, B.; Lonati, S.; Riva, C.; Boccasile, G.; Alborali, G.; Adani, F. The role of biological processes in reducing both odor impact and pathogen content during mesophilic anaerobic digestion. Sci. Total Environ. 2015, 526, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Bonetta, S.; Bonetta, S.; Ferretti, E.; Fezia, G.; Gilli, G.; Carraro, E. Agricultural reuse of the digestate from anaerobic co-digestion of organic waste: Microbiological contamination, metal hazards and fertilizing performance. Water Air Soil Pollut. 2014, 225, 2046. [Google Scholar] [CrossRef]
- Bonetta, S.; Ferretti, E.; Bonetta, S.; Fezia, G.; Carraro, E. Microbiological contamination of digested products from anaerobic co-digestion of bovine manure and agricultural by-products. Lett. Appl. Microbiol. 2011, 53, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Stutz, L.H. Risk Assessment of Input of Pathogens Residing in Co-Substrates into the River from Sewage Treatment Plant with Mesophilic Anaerobic Digestion. A Case Study of Salmonella and Campylobacter Evaluation in the Sewage Treatment Plant of Bern (ARA Bern). Bachelor’s Thesis, Life Sciences and Facility Management, Swiss Federal Institute of Technology, Zurich, Switzerland, 2015. [Google Scholar]
- Auer, A.; Burgt, N.H.V.; Abram, F.; Barry, G.; Fenton, O.; Markey, B.K.; Nolan, S.; Richards, K.; Bolton, D.; De Waal, T.; et al. Agricultural anaerobic digestion power plants in Ireland and Germany: Policy and practice. J. Sci. Food Agric. 2016, 97, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Avery, L.M.; Anchang, K.Y.; Tumwesige, V.; Strachan, N.; Goude, P.J. Potential for pathogen reduction in anaerobic digestion and biogas generation in sub-Saharan Africa. Biomass Bioenergy 2014, 70, 112–124. [Google Scholar] [CrossRef]
- Nolan, S.; Waters, N.R.; Brennan, F.; Auer, A.; Fenton, O.; Richards, K.; Bolton, D.J.; Pritchard, L.; O’Flaherty, V.; Abram, F. Toward assessing farm-based anaerobic digestate public health risks: Comparative investigation with slurry, effect of pasteurization treatments, and use of miniature bioreactors as proxies for pathogen spiking trials. Fun. Environ. Micro. 2018, 2, 1–11. [Google Scholar] [CrossRef]
- Pathmanathan, S.G.; Cardona-Castro, N.; Sanchez-Jimenez, M.M.; Correa-Ochoa, M.M.; Puthucheary, S.D.; Thong, K.L. Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene. J. Med. Microbiol. 2003, 52, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Paton, J.C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Terzi, G.; Gücükoğlu, A.; Çadirci, Ö.; Uyanik, T.; Alişarli, M. Serotyping and antibiotic susceptibility of Listeria monocytogenes isolated from ready-to-eat foods in Samsun, Turkey. Turkish J. Vet. Anim. Sci. 2015, 39, 211–217. [Google Scholar] [CrossRef]
- Dutka-Malen, S.; Evers, S.; Courvalin, P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995, 33, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Liu, C.; Finegold, S.M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 2004, 70, 6459–6465. [Google Scholar] [CrossRef] [PubMed]
- Casadei, M.A.; Ingram, R.; Skinner, R.J.; Gaze, J.E. Heat resistance of Paenibacillus polymyxa in relation to pH and acidulants. J Appl. Microbiol. 2000, 89, 801–806. [Google Scholar] [CrossRef]
- Morandi, S.; Cremonesi, P.; Silvetti, T.; Castiglioni, B.; Brasca, M. Development of a triplex real-time PCR assay for the simultaneous detection of Clostridium beijerinckii, Clostridium sporogenes and Clostridium tyrobutyricum in milk. Anaerobe 2015, 34, 44–49. [Google Scholar] [CrossRef]
- McEvoy, J.M.; Doherty, A.M.; Sheridan, J.J.; Blair, I.S.; McDowell, D.A. The prevalence of Salmonella spp. in bovine faecal, rumen and carcass samples at a commercial abattoir. J. Appl. Microbiol. 2003, 94, 693–700. [Google Scholar] [CrossRef]
- Madden, R.H.; Murray, K.A.; Gilmour, A. Carriage of four bacterial pathogens by beef cattle in Northern Ireland at time of slaughter. Lett. Appl. Microbiol. 2007, 44, 115–119. [Google Scholar] [CrossRef]
- Fox, E.; O’Mahony, T.; Clancy, M.; Dempsey, R.; O’Brien, M.; Jordan, K. Listeria monocytogenes in the Irish dairy farm environment. J. Food Prot. 2009, 72, 1450–1456. [Google Scholar] [CrossRef]
- McEvoy, J.M.; Doherty, A.M.; Sheridan, J.J.; Thomson-Carter, F.M.; Garvey, P.; McGuire, L.; Blair, I.S.; McDowell, D.A. The prevalence and spread of Escherichia coli O157:H7 at a commercial beef abattoir. J. Appl. Microbiol. 2003, 95, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.M.; McCann, M.S.; Collery, M.M.; Logan, A.; Whyte, P.; McDowell, D.A.; Duffy, G. Tracking verocytotoxigenic Escherichia coli O157, O26, O111, O103 and O145 in Irish cattle. Int. J. Food Microbiol. 2012, 153, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Froschle, B.; Messelhausser, U.; Holler, C.; Lebuhn, M. Fate of Clostridium botulinum and incidence of pathogenic clostridia in biogass processes. J. Appl. Microbiol. 2015, 119, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Colleran, E. Hygienic and sanitation requirements in biogas plants treating animal manures or mixtures of manures and other organic wastes. In Anaerobic Digestion: Making Energy and Solving Modern Waste Problems; Ørtenblad, H., Ed.; AD-NETT, Herning Municipal Authoritie: Herning, Denmark, 2000; pp. 77–86. [Google Scholar]
- Hofmann, J.; Müller, L.; Weinrich, S.; Debeer, L.; Schumacher, B.; Velghe, F.; Liebetrau, J. Assessing the effects of substrate disintegration on methane yield. Chem. Eng. Technol. 2020, 43, 47–58. [Google Scholar] [CrossRef]
- Riau, T.; De la Rubia, M.A.; Pe´rez, M. Temperature-phased anaerobic digestion (TPAD) to obtain class A biosolids: A semi-continuous study. Bioresour. Technol. 2010, 101, 2706–2712. [Google Scholar] [CrossRef] [PubMed]
- Kunte, D.P.; Yeole, T.Y.; Ranade, D.R. Inactivation of Vibrio cholera during anaerobic digestion of human night soil. Bioresour. Technol. 2000, 75, 149–151. [Google Scholar] [CrossRef]
- Termorshuizen, A.J.; Volker, D.; Blok, W.J.; ten Brummeler, E.; Hartog, B.J.; Janse, J.D.; Knol, W.; Wenneker, M. Survival of human and plant pathogens during mesophilic digestion of vegetable, fruit and garden waste. Eur. J. Soil Biol. 2003, 39, 156–171. [Google Scholar] [CrossRef]
- Santha, H.; Sandino, J.; Shrimp, G.F.; Sung, S. Performance evaluation of a sequential-batch temperature-phased anaerobic digestion (TPAD) Scheme for producing class A biosolids. Water Environ. Res. 2006, 78, 221–226. [Google Scholar] [CrossRef]
- Olsen, J.E.; Larsen, H.E. Bacterial decimation times in anaerobic digestions of animal slurries. Biol. Wastes 1987, 21, 153–168. [Google Scholar] [CrossRef]
- Cote, C.; Masse, D.I.; Quessy, S. Reduction of indicator and pathogenic microorganisms by psychrophilic anaerobic digestion in swine slurries. Bioresour. Technol. 2006, 97, 686–691. [Google Scholar] [CrossRef]
- Higgins, M.J.; Chen, Y.; Murthy, S.N.; Hendrickson, D.; Farrel, J.; Schafer, P. Reactivation and growth of non-culturable indicator bacteria in anaerobically digested biosolids after centrifuge dewatering. Water Res. 2007, 41, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Masse, D.; Gilbert, Y.; Topp, E. Pathogen removal in farm-scale psychrophillic anaerobic digesters processing swine manure. Bioresour. Technol. 2011, 102, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, F.A.; Groves, S.J.; Chambers, B.J. Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 2005, 96, 135–143. [Google Scholar] [CrossRef] [PubMed]
- McGee, P.; Bolton, D.J.; Sheridan, J.J.; Earley, B.; Leonard, N. The survival of Escherichia coli O157:H7 in slurry from cattle fed different diets. Lett. Appl. Microbiol. 2001, 32, 152–155. [Google Scholar] [CrossRef]
- Kearney, T.E.; Larkin, M.J.; Frost, J.P.; Levett, P.N. Survival of pathogenic bacteria during mesophilic anaerobic digestion of animal waste. J Appl. Microbiol. 1993, 75, 215–219. [Google Scholar] [CrossRef]
- Horan, N.J.; Fletcher, L.; Betmal, S.M.; Wilks, S.A.; Keevil, C.W. Die-off of enteric pathogens during mesophilic anaerobic digestion. Water Res. 2004, 38, 1113–1120. [Google Scholar] [CrossRef]
- Chauret, C.; Springthorpe, S.; Sattar, S. Fate of Cryptosporidium oocysts, Giardia cysts and microbial indicators during wastewater treatment and anaerobic sludge digestion. Can. J. Microbiol. 1999, 45, 257–632. [Google Scholar] [CrossRef]
- Gahan, C.; Hill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Cell. Infect. Microbiol. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Pepper, I.L.; Brooks, J.P.; Sinclair, R.G.; Gurian, P.L.; Gerba, C.P. Pathogens and indicators in United States Class B biosolids: National and historic distributions. J. Environ. Qual. 2010, 39, 2185–2190. [Google Scholar] [CrossRef]
- Viau, E.; Peccia, J. Survey of wastewater indicatorsand human pathogen genomes in biosolids produced byclass A and class B stabilization treatments. Appl. Environ. Microbiol. 2009, 75, 164–174. [Google Scholar] [CrossRef]
- Smith, S.R.; Lang, N.L.; Cheung, K.H.M.; Spanoudaki, K. Factors controlling pathogen destruction during anaerobic digestion of biowastes. Waste Manag. 2005, 25, 417–425. [Google Scholar] [CrossRef] [PubMed]

| Detection | Confirmation | ||
|---|---|---|---|
| Treatment | Selective Agar | Culture Based | Molecular |
| Salmonella spp. | |||
| buffered peptone water | modified semi-solid Rappaport Vassiliadis medium with novobiocin supplement (20 mg/L), incubated at 42 °C for 24 h | Xylose lysine deoxycholate (XLD) agar | Pathmanathan et al. [13] |
| E. coli O157 | |||
| modified tryptone soya broth (mTSB) containing cefixime (50 µg/L) and vancomycin (6 mg/L) | Immunomagnetic separation with plating on sorbitol MacConkey agar supplemented with cefixime-tellurite (CT-SMAC) | Eosin methyl blue agar and plate count agar (PCA) followed by agglutination testing using the Sifin anti-coli O157 sera test | Paton and Paton [14]. |
| L. monocytogenes | |||
| half strength Fraser broth, incubated overnight at 30 °C followed by full strength Fraser broth incubated at 37 °C for 48 h | Listeria Selective Oxford agar and Brilliance Listeria agar (BLA), incubated at 37 °C for 48 h | PCA | Terzi et al. [15] |
| E. faecalis | |||
| BBL Enterococcosel broth and plated on Slanetz and Bartley agar (SBA) incubated at 37 °C for 24 h, followed by 44 °C for an additional 24 h | Pink colonies were streaked on PCA and stabbed in rows into well-dried bile aesculin agar plates, incubated at 44 °C for 24 h. | PCA | Dutka-Malen et al. [16] |
| Clostridium spp. | |||
| Maximum recovery diluent before plating on reinforced clostridial agar (RCA) incubated anaerobically (AnaeroGen sachets in BioMérieux GENbox jars (Hampshire, UK) at 37 °C for 48 h | Columbia blood agar supplemented with 5% defibrinated horse blood | Song et al. [17] | |
| Enumeration | PCR Confirmation | |
|---|---|---|
| S. Newport | XLD, supplemented with streptomycin sulphate (1000 µL/g) | Pathmanathan et al. [13] |
| E. coli O157 | CT-SMAC | Paton and Paton [14]. |
| L. monocytogenes | BLA, supplemented with streptomycin sulphate (1000 µL/g) incubated at 37 °C for 48 h | Terzi et al. [15]. |
| E. faecalis | SBA incubated at 37 °C for 24 h, followed by 44 °C for a further 24 h | Dutka-Malen et al. [16]. |
| C. sporogenes | RCA, incubated anaerobically (AnaeroGen sachets in BioMérieux GENbox jars (Hampshire, UK) at 37 °C for 48 h | Song et al. [17] and Morandi et al. [19]. |
| Pathogen | Salmonella spp. | E. coli O157 | L. monocytogenes | E. faecalis | Clostridium spp. |
|---|---|---|---|---|---|
| Type of samples | |||||
| Pre anaerobic digestion | |||||
| food waste (13) 1 | Positive (3) 2 | negative | positive (5) | positive (11) | positive (10) |
| bovine slurry (3) | negative | negative | positive (2) | positive (3) | positive (2) |
| mixing tank (8) | positive (1) | negative | positive (8) | positive (7) | positive (8) |
| Post anaerobic digestion | |||||
| raw digestate (19) | positive (5) | positive (1) | positive (17) | positive (18) | positive (17) |
| dried digestate (2) | negative | negative | negative | positive (2) | positive (2) |
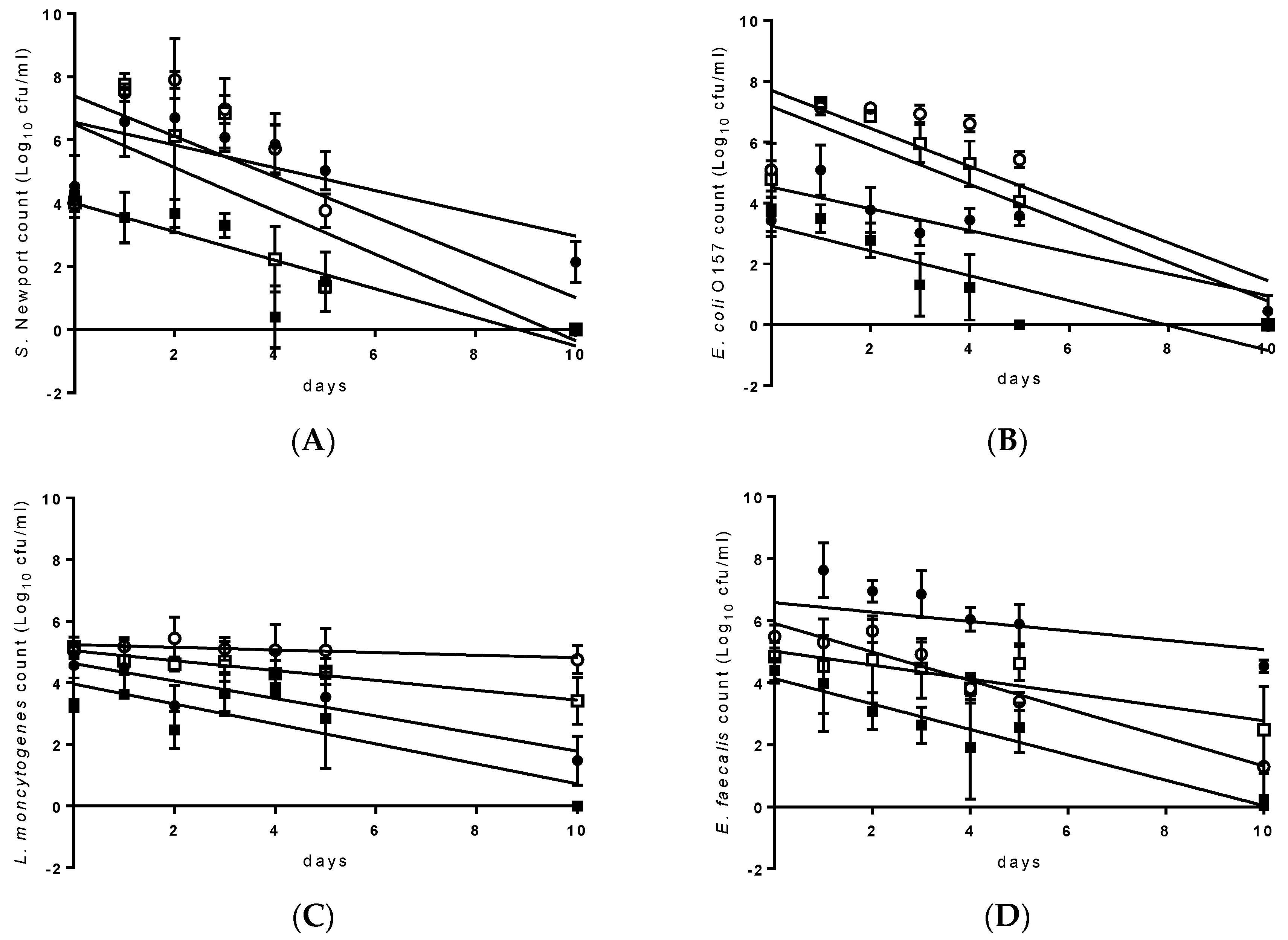
| Pathogen | Recipe | Growth | Decay Rate | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Yes/No | Period | Maximum Concentration (log10 cfu/mL) | Slope | SE | R2- Value | T90-Value (d) | n | ||
| S. Newport | 1 FW | yes | 1d | 7.8 | −0.69 | 0.110 | 0.49 | 1.5 A | 42 |
| 2 SF1 | yes | 1d | 7.3 | −0.64 | 0.089 | 0.56 | 1.6 A | 42 | |
| 3 SF2 | yes | 1d | 6.7 | −0.36 | 0.029 | 0.45 | 2.8 B | 42 | |
| 4 SGW | no | 5 NA | 6 NA | −0.45 | 0.051 | 0.66 | 2.2 B | 42 | |
| E. coli O157 | FW | yes | 1d | 7.3 | −0.64 | 0.062 | 0.77 | 1.6 A | 42 |
| SF1 | yes | 1d | 7.2 | −0.63 | 0.073 | 0.64 | 1.6 A | 42 | |
| SF2 | no | ND | NA | −0.36 | 0.044 | 0.62 | 2.8 B | 42 | |
| SGW | yes | 1d | 5.1 | −0.41 | 0.049 | 0.64 | 2.5 B | 42 | |
| L. monocytogenes | FW | no | ND | NA | −0.16 | 0.016 | 0.49 | 6.2 B | 42 |
| SF1 | no | ND | NA | 7 −0.04 | 0.027 | 0.05 | 23.5 C | 42 | |
| SF2 | no | ND | NA | −0.28 | 0.039 | 0.77 | 3.5 A | 42 | |
| SGW | no | ND | NA | −0.32 | 0.050 | 0.51 | 3.1 A | 42 | |
| E. faecalis | FW | no | ND | NA | −0.22 | 0.053 | 0.31 | 4.5 B | 42 |
| SF1 | yes | 1d | 7.6 | −0.46 | 0.030 | 0.85 | 2.2 A | 42 | |
| SF2 | yes | 1d | 7.6 | −0.15 | 0.060 | 0.14 | 6.6 C | 42 | |
| SGW | no | ND | NA | −0.41 | 0.049 | 0.63 | 2.4 A | 42 | |
| C. sporogenes | FW | yes | 3d | 7.1 | −0.13 | 0.025 | 0.38 | 8.0 C | 42 |
| SF1 | no | ND | NA | −0.15 | 0.024 | 0.50 | 6.5 B | 42 | |
| SF2 | no | ND | NA | −0.41 | 0.039 | 0.74 | 2.4 A | 42 | |
| SGW | no | ND | NA | −0.11 | 0.073 | 0.54 | 9.1 D | 42 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russell, L.; Whyte, P.; Zintl, A.; Gordon, S.; Markey, B.; de Waal, T.; Cummins, E.; Nolan, S.; O’Flaherty, V.; Abram, F.; et al. A Small Study of Bacterial Contamination of Anaerobic Digestion Materials and Survival in Different Feed Stocks. Bioengineering 2020, 7, 116. https://doi.org/10.3390/bioengineering7030116
Russell L, Whyte P, Zintl A, Gordon S, Markey B, de Waal T, Cummins E, Nolan S, O’Flaherty V, Abram F, et al. A Small Study of Bacterial Contamination of Anaerobic Digestion Materials and Survival in Different Feed Stocks. Bioengineering. 2020; 7(3):116. https://doi.org/10.3390/bioengineering7030116
Chicago/Turabian StyleRussell, Lauren, Paul Whyte, Annetta Zintl, Stephen Gordon, Bryan Markey, Theo de Waal, Enda Cummins, Stephen Nolan, Vincent O’Flaherty, Florence Abram, and et al. 2020. "A Small Study of Bacterial Contamination of Anaerobic Digestion Materials and Survival in Different Feed Stocks" Bioengineering 7, no. 3: 116. https://doi.org/10.3390/bioengineering7030116
APA StyleRussell, L., Whyte, P., Zintl, A., Gordon, S., Markey, B., de Waal, T., Cummins, E., Nolan, S., O’Flaherty, V., Abram, F., Richards, K., Fenton, O., & Bolton, D. (2020). A Small Study of Bacterial Contamination of Anaerobic Digestion Materials and Survival in Different Feed Stocks. Bioengineering, 7(3), 116. https://doi.org/10.3390/bioengineering7030116








