Application of Machine Learning in Predicting Osteogenic Differentiation of Mesenchymal Stem Cells
Abstract
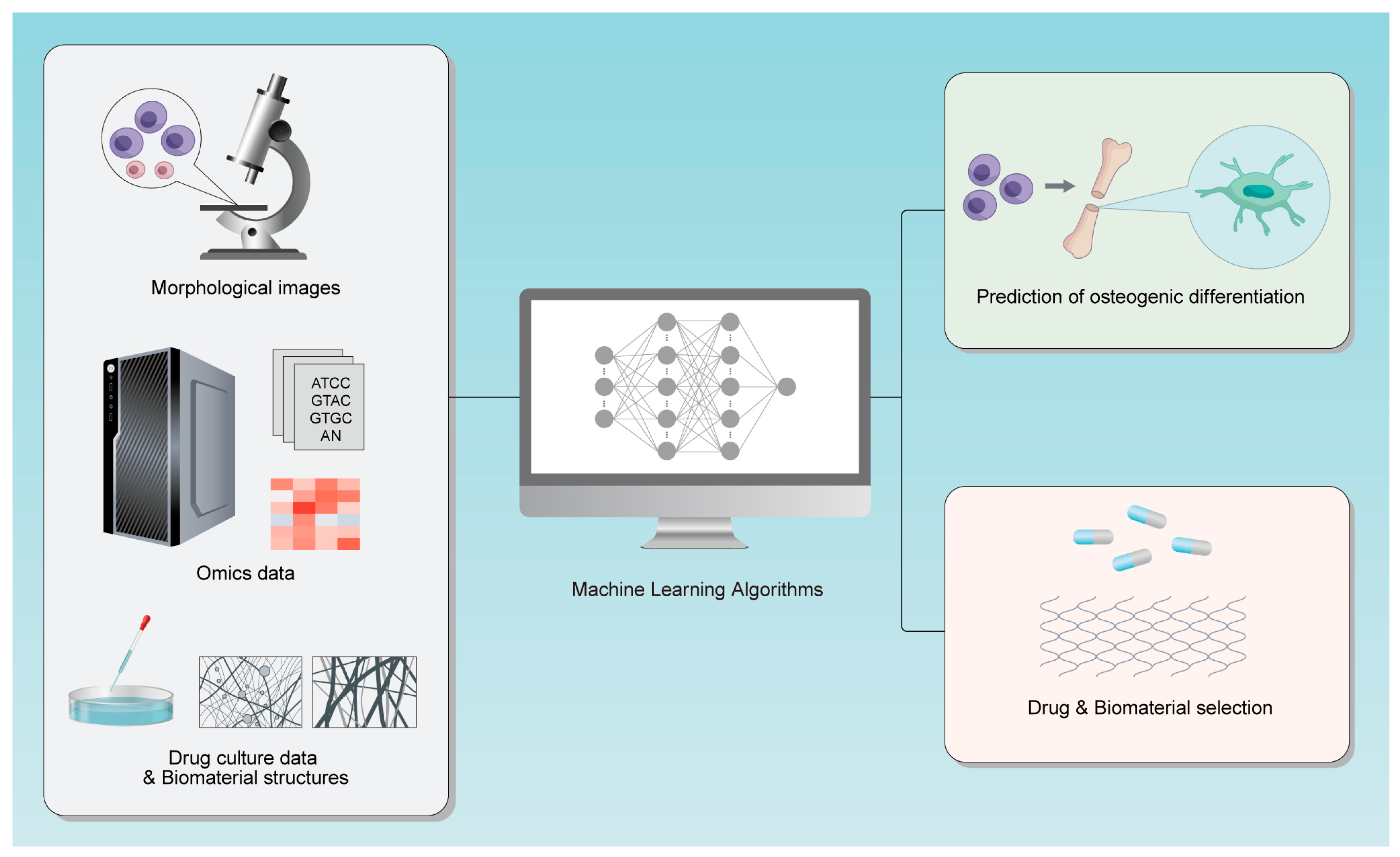
1. Introduction
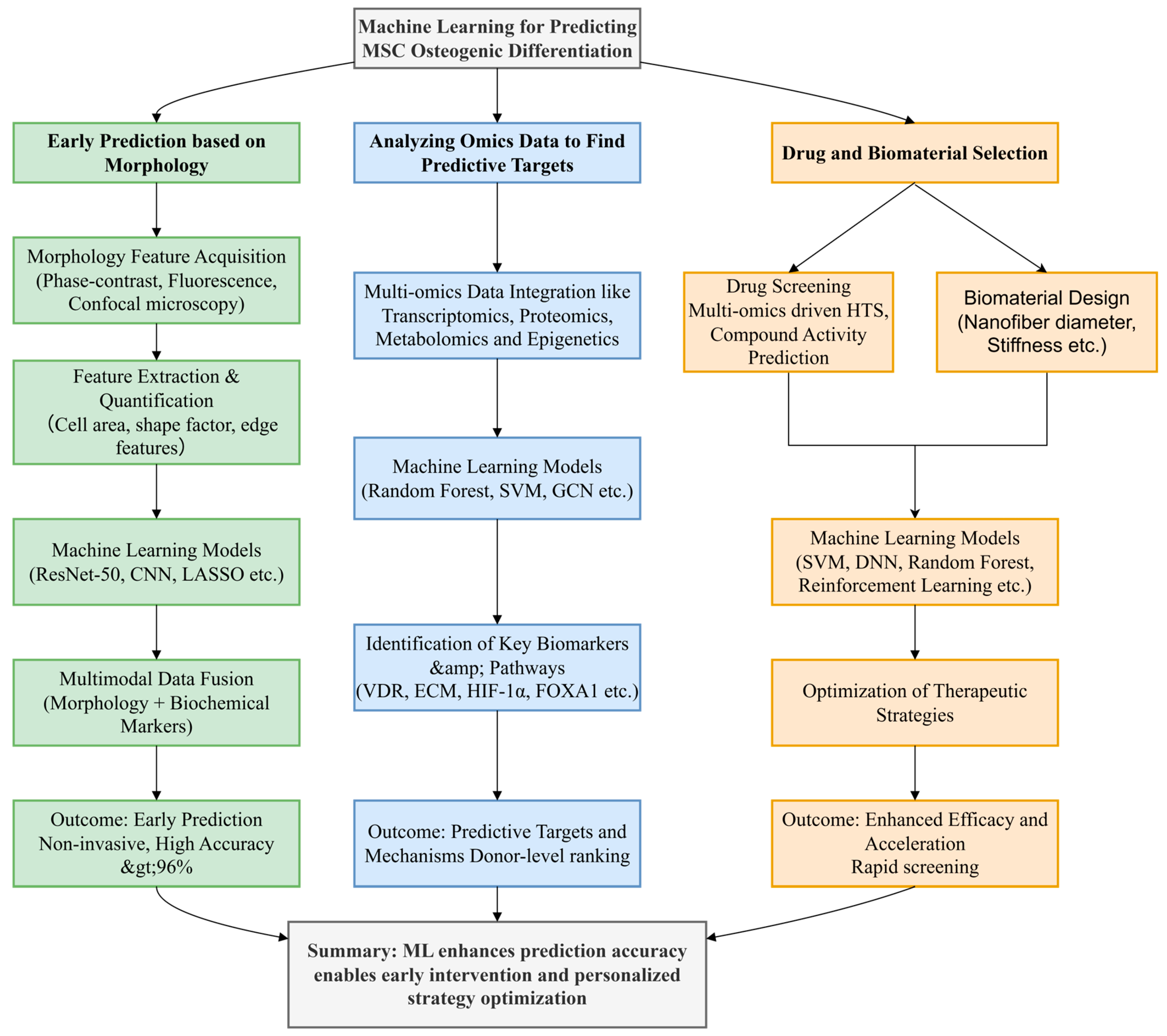
2. Method
3. Early Prediction Based on Morphology
4. Analyzing Omics Data to Find Predictive Targets
5. Drug and Biomaterial Selection
6. Conclusions
7. Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| DNN | deep neural network |
| ECM | extracellular matrix |
| GAN | generative adversarial network |
| GNN | graph neural network |
| MSC | Mesenchymal stem cell |
| PCA | Principal Component Analysis, |
| SBD | Shengxue Busui Decoction |
| scRNA-seq | single-cell RNA sequencing |
| SVM | support vector machine |
| VDR | vitamin D receptor |
References
- Sohn, H.; Oh, J. Review of bone graft and bone substitutes with an emphasis on fracture surgeries. Biomater. Res. 2019, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Burchardt, H. Biology of bone transplantation. Orthop. Clin. N. Am. 1987, 18, 187–196. [Google Scholar] [CrossRef]
- Huang, E.E.; Zhang, N.; Ganio, E.A.; Shen, H.; Li, X.; Ueno, M.; Utsunomiya, T.; Maruyama, M.; Gao, Q.; Su, N.; et al. Differential dynamics of bone graft transplantation and mesenchymal stem cell therapy during bone defect healing in a murine critical size defect. J. Orthop. Transl. 2022, 36, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Niti, A.; Koliakos, G.; Michopoulou, A. Stem Cell Therapies for Epidermolysis Bullosa Treatment. Bioengineering 2023, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Hochmann, S.; Ou, K.; Poupardin, R.; Mittermeir, M.; Textor, M.; Ali, S.; Wolf, M.; Ellinghaus, A.; Jacobi, D.; Elmiger, J.A.J.; et al. The enhancer landscape predetermines the skeletal regeneration capacity of stromal cells. Sci. Transl. Med. 2023, 15, eabm7477. [Google Scholar] [CrossRef]
- Wang, Z. Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine. Bioengineering 2023, 10, 857. [Google Scholar] [CrossRef]
- Chow, S.K.; Gao, Q.; Pius, A.; Morita, M.; Ergul, Y.; Murayama, M.; Shinohara, I.; Cekuc, M.S.; Ma, C.; Susuki, Y.; et al. The Advantages and Shortcomings of Stem Cell Therapy for Enhanced Bone Healing. Tissue Eng. Part C Methods 2024, 30, 415–430. [Google Scholar] [CrossRef]
- Shi, Q.; Song, F.; Zhou, X.; Chen, X.; Cao, J.; Na, J.; Fan, Y.; Zhang, G.; Zheng, L. Early Predicting Osteogenic Differentiation of Mesenchymal Stem Cells Based on Deep Learning Within One Day. Ann. Biomed. Eng. 2024, 52, 1706–1718. [Google Scholar] [CrossRef]
- Klontzas, M.E.; Vernardis, S.I.; Batsali, A.; Papadogiannis, F.; Panoskaltsis, N.; Mantalaris, A. Machine Learning and Metabolomics Predict Mesenchymal Stem Cell Osteogenic Differentiation in 2D and 3D Cultures. J. Funct. Biomater. 2024, 15, 367. [Google Scholar] [CrossRef]
- Feng, Z.; Su, X.; Wang, T.; Guo, S. Identification of Biomarkers That Modulate Osteogenic Differentiation in Mesenchymal Stem Cells Related to Inflammation and Immunity: A Bioinformatics-Based Comprehensive Study. Pharmaceuticals 2022, 15, 1094. [Google Scholar] [CrossRef]
- Han, J.; Chang, H.; Giricz, O.; Lee, G.Y.; Baehner, F.L.; Gray, J.W.; Bissell, M.J.; Kenny, P.A.; Parvin, B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput. Biol. 2010, 6, e1000684. [Google Scholar] [CrossRef]
- Seal, S.; Trapotsi, M.; Spjuth, O.; Singh, S.; Carreras-Puigvert, J.; Greene, N.; Bender, A.; Carpenter, A.E. Cell Painting: A decade of discovery and innovation in cellular imaging. Nat. Methods 2025, 22, 254–268. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, W.; Masson, A.; Li, Y. Cell signaling and transcriptional regulation of osteoblast lineage commitment, differentiation, bone formation, and homeostasis. Cell Discov. 2024, 10, 71. [Google Scholar] [CrossRef]
- Mai, M.; Luo, S.; Fasciano, S.; Oluwole, T.E.; Ortiz, J.; Pang, Y.; Wang, S. Morphology-based deep learning approach for predicting adipogenic and osteogenic differentiation of human mesenchymal stem cells (hMSCs). Front. Cell Dev. Biol. 2023, 11, 1329840. [Google Scholar] [CrossRef]
- Sasaki, H.; Takeuchi, I.; Okada, M.; Sawada, R.; Kanie, K.; Kiyota, Y.; Honda, H.; Kato, R. Label-free morphology-based prediction of multiple differentiation potentials of human mesenchymal stem cells for early evaluation of intact cells. PLoS ONE 2014, 9, e93952. [Google Scholar] [CrossRef]
- Hyder, H.; Baloch, G.; Batool, A.; Kim, Y.; Byun, Y. A Robust Deep Learning Framework for Mitigating Label Noise With Dual Selective Attention. IEEE Access 2025, 13, 115604–115626. [Google Scholar] [CrossRef]
- Lee, J.C.; Byeon, K.; Song, B.; Kim, K.; Kwak, J.T. DIOR-ViT: Differential ordinal learning Vision Transformer for cancer classification in pathology images. Med. Image Anal. 2025, 105, 103708. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, F.; Takeuchi, I.; Agata, H.; Kagami, H.; Shiono, H.; Kiyota, Y.; Honda, H.; Kato, R. Morphology-based prediction of osteogenic differentiation potential of human mesenchymal stem cells. PLoS ONE 2013, 8, e55082. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, F.; Takeuchi, I.; Agata, H.; Kagami, H.; Shiono, H.; Kiyota, Y.; Honda, H.; Kato, R. Characterization of time-course morphological features for efficient prediction of osteogenic potential in human mesenchymal stem cells. Biotechnol. Bioeng. 2014, 111, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wei, Z.; Yu, W.; Yin, R.; Yuan, Y.; Li, B.; Tang, Z.; Lu, Y.; Yang, Y. Spatial transcriptomics prediction from histology jointly through Transformer and graph neural networks. Brief Bioinform 2022, 23, bbac297. [Google Scholar] [CrossRef]
- Kong, Y.; Ao, J.; Chen, Q.; Su, W.; Zhao, Y.; Fei, Y.; Ma, J.; Ji, M.; Mi, L. Evaluating Differentiation Status of Mesenchymal Stem Cells by Label-Free Microscopy System and Machine Learning. Cells 2023, 12, 1524. [Google Scholar] [CrossRef]
- Zhang, Z.; Gong, L.; Li, M.; Wei, G.; Liu, Y. The osteogenic differentiation of human bone marrow stromal cells induced by nanofiber scaffolds using bioinformatics. BBA-Mol. Basis Dis. 2021, 1867, 166245. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, S.; Chu, X.; Reiter, J.; Gao, H.; McGuire, P.; Yu, X.; Xuei, X.; Liu, Y.; Wan, J.; et al. Osteogenic Differentiation Potential of Mesenchymal Stem Cells Using Single Cell Multiomic Analysis. Genes 2023, 14, 1871. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ping, X.; Guo, Y.; Heng, B.C.; Wang, Y.; Meng, Y.; Jiang, S.; Wei, Y.; Lai, B.; Zhang, X.; et al. Assessing Biomaterial-Induced Stem Cell Lineage Fate by Machine Learning-Based Artificial Intelligence. Adv. Mater. 2023, 35, e2210637. [Google Scholar] [CrossRef]
- Shen, G.; Ren, H.; Shang, Q.; Zhao, W.; Zhang, Z.; Yu, X.; Tang, K.; Tang, J.; Yang, Z.; Liang, D.; et al. Foxf1 knockdown promotes BMSC osteogenesis in part by activating the Wnt/beta-catenin signalling pathway and prevents ovariectomy-induced bone loss. Ebiomedicine 2020, 52, 102626. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, P.; Wang, J.; Lv, H.; Han, J.; Hou, Z.; Xu, R.; Chen, W. Advances in spatial transcriptomics and its application in the musculoskeletal system. Bone Res. 2025, 13, 21–54. [Google Scholar] [CrossRef]
- Chen, J.; Kuang, S.; Cen, J.; Zhang, Y.; Shen, Z.; Qin, W.; Huang, Q.; Wang, Z.; Gao, X.; Huang, F.; et al. Multiomics profiling reveals VDR as a central regulator of mesenchymal stem cell senescence with a known association with osteoporosis after high-fat diet exposure. Int. J. Oral. Sci. 2024, 16, 41. [Google Scholar] [CrossRef]
- Feng, K.; Yu, M.; Lou, X.; Wang, D.; Wang, L.; Ren, W. Multi-omics analysis of bone marrow mesenchymal stem cell differentiation differences in osteoporosis. Genomics 2023, 115, 110668. [Google Scholar] [CrossRef]
- Kamimoto, K.; Stringa, B.; Hoffmann, C.M.; Jindal, K.; Solnica-Krezel, L.; Morris, S.A. Dissecting cell identity via network inference and in silico gene perturbation. Nature 2023, 614, 742–751. [Google Scholar] [CrossRef]
- Chiarella, E.; Aloisio, A.; Scicchitano, S.; Lucchino, V.; Montalcini, Y.; Galasso, O.; Greco, M.; Gasparini, G.; Mesuraca, M.; Bond, H.M.; et al. ZNF521 Represses Osteoblastic Differentiation in Human Adipose-Derived Stem Cells. Int. J. Mol. Sci. 2018, 19, 4095. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Duffy, M.P.; Ahn, K.J.; Sussman, J.H.; Pang, M.; Smith, D.; Duncan, G.; Zhang, I.; Huang, J.; Lin, Y.; et al. Mapping the cellular biogeography of human bone marrow niches using single-cell transcriptomics and proteomic imaging. Cell 2024, 187, 3120–3140. [Google Scholar] [CrossRef]
- Piña, J.O.; Raju, R.; Roth, D.M.; Winchester, E.W.; Chattaraj, P.; Kidwai, F.; Faucz, F.R.; Iben, J.; Mitra, A.; Campbell, K.; et al. Multimodal spatiotemporal transcriptomic resolution of embryonic palate osteogenesis. Nat. Commun. 2023, 14, 5687. [Google Scholar] [CrossRef]
- Liu, M.; Ye, J.; Wu, R.; Luo, D.; Huang, T.; Dai, D.; Wang, K.; Du, Y.; Ou, J. Shengxue Busui Decoction activates the PI3K/Akt and VEGF pathways, enhancing vascular function and inhibiting osteocyte apoptosis to combat steroid-induced femoral head necrosis. Front. Pharmacol. 2025, 15, 1506594. [Google Scholar] [CrossRef]
- Hitora, Y.; Hokaguchi, M.; Sadahiro, Y.; Higaki, T.; Tsukamoto, S. Machine Learning Accelerates Screening of Osteoclast Differentiation Inhibitors from Natural Products. J. Nat. Prod. 2024, 87, 2393–2397. [Google Scholar] [CrossRef]
- Wang, L.; Lee, Y.; Bai, C.; Chiang, H.; Wang, H.; Yen, B.L.; Yen, M. A Rapid and Highly Predictive in vitro Screening Platform for Osteogenic Natural Compounds Using Human Runx2 Transcriptional Activity in Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2020, 8, 607383. [Google Scholar] [CrossRef] [PubMed]
- Fukuyasu, S.; Kayashima, H.; Moribayashi, A.; Matsuoka, S.; Nagasaki, A.; Okawa, H.; Yatani, H.; Saeki, M.; Egusa, H. Cell-Based Double-Screening Method to Identify a Reliable Candidate for Osteogenesis-Targeting Compounds. Biomedicines 2022, 10, 426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, J.; Zhang, G.; Wang, Y.; Si, S.; Chen, L.; Wang, Z. Small molecule T63 suppresses osteoporosis by modulating osteoblast differentiation via BMP and WNT signaling pathways. Sci. Rep. 2017, 7, 10397. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.E.; Cazzola, C.N.; Coleman, C.M. Design considerations when creating a high throughput screen-compatible in vitro model of osteogenesis. Slas Discov. 2024, 29, 100184. [Google Scholar] [CrossRef]
- Ye, C.; Ho, D.J.; Neri, M.; Yang, C.; Kulkarni, T.; Randhawa, R.; Henault, M.; Mostacci, N.; Farmer, P.; Renner, S.; et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun. 2018, 9, 4307. [Google Scholar] [CrossRef]
- de Melo, E.L.; Miranda, J.M.; Lima, V.B.D.S.; Gaião, W.D.C.; Tostes, B.D.V.A.; Rodrigues, C.G.; Bezerra Da Silva, M.; Júnior, S.A.; Pontes Perger, E.L.; Bispo, M.E.A.; et al. Effect of laser photobiomodulation combined with hydroxyapatite nanoparticles on the osteogenic differentiation of mesenchymal stem cells using artificial intelligence: An in vitro study. PLoS ONE 2024, 19, e0313787. [Google Scholar] [CrossRef]
- Wang, Z.; Dabaja, R.; Chen, L.; Banu, M. Machine learning unifies flexibility and efficiency of spinodal structure generation for stochastic biomaterial design. Sci. Rep. 2023, 13, 5414. [Google Scholar] [CrossRef]
- Lin, S.; Zhuang, Y.; Chen, K.; Lu, J.; Wang, K.; Han, L.; Li, M.; Li, X.; Zhu, X.; Yang, M.; et al. Osteoinductive biomaterials: Machine learning for prediction and interpretation. Acta Biomater. 2024, 187, 422–433. [Google Scholar] [CrossRef]
- Liu, Y.Y.F.; Lu, Y.; Oh, S.; Conduit, G.J. Machine learning to predict mesenchymal stem cell efficacy for cartilage repair. Plos Comput. Biol. 2020, 16, e1008275. [Google Scholar] [CrossRef]
- Mesuraca, M.; Nisticò, C.; Lombardo, N.; Piazzetta, G.L.; Lobello, N.; Chiarella, E. Cellular and Biochemical Characterization of Mesenchymal Stem Cells from Killian Nasal Polyp. Int. J. Mol. Sci. 2022, 23, 13214. [Google Scholar] [CrossRef]

| Model | Principle | Advantages | Limitations | Application Scenarios | Performance Metrics (References) |
|---|---|---|---|---|---|
| ResNet-50 | Alleviates gradient vanishing in deep networks via residual blocks and automatically extracts spatial features (e.g., cell edges, textures) from images. | High accuracy (AUC > 0.96); enables early prediction (within 24 h); suitable for high-resolution images. | High computational cost; requires large training datasets; sensitive to imaging parameters. | Morphological image analysis (e.g., live-cell imaging, bright-field images). | AUC > 0.96 (Mai et al.); Accuracy > 96% [8] |
| LASSO | Uses L1 regularization for feature selection to retain key morphological features (e.g., cell area, shape factor) associated with osteogenic differentiation. | Reduces overfitting; suitable for small sample sizes; non-invasive (avoids cell destruction). | Cannot handle high-dimensional image data; relies on manual feature extraction; lower accuracy than deep learning models. | Morphological parameter analysis (e.g., quantitative cell morphology indices). | Accuracy: 82% [15] |
| Ridge Regression | Optimizes image acquisition and analysis methods, combining biochemical markers (e.g., ALP activity, calcium deposition) to build prediction models. | Enables early prediction (3-day morphological features predict 3-week results); improves reliability by integrating biochemical markers. | Requires validation with biochemical markers; depends on consistent image acquisition. | Osteogenic prediction combining morphology and biochemical markers. | Accurately predicts 3-week osteogenic differentiation results [19] |
| Generative Adversarial Network (GAN) | Enhances cell image data through adversarial training of a generator and discriminator, improving model performance in small-sample scenarios. | Solves overfitting in small-sample cases; improves model generalization ability. | Requires high-quality generated data; complex training process. | Small-sample morphological analysis (e.g., limited cell image data). | Accuracy > 85% [22] |
| Random Walk | random walk on PPI networks to screen core genes. | Uncovers MSC heterogeneity; identifies core osteogenic regulatory genes; constructs gene regulatory networks. | Requires scRNA-seq data; depends on PPI network accuracy. | Transcriptomic data processing (e.g., scRNA-seq analysis). | Identifies osteogenic regulatory genes (e.g., FOXA1) [24] |
| Cross-modal Transformer | Fuses RNA-seq (transcriptomics) and TMT (proteomics) data, capturing time delays between gene expression and protein activity via self-attention mechanisms. | Reveals post-transcriptional regulatory mechanisms; integrates multi-omics data. | Requires multi-omics data; high computational complexity. | Transcriptomics-proteomics integration analysis. | Determines 24 h delay between ALP gene expression and protein activity [21] |
| Support Vector Machine (SVM) | SVM for processing proteomic data to identify differential proteins | Identifies differential proteins in ECM pathways; predicts early metabolic markers. | Requires proteomic/metabolomic data; poor model interpretability. | Proteomic/metabolomic analysis (e.g., data from MSCs of osteoporosis patients). | Identifies 205 differential proteins in ECM pathways (Feng et al.); accuracy: 89% [9] |
| Random Forest | Integrates multiple decision trees to analyze metabolomic data (e.g., lactate, ATP levels) and correlate metabolites with osteogenic differentiation efficiency. | Resists overfitting; suitable for multi-feature data; identifies early metabolic markers. | Poor model interpretability; long computation time. | Metabolomic data screening (e.g., metabolomic analysis of MSCs in 2D/3D cultures). | Accuracy: 89% [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, H.; Zhou, Z.; Yang, Y.; Lin, K.; Zhou, C.; Wang, X. Application of Machine Learning in Predicting Osteogenic Differentiation of Mesenchymal Stem Cells. Bioengineering 2025, 12, 1089. https://doi.org/10.3390/bioengineering12101089
Mao H, Zhou Z, Yang Y, Lin K, Zhou C, Wang X. Application of Machine Learning in Predicting Osteogenic Differentiation of Mesenchymal Stem Cells. Bioengineering. 2025; 12(10):1089. https://doi.org/10.3390/bioengineering12101089
Chicago/Turabian StyleMao, Hanyue, Zheng Zhou, Ying Yang, Kunlu Lin, Chuyao Zhou, and Xiaoyan Wang. 2025. "Application of Machine Learning in Predicting Osteogenic Differentiation of Mesenchymal Stem Cells" Bioengineering 12, no. 10: 1089. https://doi.org/10.3390/bioengineering12101089
APA StyleMao, H., Zhou, Z., Yang, Y., Lin, K., Zhou, C., & Wang, X. (2025). Application of Machine Learning in Predicting Osteogenic Differentiation of Mesenchymal Stem Cells. Bioengineering, 12(10), 1089. https://doi.org/10.3390/bioengineering12101089






