Automatic Segmentation of Bone Marrow Lesions on MRI Using a Deep Learning Method
Abstract
1. Introduction
2. Methods
2.1. Dataset
2.2. BML Segmentation Flowchart
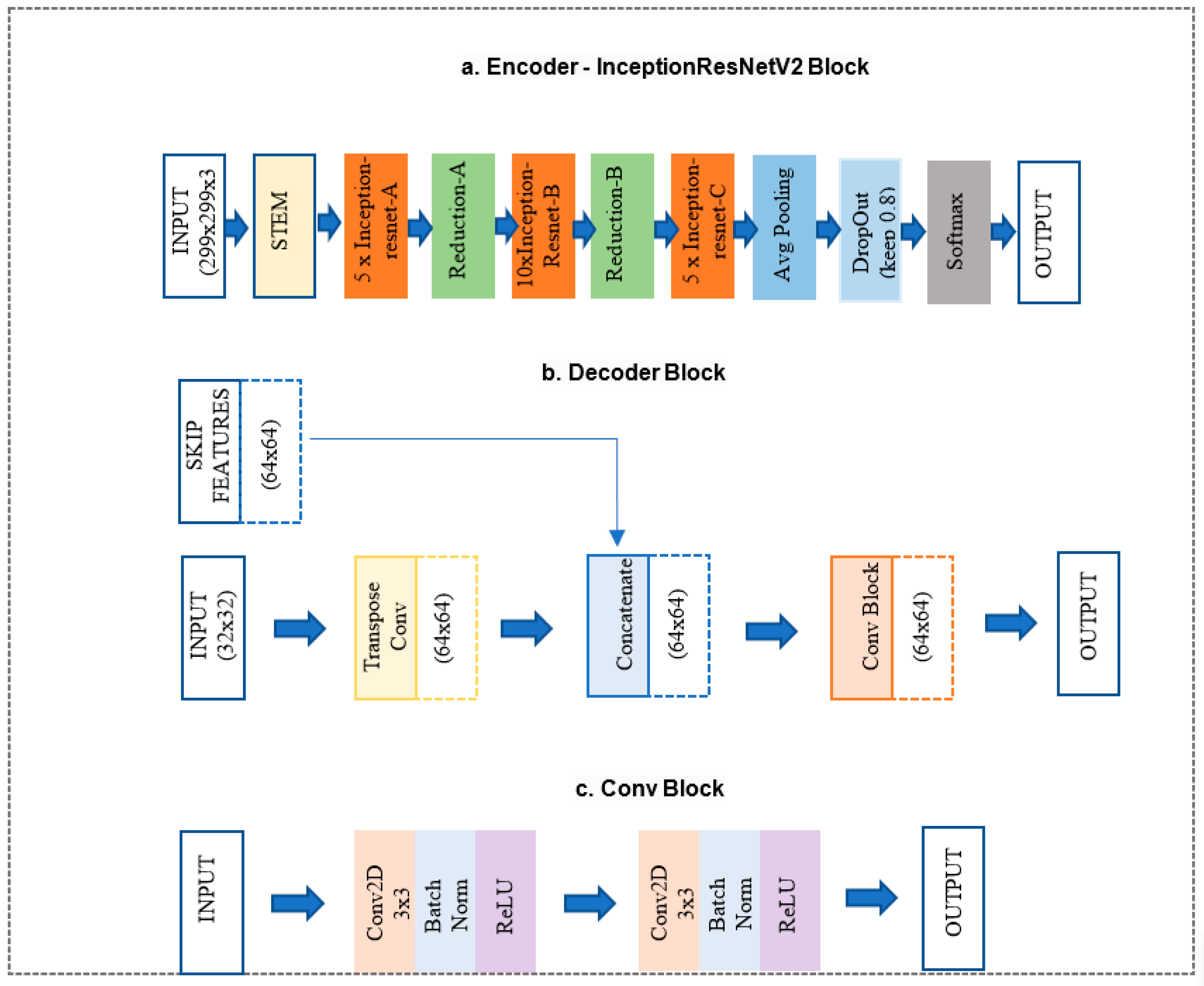
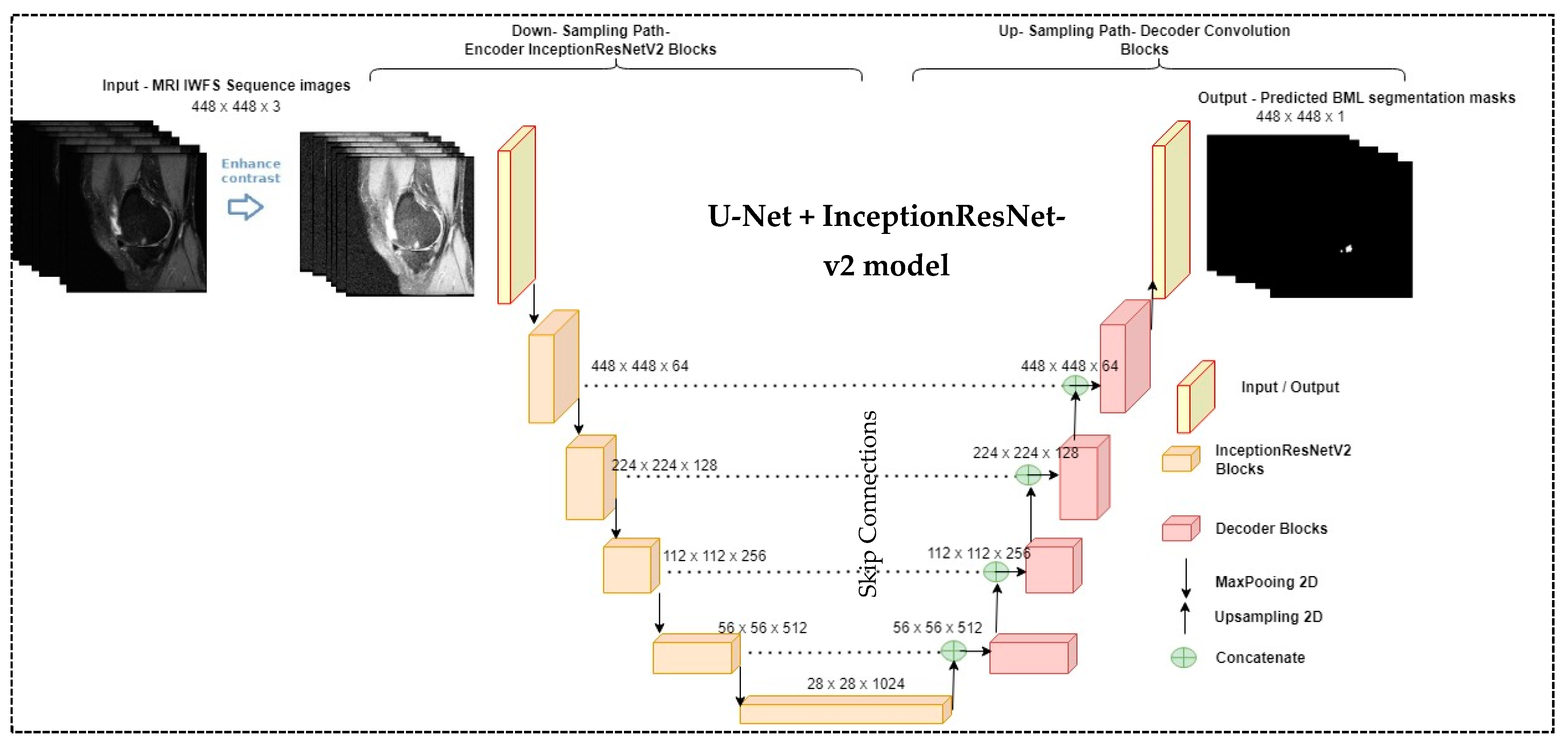
2.3. The Proposed Segmentation Model (U-Net + InceptionResNet-v2 Network)
- InceptionResNet-v2 block (Figure 3a) is a convolutional neural network trained on more than a million images from the ImageNet database. The network is 164 layers deep and can classify images into 1000 object categories. Therefore, the network has learned rich feature representations for a wide range of images. The network accepts an image input size of 299 × 299, and the output is a list of estimated class probabilities, as shown in Figure 3a. This block replaced the proposed model’s original U-Net encoder and bottleneck path. Here, we used the pre-trained weights from ImageNet.
- The decoder block (Figure 3b) consists of a 2 × 2 transpose convolution layer followed by the skip connection taken from the InceptionResNet-v2 block encoder, followed by a convolution block (Figure 3c). The convolution block consists of two 3 × 3 convolution layers. A batch normalization layer and a ReLU activation function follow each convolution layer. This block replaced the original U-Net model decoder convolution layer. The Decoder block will take input from the previous block and skip the connection feature map from the encoder to regenerate the segmentation mask using the convolution block.
2.4. Loss Function
3. Experiment and Results
3.1. Experiment Setup
3.2. Evaluation Metrics
3.3. Results and Comparison with Other Methods
4. Discussion
- The proposed method is fully automatic with no requirement for human intervention.
- It produces exact BML volume as a quantitative output of the pipeline, which is different from previous methods that generated approximate measures or categorical output, such as manual linear measurement (e.g., the greatest diameter [38], approximate BML volume [12]) or semi-quantitative scoring methods (e.g., BLOKS [7], WORMS [6]).
- It does not require treating different BML sizes differently (e.g., treating larger BML differently for the flow to work [20]). Our method can automatically learn the contextual information about the underlined structures by training different label data variants when the input images pass through the model network. This helps us to achieve full automation and precision for BML segmentation, to facilitate the future incorporation of BML features into the diagnosis model for knee OA.
- Lastly, our method can provide both the 2D slice-level segmentation masks and the 3D volume of BML. This enables evaluation from different levels and perspectives to better understand the model’s performance. The 2D masks may be further used to analyze the shape of BML besides computing the volume only.
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Chronic Disease Fact Sheets-Arthritis. Available online: https://www.cdc.gov/chronicdisease/resources/publications/factsheets/arthritis.htm (accessed on 17 February 2023).
- Centers for Disease Control and Prevention. National Statistics. Available online: https://www.cdc.gov/arthritis/data_statistics/national-statistics.html (accessed on 17 February 2023).
- Hootman, J.M.; Helmick, C.G.; Barbour, K.E.; Theis, K.A.; Boring, M.A. Updated projected prevalence of self-reported doctor-diagnosed arthritis and arthritis-attributable activity limitation among US adults, 2015–2040. Arthritis Rheumatol. 2016, 68, 1582–1587. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Guermazi, A.; Lo, G.H.; Grainger, A.J.; Conaghan, P.G.; Boudreau, R.M.; Roemer, F.W. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthr. Cartil. 2011, 19, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Driban, J.B.; Price, L.L.; Lo, G.H.; Pang, J.; Hunter, D.J.; Miller, E.; Ward, R.J.; Eaton, C.B.; Lynch, J.A.; McAlindon, T.E. Evaluation of bone marrow lesion volume as a knee osteoarthritis biomarker-longitudinal relationships with pain and structural changes: Data from the Osteoarthritis Initiative. Arthritis Res. Ther. 2013, 15, R112. [Google Scholar] [CrossRef] [PubMed]
- Peterfy, C.; Guermazi, A.; Zaim, S.; Tirman, P.; Miaux, Y.; White, D.; Kothari, M.; Lu, Y.; Fye, K.; Zhao, S.; et al. Whole-Organ Magnetic Resonance Imaging Score (WORMS) of the knee in osteoarthritis. Osteoarthr. Cartil. 2004, 12, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Lo, G.H.; Gale, D.; Grainger, A.J.; Guermazi, A.; Conaghan, P.G. The reliability of a new scoring system for knee osteoarthritis MRI and the validity of bone marrow lesion assessment: BLOKS (Boston–Leeds Osteoarthritis Knee Score). Ann. Rheum. Dis. 2008, 67, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Kornaat, P.R.; Ceulemans, R.Y.T.; Kroon, H.M.; Riyazi, N.; Kloppenburg, M.; Carter, W.O.; Woodworth, T.G.; Bloem, J.L. MRI assessment of knee osteoarthritis: Knee Osteoarthritis Scoring System (KOSS)—Inter-observer and intra-observer repro-ducibility of a compartment-based scoring system. Skelet. Radiol. 2004, 34, 95–102. [Google Scholar] [CrossRef]
- Preiswerk, F.; Sury, M.S.; Wortman, J.R.; Neumann, G.; Wells, W.; Duryea, J. Fast quantitative bone marrow lesion measurement on knee MRI for the assessment of osteoarthritis. Osteoarthr. Cartil. Open 2022, 4, 100234. [Google Scholar] [CrossRef] [PubMed]
- Felson, D.; Parkes, M.; Marjanovic, E.; Callaghan, M.; Gait, A.; Cootes, T.; Lunt, M.; Oldham, J.; Hutchinson, C. Bone marrow lesions in knee osteoarthritis change in 6–12 weeks. Osteoarthr. Cartil. 2012, 20, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, S.; Roman-Blas, J.A.; Largo, R.; Herrero-Beaumont, G. Subchondral bone as a key target for osteoarthritis treatment. Biochem. Pharmacol. 2012, 83, 315–323. [Google Scholar] [CrossRef]
- Driban, J.B.; Lo, G.H.; Lee, J.Y.; Ward, R.J.; Miller, E.; Pang, J.; Price, L.L.; McAlindon, T.E. Quantitative bone marrow lesion size in oste-oarthritic knees correlates with cartilage damage and predicts longitudinal cartilage loss. BMC Musculoskelet. Disord. 2011, 12, 217. [Google Scholar] [CrossRef]
- Kothari, A.; Guermazi, A.; Chmiel, J.S.; Dunlop, D.; Song, J.; Almagor, O.; Marshall, M.; Cahue, S.; Prasad, P.; Sharma, L.; et al. Within-subregion relationship between bone marrow lesions and subsequent cartilage loss in knee osteoar-thritis. Arthritis Care Res. 2010, 62, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Roemer, F.W.; Guermazi, A.; Javaid, M.K.; Lynch, J.A.; Niu, J.; Zhang, Y.; Felson, D.T.; Lewis, C.E.; Torner, J.; Nevitt, M.C.; et al. Change in MRI-detected subchondral bone marrow lesions is associated with cartilage loss: The MOST Study. A longitudinal multicentre study of knee osteoarthritis. Ann. Rheum. Dis. 2009, 68, 1461–1465. [Google Scholar] [PubMed]
- Roemer, F.; Neogi, T.; Nevitt, M.; Felson, D.; Zhu, Y.; Zhang, Y.; Lynch, J.; Javaid, M.; Crema, M.D.; Torner, J.; et al. Subchondral bone marrow lesions are highly associated with, and predict subchondral bone attrition longitudinally: The MOST study. Osteoarthr. Cartil. 2010, 18, 47–53. [Google Scholar] [CrossRef]
- Yusuf, E.; Kortekaas, M.C.; Watt, I.; Huizinga, T.W.J.; Kloppenburg, M. Do knee abnormalities visualised on MRI explain knee pain in knee osteoarthritis? A systematic review. Ann. Rheum. Dis. 2010, 70, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.; Anbeek, P.; Yang, K.A.; Vincken, K.; Viergever, M.; Castelein, R.; Saris, D. Validation of a Novel Semiautomated Segmentation Method for MRI Detection of Cartilage-Related Bone Marrow Lesions. Cartilage 2010, 1, 328–334. [Google Scholar] [CrossRef]
- Pang, J.; Driban, J.B.; Destenaves, G.; Miller, E.; Lo, G.H.; Ward, R.J.; Price, L.L.; A Lynch, J.; Eaton, C.B.; Eckstein, F.; et al. Quantification of bone marrow lesion volume and volume change using semi-automated segmentation: Data from the osteoarthritis initiative. BMC Musculoskelet. Disord. 2013, 14, 3. [Google Scholar] [CrossRef]
- Nielsen, F.K.; Egund, N.; Peters, D.; Jurik, A.G. Measurement of bone marrow lesions by MR imaging in knee osteoarthritis using quantitative segmentation methods–a reliability and sensitivity to change analysis. BMC Musculoskelet. Disord. 2014, 15, 447. [Google Scholar] [CrossRef] [PubMed]
- Dodin, P.; Abram, F.; Pelletier, J.-P.; Martel-Pelletier, J. A fully automated system for quantification of knee bone marrow lesions using MRI and the osteoarthritis initiative cohort. J. Biomed. Graph. Comput. 2012, 3, 51. [Google Scholar] [CrossRef]
- Stout, A.C.; Barbe, M.F.; Eaton, C.B.; Amin, M.; Al-Eid, F.; Price, L.L.; Lu, B.; Lo, G.H.; Zhang, M.; Pang, J.; et al. Inflammation and glucose homeostasis are associated with specific structural features among adults without knee osteoarthritis: A cross-sectional study from the osteoarthritis initiative. BMC Musculoskelet. Disord. 2018, 19, 1. [Google Scholar] [CrossRef]
- Anandarajah, A.P.; Schwarz, E.M.; Totterman, S.; Monu, J.; Feng, C.Y.; Shao, T.; A Haas-Smith, S.; Ritchlin, C.T. The effect of etanercept on osteoclast precursor frequency and enhancing bone marrow oedema in patients with psoriatic arthritis. Ann. Rheum. Dis. 2008, 67, 296–301. [Google Scholar] [CrossRef]
- Roemer, F.; Khrad, H.; Hayashi, D.; Jara, H.; Ozonoff, A.; Fotinos-Hoyer, A.; Guermazi, A. Volumetric and semiquantitative assessment of MRI-detected subchondral bone marrow lesions in knee osteoarthritis: A comparison of contrast-enhanced and non-enhanced imaging. Osteoarthr. Cartil. 2010, 18, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Osteoarthritis Initiative Releases First Data. Available online: https://www.nih.gov/news-events/news-releases/osteoarthritis-initiative-releases-first-data (accessed on 17 February 2023).
- OAI. The Osteoarthritis Initiative. Available online: https://nda.nih.gov/oai (accessed on 17 February 2023).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. Available online: https://arxiv.org/abs/1602.07261 (accessed on 17 February 2023).
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Chollet, F. Keras. Available online: https://keras.io (accessed on 17 February 2023).
- TensorFlow. Available online: https://www.tensorflow.org/about/bib (accessed on 17 February 2023).
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- OpenCV. Available online: https://opencv.org/ (accessed on 17 February 2023).
- Kirch, W. (Ed.) Pearson’s Correlation Coefficient; Springer: Dordrecht, The Netherlands, 2008; p. 1090. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Granada, Spain, 20 September 2018. [Google Scholar]
- Zunair, H.; Hamza, A.B. Sharp U-Net: Depthwise Convolutional Network for Biomedical Image Segmentation. Comput. Biol. Med. 2021, 136, 104699. [Google Scholar] [CrossRef] [PubMed]
- Abedalla, A.; Abdullah, M.; Al-Ayyoub, M.; Benkhelifa, E. The 2ST-UNet for Pneumothorax Segmentation in Chest X-Rays using ResNet34 as a Backbone for U-Net. arXiv 2020, arXiv:2009.02805. [Google Scholar]
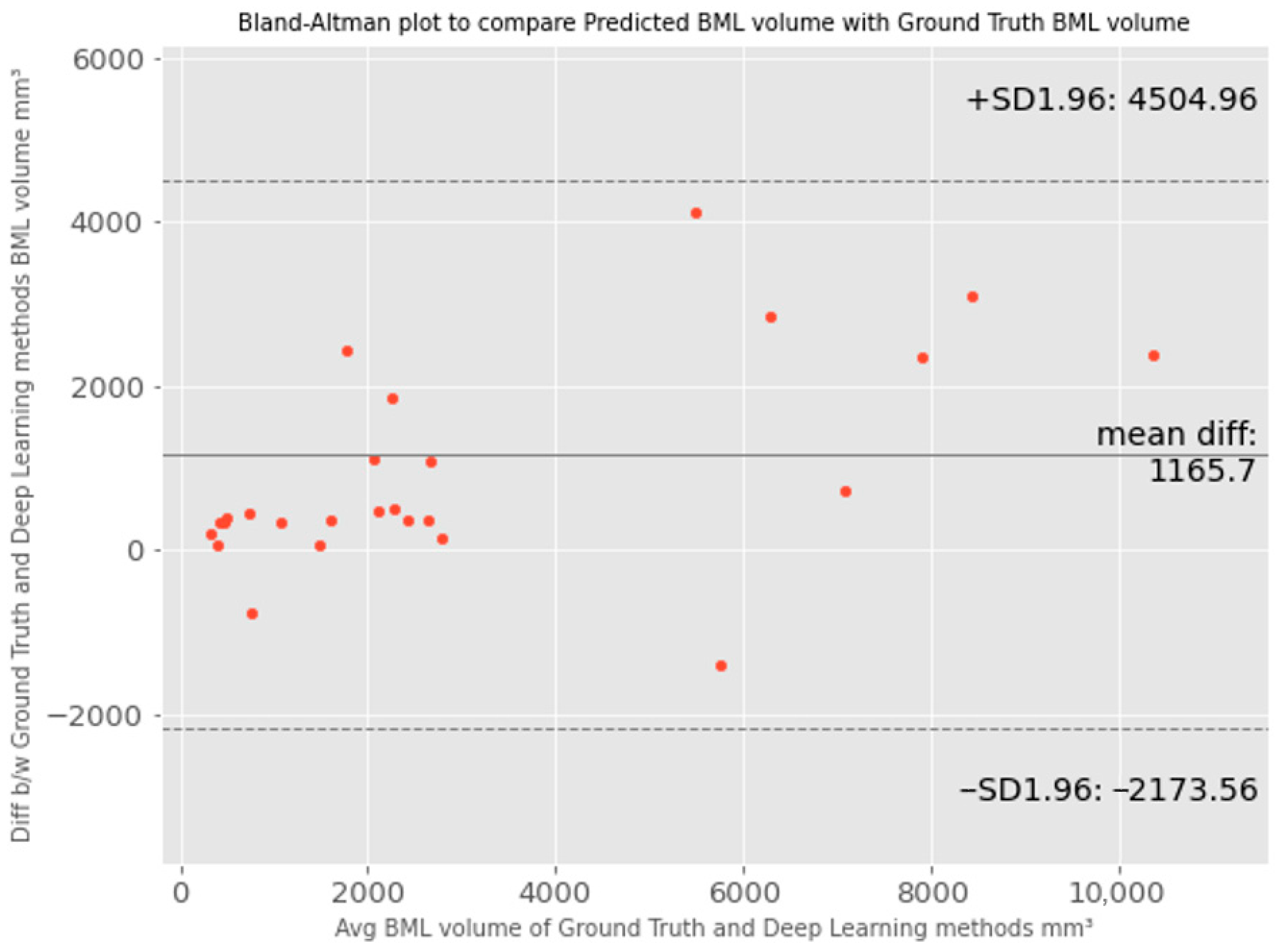
- Altman, D.G.; Bland, J.M. Measurement in medicine: The analysis of method comparison studies. Statistician 1983, 32, 307–317. [Google Scholar] [CrossRef]
- Raynauld, J.-P.; Martel-Pelletier, J.; Berthiaume, M.-J.; Abram, F.; Choquette, D.; Haraoui, B.; Beary, J.F.; A Cline, G.; Meyer, J.M.; Pelletier, J.-P. Correlation between bone lesion changes and cartilage volume loss in patients with osteoarthritis of the knee as assessed by quantitative magnetic resonance imaging over a 24-month period. Ann. Rheum. Dis. 2008, 67, 683–688. [Google Scholar] [CrossRef]
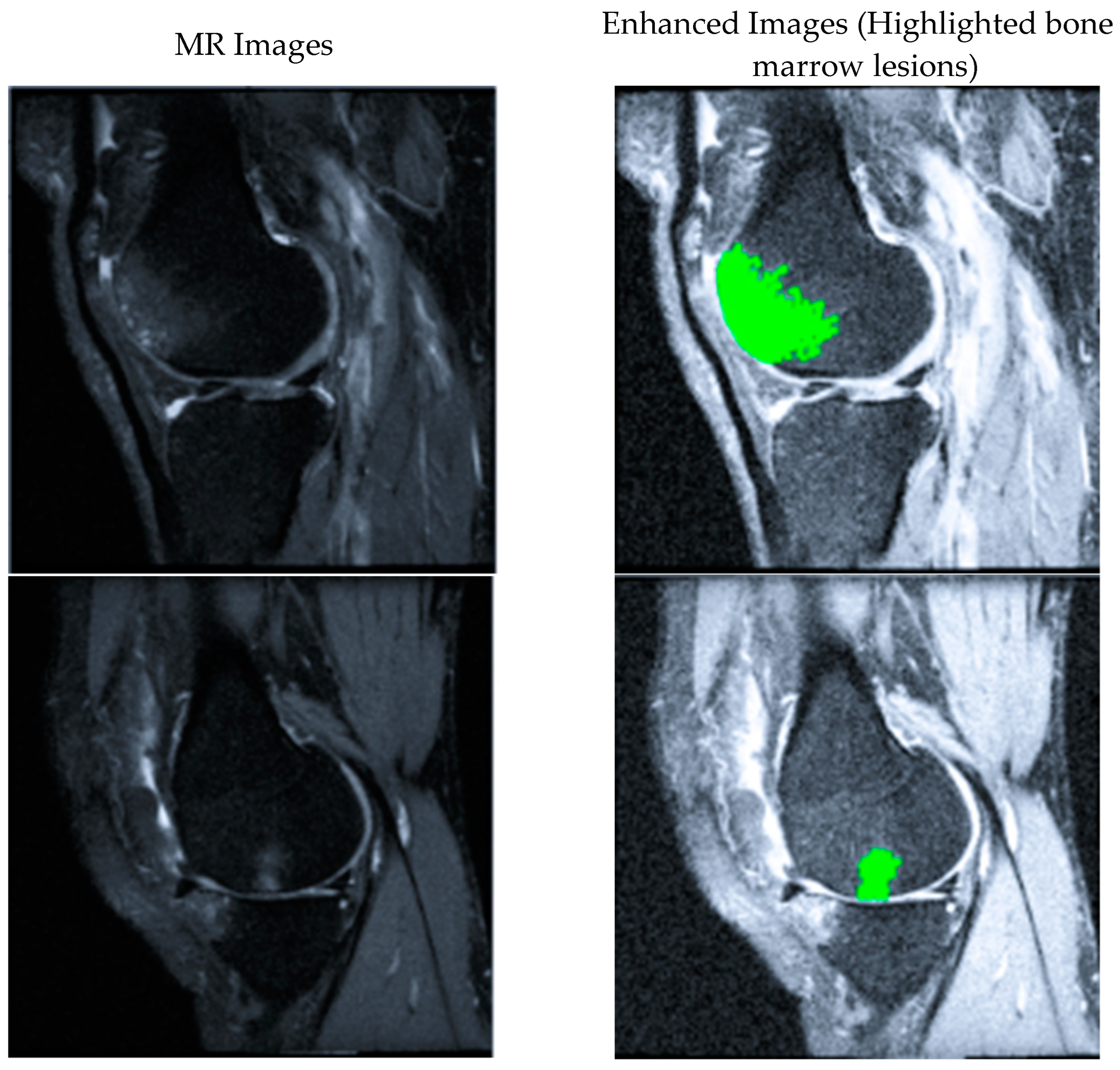


| Set | Number of Images | Subjects |
|---|---|---|
| Training | 1034 | 210 |
| Validation | 194 | 45 |
| Testing | 209 | 45 |
| Total | 1437 | 300 |
| Model | r (p < 0.05) | 2D DSC | 3D DSC |
|---|---|---|---|
| U-Net-based BML segmentation method [9] | 0.96 | 0.56 | 0.51 |
| U-Net++ [34] | 0.96 | 0.59 | 0.58 |
| Sharp U-Net [35] | 0.96 | 0.57 | 0.53 |
| U-Net + ResNet34 [36] | 0.98 | 0.66 | 0.59 |
| U-Net + InceptionResNetV2 (proposed) | 0.98 | 0.68 | 0.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponnusamy, R.; Zhang, M.; Wang, Y.; Sun, X.; Chowdhury, M.; Driban, J.B.; McAlindon, T.; Shan, J. Automatic Segmentation of Bone Marrow Lesions on MRI Using a Deep Learning Method. Bioengineering 2024, 11, 374. https://doi.org/10.3390/bioengineering11040374
Ponnusamy R, Zhang M, Wang Y, Sun X, Chowdhury M, Driban JB, McAlindon T, Shan J. Automatic Segmentation of Bone Marrow Lesions on MRI Using a Deep Learning Method. Bioengineering. 2024; 11(4):374. https://doi.org/10.3390/bioengineering11040374
Chicago/Turabian StylePonnusamy, Raj, Ming Zhang, Yue Wang, Xinyue Sun, Mohammad Chowdhury, Jeffrey B. Driban, Timothy McAlindon, and Juan Shan. 2024. "Automatic Segmentation of Bone Marrow Lesions on MRI Using a Deep Learning Method" Bioengineering 11, no. 4: 374. https://doi.org/10.3390/bioengineering11040374
APA StylePonnusamy, R., Zhang, M., Wang, Y., Sun, X., Chowdhury, M., Driban, J. B., McAlindon, T., & Shan, J. (2024). Automatic Segmentation of Bone Marrow Lesions on MRI Using a Deep Learning Method. Bioengineering, 11(4), 374. https://doi.org/10.3390/bioengineering11040374






