Ensemble Fusion Models Using Various Strategies and Machine Learning for EEG Classification
Abstract
1. Introduction
- (a)
- The first proposed ensemble technique utilizes an equidistant assessment and ranking determination mode for the classification of EEG signals;
- (b)
- The second proposed ensemble technique utilizes the concept of Infinite Independent Component Analysis (I-ICA) and multiple classifiers with a majority voting concept;
- (c)
- The third proposed ensemble technique utilizes the Genetic Algorithm (GA)-based feature selection technique and bagging SVM-based classification model;
- (d)
- The fourth proposed ensemble technique utilizes the concept of Hilbert Huang Transform (HHT) and multiple classifiers with GA-based multiparameter optimization;
- (e)
- The fifth proposed ensemble technique utilizes the concept of Factor analysis with an Ensemble layer K nearest neighbor (KNN) classifier.
2. Proposed Ensemble Techniques
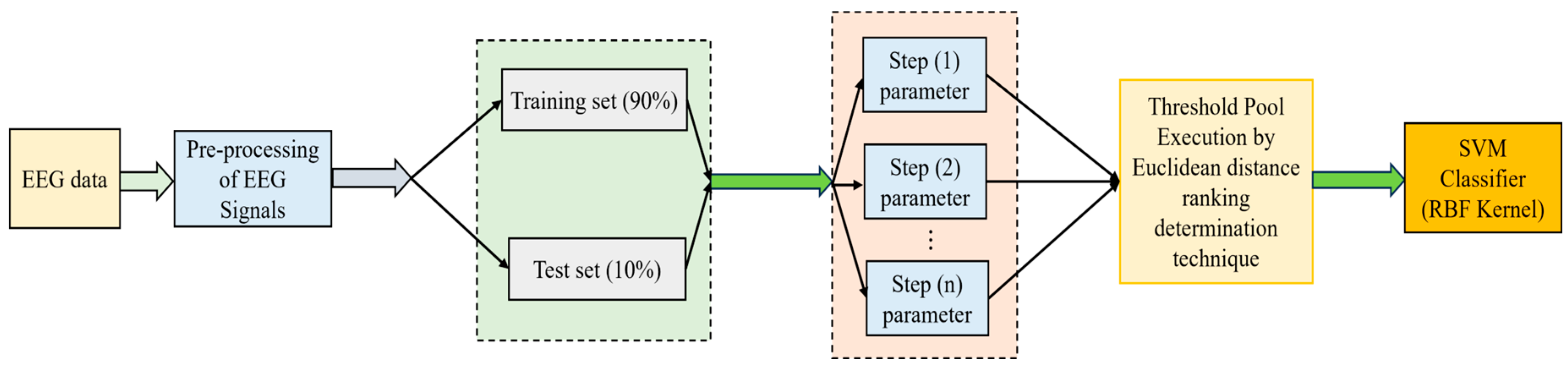
2.1. Proposed Technique 1: Ensemble Hybrid Model Using Equidistant Assessment and Ranking Determination Method
- (a)
- Equidistant assessment of the basic model parameters;
- (b)
- K-means clustering with ranking assessment and determination is utilized for ensemble pruning;
- (c)
- The final prediction result is voted on with the help of the divide-and-conquer strategy.
2.1.1. Equidistant Assessment of the Model Parameters
| Algorithm 1: Equidistant assessment optimization |
| Input: model and parameter (“k, 2, 16, 4”) |
| Disintegrate and obtain each step value. |
| for i = 1 to step do |
| Add every step task into the thread pool. |
| end for |
| Train the model |
| Save in equidistant mode. |
2.1.2. Evaluation Assessment for Ranking Determination
2.1.3. Design of Ranking Determination Method
2.1.4. Ensemble Hybrid Technique
2.1.5. Feature Selection
2.2. Proposed Technique 2: Ensemble Hybrid Model Using Infinite I-ICA and Multiple Classifiers with Majority Voting Concept
2.2.1. Feature Extraction and Selection Using Infinite ICA
2.2.2. Random Subspace Ensemble Learning Classification
2.2.3. Ensemble Learning in a Random Manner
2.2.4. Random Ensemble Learning by Hybrid Classifiers
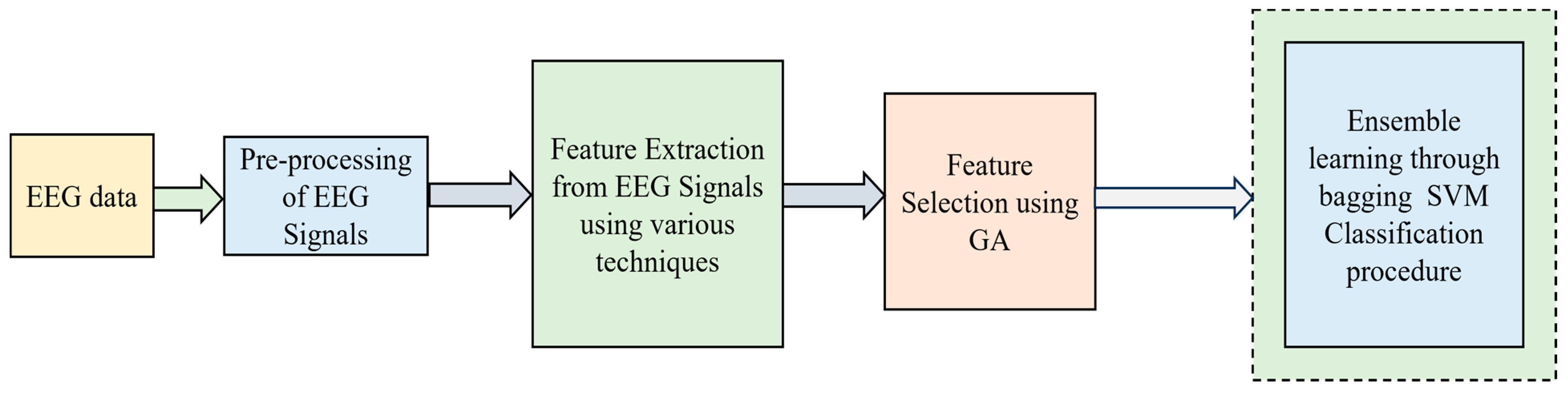
2.3. Proposed Technique 3: Ensemble Hybrid Model with GA-Based Feature Selection and Bagging SVM-Based Classification Model
2.3.1. Genetic Algorithm
- (1)
- The modeling of every feature is performed as a gene, and almost all the features are like chromosomes, as they share a similar length to the features. A varied subset of features is indicated by every chromosome;
- (2)
- The highest evolution algebra is set. The initial population is created that includes all the individuals;
- (3)
- Every individual is projected as ;
- (4)
- In every chromosome, the number “1” is generated randomly, and then the random assignment of these chromosomes is performed so that a varied number of features can be clearly represented;
- (5)
- The evaluation value of fitness is tested, and the main intention of feature selection is utilized with fewer features, so that a good classification rate can be achieved;
- (6)
- The feature subset input and the classification accuracy are used to evaluate the fitness function for every individual and are represented as follows:where the recognition accuracy is represented by , and indicates the number of features. The features traced in the feature subset are used when the classifiers are trained;
- (7)
- To select the operators, roulette is used, implying that based on fitness ratio, the chromosomes are selected. The probability of chromosomes is represented as follows:where indicates the reciprocal of fitness value, and the population size is indicated by ;
- (8)
- The single-point cross technique is utilized, and two individuals are chosen with similar probabilities from . Unless a new group is formed, this process is repeated;
- (9)
- Based on a particular mutative probability, the value of every individual is randomly changed, and a new generation of groups is created, such as ;
- (10)
- Check whether the termination condition is satisfied or not. If the condition is satisfied, the entire operation stops as the best solution with a high fitness value is obtained as output. Otherwise, step 2 is repeated once again.
2.3.2. Ensemble Learning through Bagging Procedure
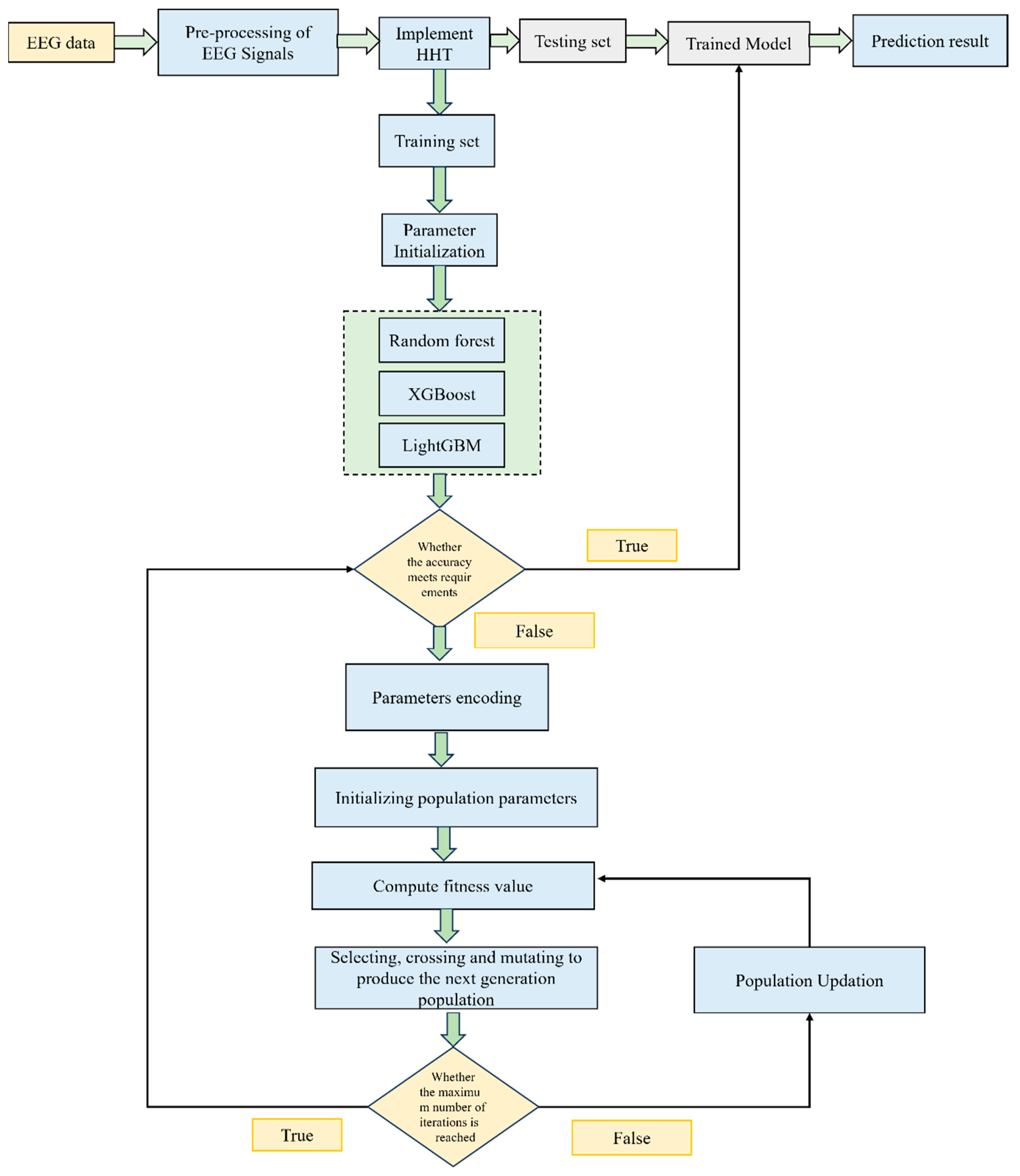
2.4. Proposed Technique 4: Ensemble Hybrid Model with HHT and Multiple Classifiers with GA-Based Multiparameter Optimization
2.4.1. Random Forest Regression Model
2.4.2. LightGBM Model
2.4.3. XGBoost Model
2.4.4. Ensemble Technique Dependent on GA-Based Multiparameter Optimization
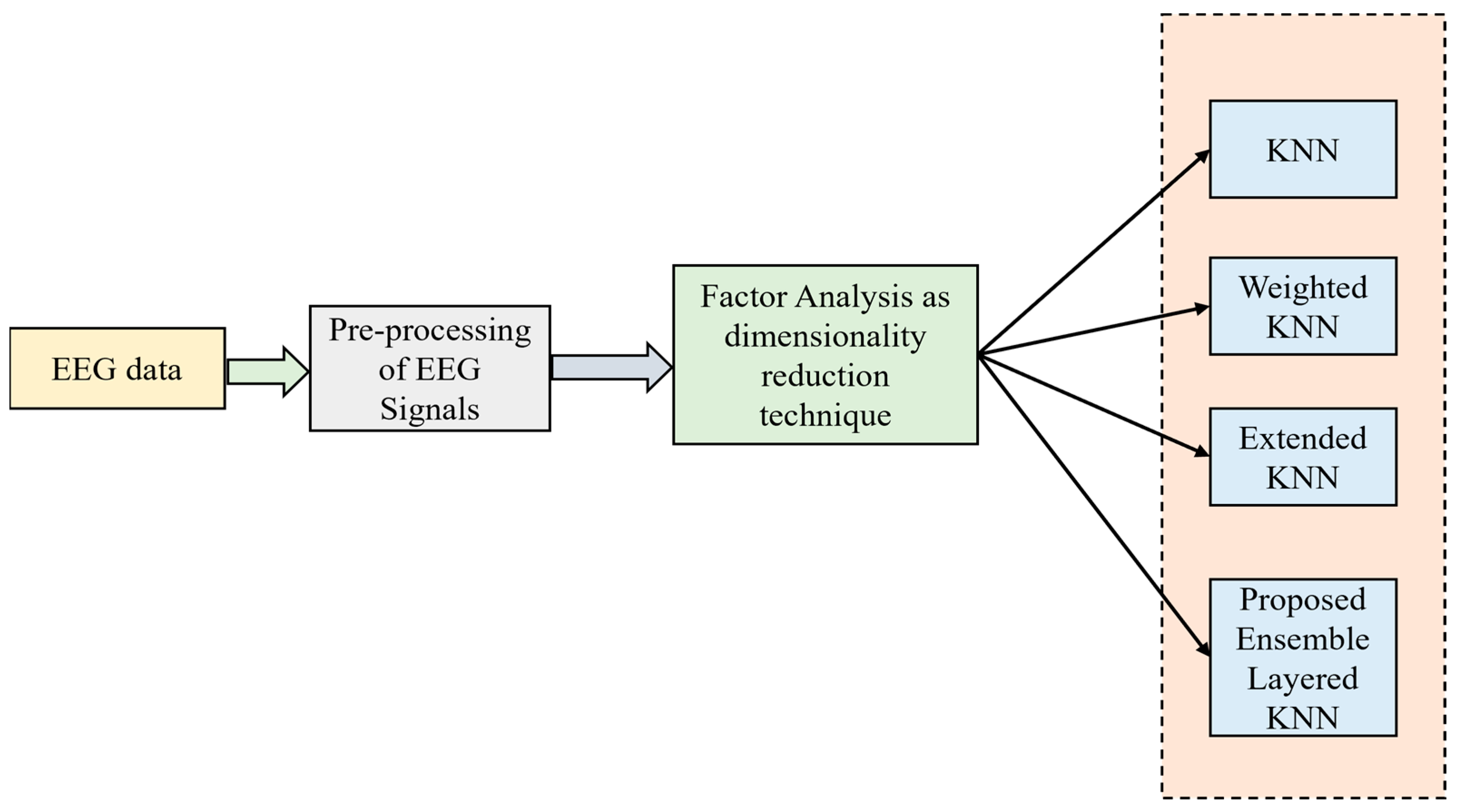
2.5. Proposed Technique 5: Ensemble Hybrid Model with Factor Analysis Concept and Ensemble-Layered KNN Classifier
2.5.1. Factor Analysis
2.5.2. Layered K-Nearest Neighbor Classifier
3. Results and Discussion
4. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patidar, S.; Pachori, R.B.; Upadhyay, A.; Acharya, U.R. An integrated alcoholic index using tunable-Q wavelet transform based features extracted from EEG signals for diagnosis of alcoholism. Appl. Soft Comput. 2017, 50, 71–78. [Google Scholar] [CrossRef]
- Durongbhan, P.; Zhao, Y.; Chen, L.; Zis, P.; De Marco, M.; Unwin, Z.C.; Venneri, A.; He, X.; Li, S.; Zhao, Y.; et al. A dementia classification framework using frequency and time-frequency features based on EEG signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.K.; Bajaj, V.; Acharya, U.R. Detection of Parkinson’s disease using automated tunable Q wavelet transform technique with EEG signals. Biocybern. Biomed. Eng. 2021, 41, 679–689. [Google Scholar] [CrossRef]
- Oh, S.L.; Hagiwara, Y.; Raghavendra, U.; Yuvaraj, R.; Arunkumar, N.; Murugappan, M.; Acharya, U.R. A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput. Appl. 2020, 32, 10927–10933. [Google Scholar] [CrossRef]
- Saeedi, M.; Saeedi, A.; Maghsoudi, A. Major depressive disorder assessment via enhanced k-nearest neighbor method and EEG signals. Phys. Eng. Sci. Med. 2020, 43, 1007–1018. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Singh, L.; Pachori, R.B. Identification of epileptic seizures from scalp EEG signals based on TQWT. Adv. Intell. Syst. Comput. 2019, 748, 209–221. [Google Scholar]
- Yuan, S.Y.; Liu, J.M. The prescription rule of traditional Chinese medicine for epilepsy by data mining. Chin. J. Integr. Med. Cardio-Cerebrovasc. Dis. 2021, 19, 4044–4049. [Google Scholar]
- Samiee, K.; Kovacs, P.; Gabbouj, M. Epileptic seizure classification of EEG time-series using rational discrete short-time Fourier transform. IEEE Trans. Biomed. Eng. 2014, 62, 541–552. [Google Scholar] [CrossRef]
- Peker, M.; Sen, B.; Delen, D. A novel method for automated diagnosis of epilepsy using complex-valued classifiers. IEEE J. Biomed. Health Inform. 2016, 20, 108–118. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Pachori, R.B.; Kanhangad, V.; Panigrahi, B.K. Automated diagnosis of epilepsy using key-point-based local binary pattern of EEG signals. IEEE J. Biomed. Health Inform. 2016, 21, 888–896. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.-D.; Luo, M.-L.; Li, K.; Yang, X.-F.; Guo, Q. Epileptic seizure classification of EEGs using time–frequency analysis based multiscale radial basis functions. IEEE J. Biomed. Health Inform. 2017, 22, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, M.U.; Rashad, A.; Basalamah, A.M. Tariq Detection of Epilepsy Seizures in Neo-Natal EEG Using LSTM Architecture. IEEE Access 2019, 7, 179074–179085. [Google Scholar] [CrossRef]
- Wang, Z.; Na, J.; Zheng, B. An improved kNN classifier for epilepsy diagnosis. IEEE Access 2020, 8, 100022–100030. [Google Scholar] [CrossRef]
- Darjani, N.; Omranpour, H. Phase space elliptic density feature for epileptic EEG signals classification using metaheuristic optimization method. Knowl.-Based Syst. 2020, 205, 106276. [Google Scholar] [CrossRef]
- Carvalho, V.R.; Moraes, M.F.; Braga, A.P.; Mendes, E.M. Evaluating three different adaptive decomposition methods for EEG signal seizure detection and classification. bioRxiv 2019. [Google Scholar] [CrossRef]
- Prabakar, S.K.; Lee, S.-W. ENIC: Ensemble and Nature Inclined Classification with Sparse Depiction based Deep and Transfer Learning for Biosignal Classification. Appl. Soft Comput. 2022, 117, 108416. [Google Scholar] [CrossRef]
- Prabhakar, S.K.; Lee, S.-W. Improved Sparse Representation with Robust Hybrid Feature Extraction Models and Deep Learning for EEG Classification. Expert Syst. Appl. 2022, 198, 116783. [Google Scholar] [CrossRef]
- Prabhakar, S.K.; Lee, S.-W. SASDL and RBATQ: Sparse Autoencoder with Swarm based Deep Learning and Reinforcement based Q-learning for EEG Classification. IEEE Open J. Eng. Med. Biol. 2022, 3, 58–68. [Google Scholar] [CrossRef]
- Prabhakar, S.K.; Ju, Y.-G.; Rajaguru, H.; Won, D.O. Sparse measures with swarm-based pliable hidden Markov model and deep learning for EEG classification. Front. Comput. Neurosci. 2022, 16, 1016516. [Google Scholar] [CrossRef]
- Prabhakar, S.K.; Won, D.-O. Performance Comparison of Bio-inspired and Learning Based Clustering Analysis with Machine Learning Techniques for Classification of EEG Signals. Front. Artif. Intell. 2022, 16, 1016516. [Google Scholar] [CrossRef]
- Satapathy, S.K.; Dehuri, S.; Jagadev, A.K. EEG signal classification using PSO trained RBF neural network for epilepsy identification. Inform. Med. Unlocked 2017, 6, 1156269. [Google Scholar] [CrossRef]
- Gong, C.; Zhang, X.; Niu, Y. Identification of Epilepsy from Intracranial EEG Signals by Using Different Neural Network Models. Comput. Biol. Chem. 2020, 87, 107310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, Y.; Yang, P.O.; Chen, W.; Lo, B. Epilepsy seizure prediction on EEG using common spatial pattern and convolutional neural network. IEEE J. Biomed. Health Inform. 2020, 24, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Taran, S.; Bajaj, V. Clustering variational mode decomposition for identification of focal EEG signals. IEEE Sens. Lett. 2018, 2, 7001304. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, G.; Xie, Z.; Shang, W. A Unified Framework and Method for EEG-Based Early Epileptic Seizure Detection and Epilepsy Diagnosis. IEEE Access 2020, 8, 20080–20092. [Google Scholar] [CrossRef]
- Sharma, R.; Sircar, P.; Pachori, R.B. Automated focal EEG signal detection based on third order cumulant function. Biomed. Signal Process. Control 2020, 58, 101856. [Google Scholar] [CrossRef]
- Lopez, S.; Suarez, G.; Jungreis, D.; Obeid, I.; Picone, J. Automated Identification of Abnormal Adult EEGs. In Proceedings of the 2015 IEEE Signal Processing in Medicine and Biology Symposium, Philadelphia, PA, USA, 12 December 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–5. [Google Scholar]
- Yıldırım, Ö.; Baloglu, U.B.; Acharya, U.R. A deep convolutional neural network model for automated identification of abnormal EEG signals. Neural Comput. Appl. 2018, 32, 15857–15868. [Google Scholar] [CrossRef]
- Sharma, M.; Patel, S.; Acharya, U.R. Automated detection of abnormal EEG signals using localized wavelet filter banks. Pattern Recognit. Lett. 2020, 133, 188–194. [Google Scholar] [CrossRef]
- Gemein, L.A.; Schirrmeister, R.T.; Chrabąszcz, P.; Wilson, D.; Boedecker, J.; Schulze-Bonhage, A.; Hutter, F.; Ball, T. Machine-learning-based diagnostics of EEG pathology. NeuroImage 2020, 220, 117021. [Google Scholar] [CrossRef]
- Alhussein, M.; Muhammad, G.; Hossain, M.S. EEG pathology detection based on deep learning. IEEE Access 2019, 7, 27781–27788. [Google Scholar] [CrossRef]
- Amin, S.U.; Hossain, M.S.; Muhammad, G.; Alhussein, M.; Rahman, M.A. Cognitive smart healthcare for pathology detection and monitoring. IEEE Access 2019, 7, 10745–10753. [Google Scholar] [CrossRef]
- Albaqami, H.; Hassan, G.M.; Subasi, A.; Datta, A. Automatic detection of abnormal EEG signals using wavelet feature extraction and gradient boosting decision tree. arXiv 2020, arXiv:2012.10034. [Google Scholar] [CrossRef]
- Tibor Schirrmeister, R.; Gemein, L.; Eggensperger, K.; Hutter, F.; Ball, T. Deep learning with convolutional neural networks for decoding and visualization of eeg pathology. arXiv 2017, arXiv:1708.08012. [Google Scholar]
- Roy, S.; Kiral-Kornek, I.; Harrer, S. ChronoNet: A Deep Recurrent Neural Network for Abnormal EEG Identification. In Proceedings of the 17th Conference on Artificial Intelligence in Medicine, AIME 2019, Poznan, Poland, 26–29 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 47–56. [Google Scholar]
- Tuncer, T.; Dogan, S.; Acharya, U.R. Automated EEG signal classification using chaotic local binary pattern. Expert Syst. Appl. 2021, 182, 115175. [Google Scholar] [CrossRef]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Banos, O.; Damas, M.; Pomares, H.; Rojas, F.; Delgado-Marquez, B.; Valenzuela, O. Human activity recognition based on a sensor weighting hierarchical classifier. Soft Comput. 2013, 17, 333–343. [Google Scholar] [CrossRef]
- Liu, F.; Juan, Y.; Yao, L. Research on the number of clusters in K-means clustering algorithm. Electron. Des. Eng. 2017, 25, 9–13. [Google Scholar]
- Feng, W.; Zhu, Q.; Zhuang, J.; Yu, S. An expert recommendation algorithm based on Pearson correlation coefficient and FP-growth. Clust. Comput. 2018, 22, 7401–7412. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Oja, E. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Sathwika, G.J.; Bhattacharya, A. Prediction of cardiovascular disease (CVD) using ensemble learning algorithms. In Proceedings of the 5th Joint International Conference on Data Science & Management of Data (9th ACM IKDD CODS and 27th COMAD), Bangalore, India, 8–10 January 2022; pp. 292–293. [Google Scholar]
- Ma, C.; Du, X.; Cao, L. Improved KNN algorithm for fine-grained classification of encrypted network flow. Electronics 2020, 9, 324. [Google Scholar] [CrossRef]
- Moz, M.; Pato, M.V. A genetic algorithm approach to a nurse rerostering problem. Comput. Oper. Res. 2007, 34, 667–691. [Google Scholar] [CrossRef]
- Jung, S.; Moon, J.; Park, S.; Rho, S.; Baik, S.W.; Hwang, E. Bagging ensemble of multilayer perceptron for missing electricity consumption data imputation. Sensors 2020, 20, 1772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.R.; Ma, S.; Safak, E.; Hartzell, S. Hilbert-Huang transform analysis of dynamic and earthquake motion recordings. J. Eng. Mech. 2003, 129, 861–875. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random Forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Zheng, K.; Wang, L.; You, Z.-H. CGMDA: An approach to predict and validate microRNA-disease associations by utilizing chaos game representation and LightGBM. IEEE Access 2019, 7, 133314–133323. [Google Scholar] [CrossRef]
- Asselman, A.; Khaldi, M.; Aammou, S. Enhancing the prediction of student performance based on the machine learning XGBoost algorithm. Interact. Learn. Environ. 2021, 31, 3360–3379. [Google Scholar] [CrossRef]
- Rubin, D.B.; Thayer, D.T. EM algorithms for ML factor analysis. Psychometrika 1982, 47, 69–76. [Google Scholar] [CrossRef]
- Liu, L.; Su, J.; Liu, X.; Chen, R.; Huang, K.; Deng, R.H.; Wang, X. Toward highly secure yet efficient KNN classification scheme on outsourced cloud data. IEEE Internet Things J. 2019, 6, 9841–9852. [Google Scholar] [CrossRef]
- Obeid, I.; Picone, J. The temple university hospital EEG data corpus. Front. Neurosci. 2016, 10, 196. [Google Scholar] [CrossRef]


| Techniques Proposed | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| Ensemble hybrid model using equidistant assessment and ranking determination method with GA-based feature selection method and SVM Classifier. | 85.45 | 86.45 | 85.95 |
| Ensemble hybrid model using equidistant assessment and ranking determination method with ACO-based feature selection method and SVM Classifier. | 84.34 | 83.34 | 83.84 |
| Ensemble hybrid model using equidistant assessment and ranking determination method with PSO-based feature selection method and SVM Classifier. | 85.34 | 85.46 | 85.4 |
| Ensemble hybrid model using equidistant assessment and ranking determination method with GSO-based feature selection method and SVM Classifier. | 86.36 | 87.51 | 86.93 |
| Ensemble hybrid model using equidistant assessment and ranking determination method with proposed ESCD-based feature selection technique and SVM Classifier. | 88.98 | 90.99 | 89.98 |
| Techniques Proposed | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| I-ICA with SVM classifier | 86.23 | 85.34 | 85.78 |
| I-ICA with MLP classifier | 82.34 | 83.43 | 82.88 |
| I-ICA with EKNN classifier | 87.65 | 88.32 | 87.98 |
| I-ICA with random ensemble learning by hybrid classifiers | 89.99 | 89.01 | 89.5 |
| Techniques Proposed | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| GA with Linear SVM | 83.45 | 82.34 | 82.89 |
| GA with Polynomial SVM | 85.45 | 85.01 | 85.23 |
| GA with Radial Basis Function Kernel SVM | 87.77 | 86.99 | 87.38 |
| GA with bagging SVM | 88.01 | 88.29 | 88.15 |
| Techniques Proposed | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| Ensemble hybrid model with HHT and RF classifier with GA-based multiparameter optimization | 88.03 | 87.91 | 87.97 |
| Ensemble hybrid model with HHT and LightGBM classifier with GA-based multiparameter optimization | 86.23 | 84.23 | 85.23 |
| Ensemble hybrid model with HHT and XGBoost classifier with GA-based multiparameter optimization | 87.23 | 87.11 | 87.17 |
| Ensemble hybrid model with HHT and multiple classifiers with GA-based multiparameter optimization | 90.01 | 89.91 | 89.96 |
| Techniques Proposed | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| Factor analysis with KNN ensemble hybrid model | 82.21 | 80.23 | 81.22 |
| Factor analysis with Weighted KNN ensemble hybrid model | 83.34 | 83.45 | 83.39 |
| Factor analysis with Extended KNN ensemble hybrid model | 84.45 | 85.43 | 84.94 |
| Factor analysis with Proposed ensemble-layered KNN hybrid model | 88.21 | 89.01 | 88.61 |
| References | Techniques Used | Number of Channels Used | Classification Accuracy (%) |
|---|---|---|---|
| Lopez et al. [27] | Ensemble learning with KNN and RF | 4 | 68.30 |
| Sharma et al. [29] | Nonlinear features with SVM | 4 | 79.34 |
| Yildrim et al. [28] | Deep CNN | 4 | 79.34 |
| Gemein et al. [30] | Handcrafted features | 21 | 85.9 |
| Alhussein et al. [31] | Deep learning | 21 | 89.13 |
| Amin et al. [32] | AlexNet and SVM | 21 | 87.32 |
| Albaqami et al. [33] | Boosting concept | 21 | 87.68 |
| Schirrmeister et al. [34] | Deep learning | 24 | 85.4 |
| Roy et al. [35] | Chrononet | 24 | 86.57 |
| Tuncer et al. [36] | Concept of Chaotic Local binary pattern with iterative minimum redundancy maximum relevancy | PZ Channel | 98.19 |
| Proposed works 1 | Ensemble hybrid model using equidistant assessment and ranking determination method with proposed ESCD-based feature selection technique and SVM classifier | 24 | 89.98 |
| Proposed works 2 | I-ICA with random ensemble learning by hybrid classifiers. | 24 | 89.5 |
| Proposed works 3 | GA with bagging SVM | 24 | 88.15 |
| Proposed works 4 | Ensemble hybrid model with HHT and multiple classifiers with GA-based multiparameter optimization | 24 | 89.96 |
| Proposed works 5 | Factor analysis with Proposed Ensemble-layered KNN hybrid model. | 24 | 88.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prabhakar, S.K.; Lee, J.J.; Won, D.-O. Ensemble Fusion Models Using Various Strategies and Machine Learning for EEG Classification. Bioengineering 2024, 11, 986. https://doi.org/10.3390/bioengineering11100986
Prabhakar SK, Lee JJ, Won D-O. Ensemble Fusion Models Using Various Strategies and Machine Learning for EEG Classification. Bioengineering. 2024; 11(10):986. https://doi.org/10.3390/bioengineering11100986
Chicago/Turabian StylePrabhakar, Sunil Kumar, Jae Jun Lee, and Dong-Ok Won. 2024. "Ensemble Fusion Models Using Various Strategies and Machine Learning for EEG Classification" Bioengineering 11, no. 10: 986. https://doi.org/10.3390/bioengineering11100986
APA StylePrabhakar, S. K., Lee, J. J., & Won, D.-O. (2024). Ensemble Fusion Models Using Various Strategies and Machine Learning for EEG Classification. Bioengineering, 11(10), 986. https://doi.org/10.3390/bioengineering11100986






