Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models
Abstract
1. Introduction
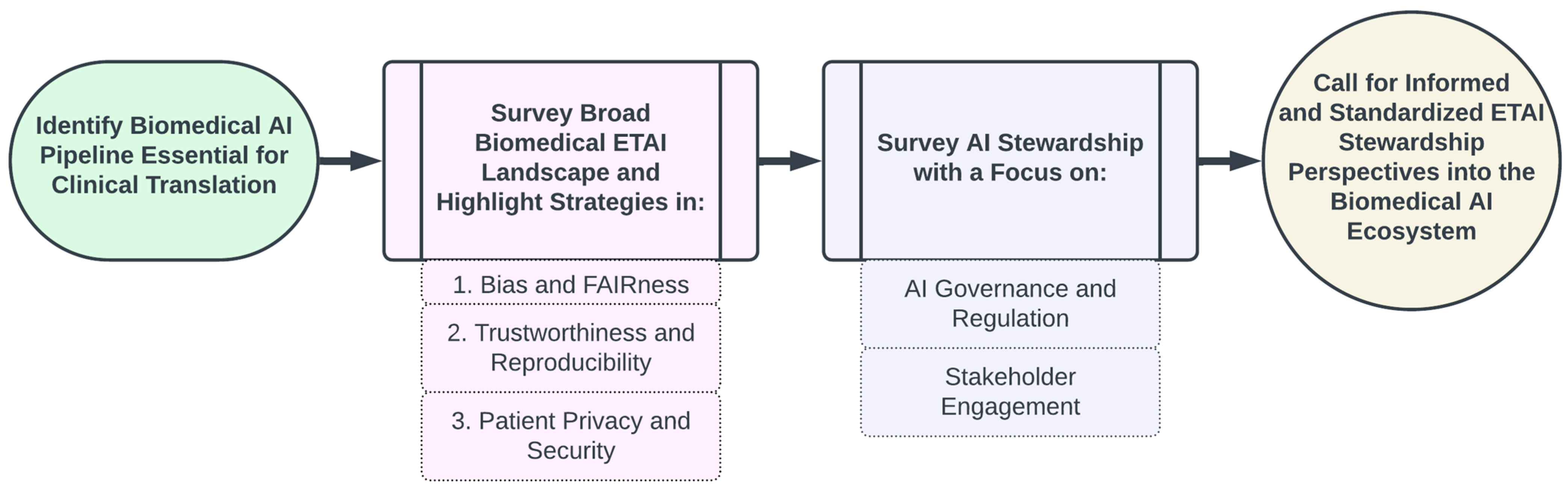
- We outline the essential components of an AI Ecosystem for integrating biomedical AI technologies into clinical and/or translational settings;
- We examine the current landscape of ethical considerations and ethical practices broadly applicable to biomedical AI and with applications to FM development and deployment, highlighting critical challenges and existing mitigation strategies across three key areas:
- Mitigating Bias and Enhancing Fairness
- Ensuring Trustworthiness and Reproducibility
- Safeguarding Patient Privacy and Security;
- We examine pivotal government and scholarly publications that chart the present guidelines and future directions for AI stewardship, emphasizing two main components:
- AI Governance and Regulation
- Stakeholder Engagement;
- Finally, we discuss a unified perspective of how the principles of ethical and trustworthy AI and stewardship in AI integrate into the ecosystem.
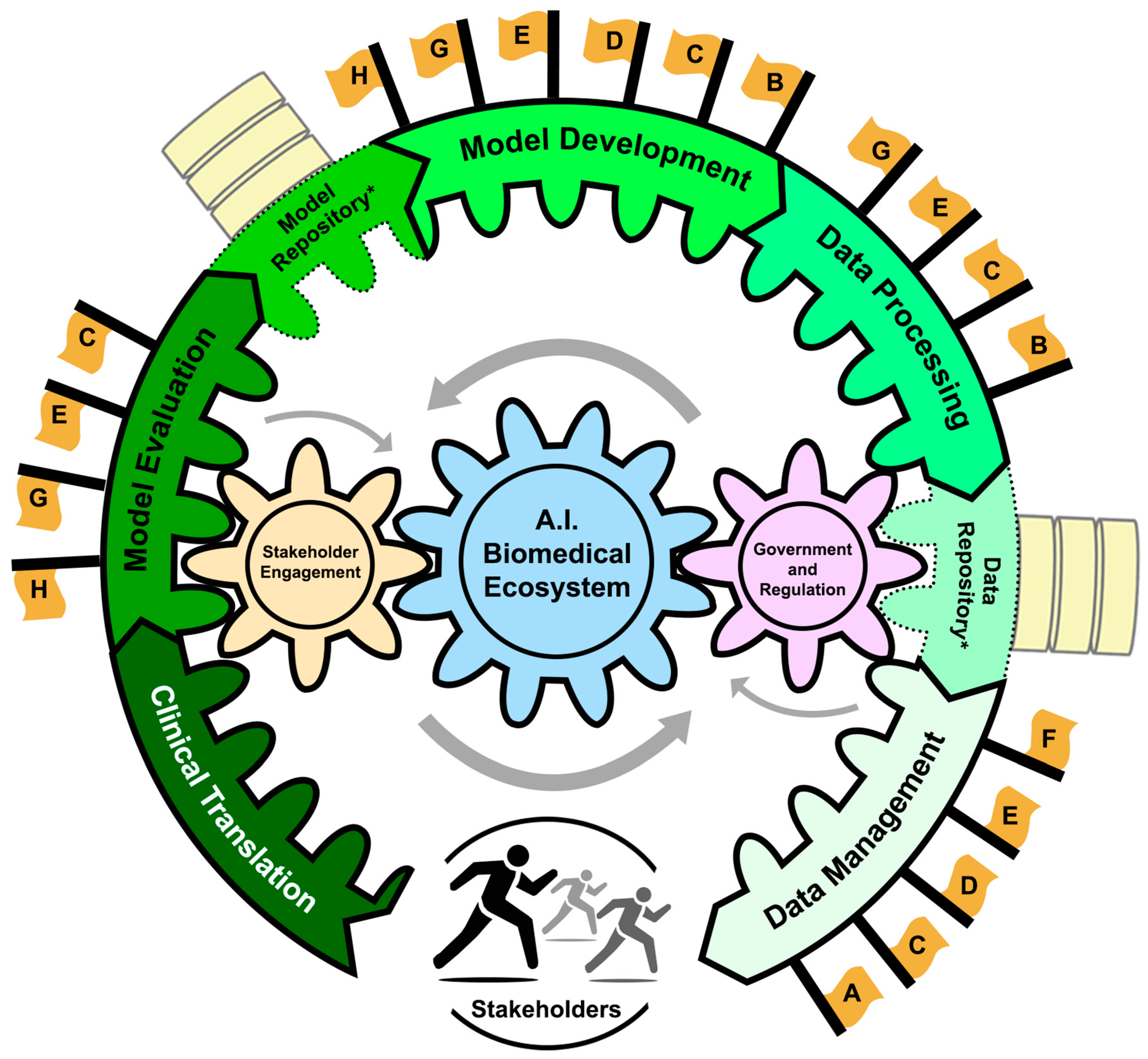
2. AI Ecosystem in Biomedicine
- -
- Data Lifecycle Management: The collection, dissemination, and curation of vast amounts of diverse biomedical data.
- -
- Data Repositories: Centralized systems that store, validate, and distribute data, promoting transparent and reproducible AI technologies.
- -
- Data Processing: Cleaning, annotating, and structuring data to make them AI-ready. Model Development: The development and training of FMs that can then be utilized for various downstream tasks such as hypothesis generation, explanation, causal reasoning, and clinical decision support.
- -
- Model Repositories: Centralized storage for managing and sharing AI models to promote accessibility and collaboration amongst stakeholders. Centralized storage for managing and sharing AI models to promote accessibility and collaboration amongst stakeholders.
- -
- Model Evaluation: The assessment of model performance and reliability prior to deployment in biomedical settings.
- -
- Clinical Translation: Operationalizing FMs in a clinical setting to enhance patient care.
- -
- AI Governance and Regulation: Established legal and ethical standards that enforce compliance through both government processes and committee/board regulation.
- -
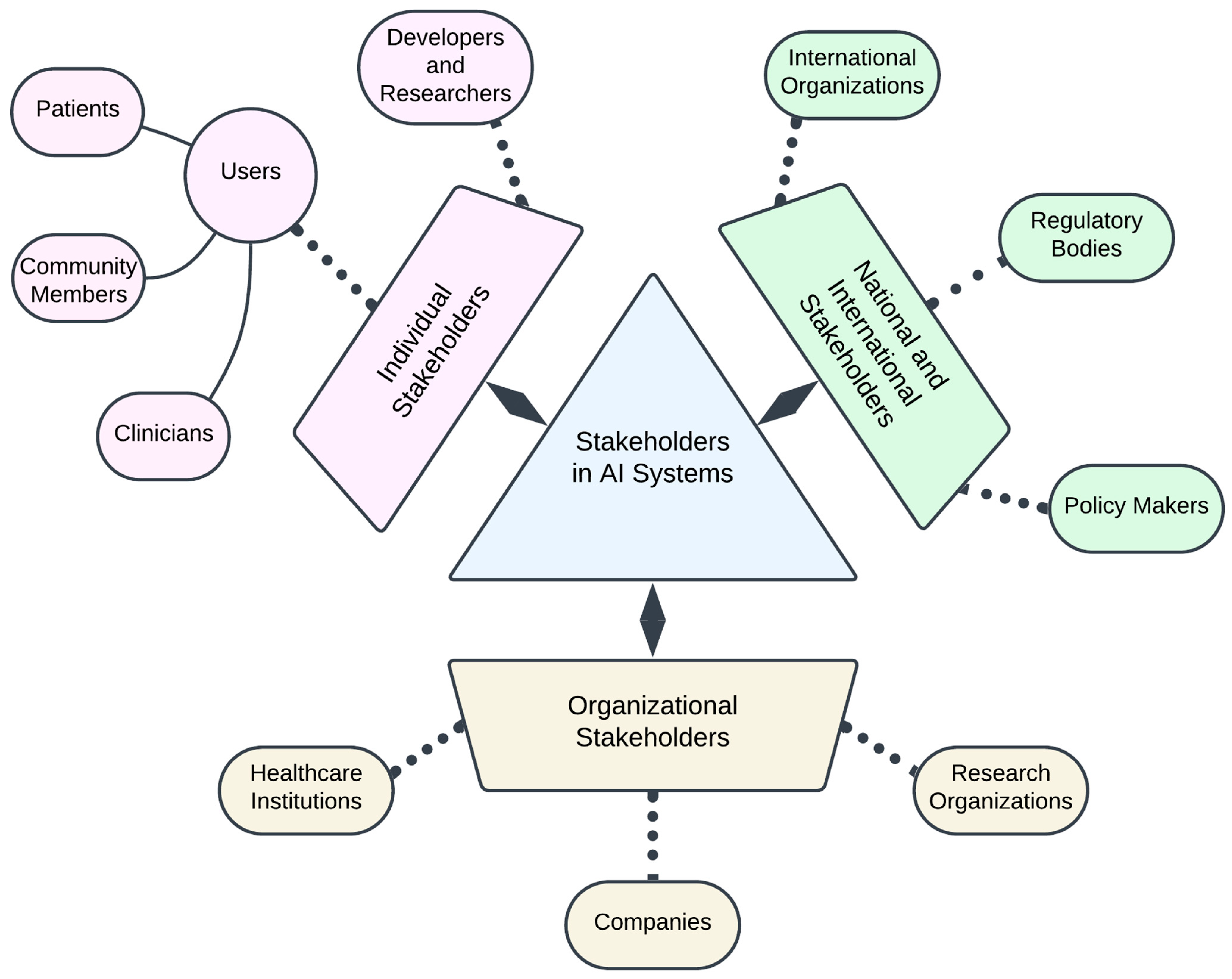
- Stakeholder Engagement: Diverse communities and individuals contributing to and affected by AI in biomedicine. Engagement refers to the active participation of these groups in the entire pipeline of AI from bench to bedside in both an industrial and healthcare setting.
3. Ethical Considerations in the AI Pipeline for Foundation Models
3.1. Mitigating Bias and Enhancing Fairness
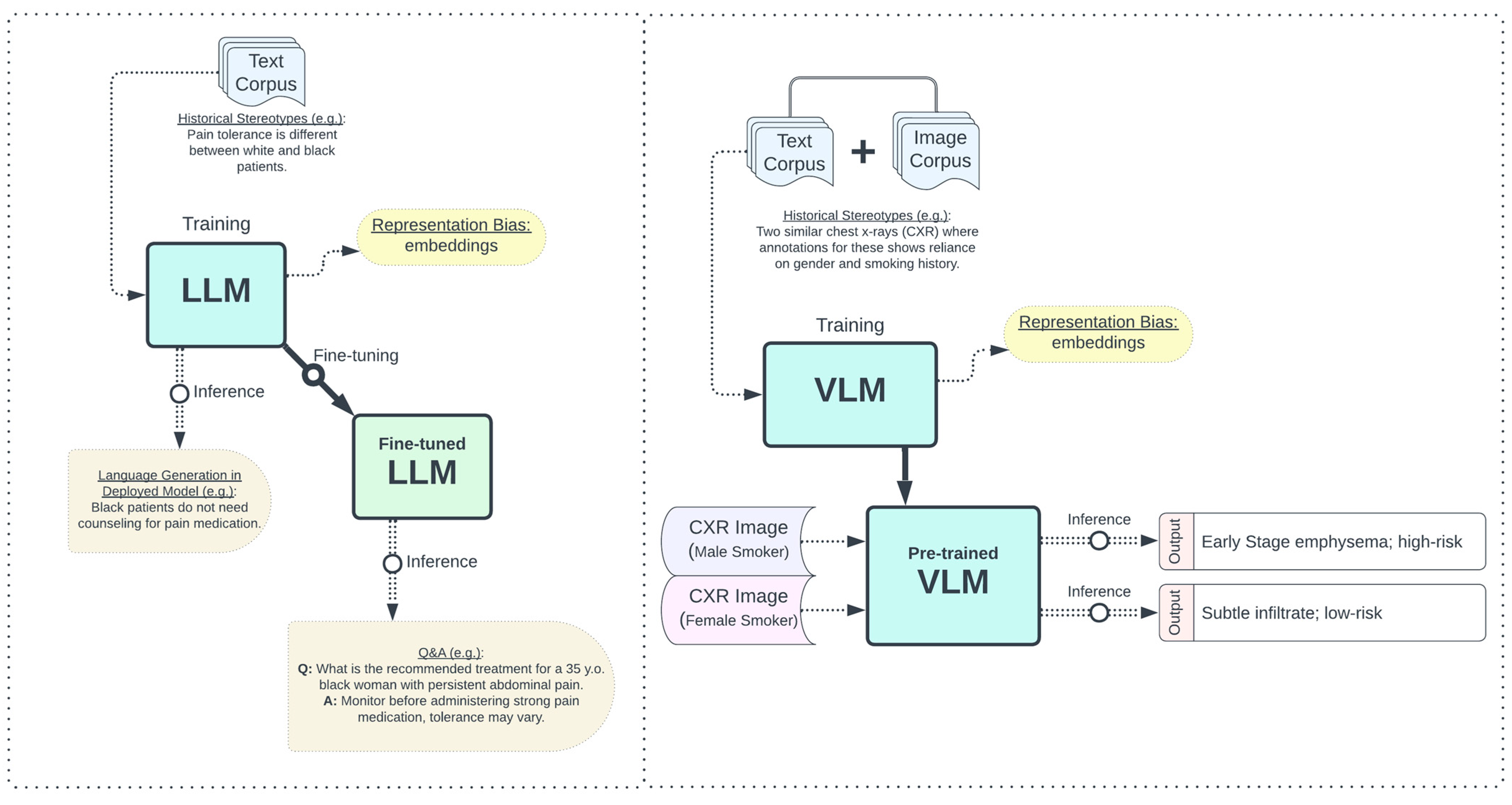
3.1.1. Social AI Bias: Underrepresentation Bias
3.1.2. Social AI Bias: Stereotypical Biases
3.2. Ensuring Trustworthiness and Reproducibility
3.2.1. Essential Data Lifecycle Concepts
3.2.2. Interpretability and Explainability
3.2.3. Enhancing AI Reliability with Human-in-the-Loop
3.2.4. Transparency and Reproducibility
3.3. Safeguarding Patient Privacy and Security
3.3.1. Essential Data Lifecycle Concepts
3.3.2. Protecting Patient Privacy in Cloud Storage and Computation
3.3.3. Patient Re-Identification and Membership Inference Attacks
3.3.4. Memorization of Patient Data
4. AI Stewardship
4.1. AI Governance and Regulation
Landscape and Integration of AI Governance and Regulation
4.2. Stakeholder Engagement
Co-Design
5. Concluding Unified Perspective
5.1. AI Lifecycle and AI Pipeline in the AI Biomedical Ecosystem
5.2. Standardizing Bias Mitigation, Trustworthiness and Reproducibility, and Privacy and Security
5.3. Call for Continuous AI Stewardship and Harmonious AI Governance in the AI Lifecycle
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Large Scale Machine Learning Systems. Available online: https://www.kdd.org/kdd2016/topics/view/large-scale-machine-learning-systems (accessed on 21 July 2024).
- Awais, M.; Naseer, M.; Khan, S.; Anwer, R.M.; Cholakkal, H.; Shah, M.; Yang, M.-H.; Khan, F.S. Foundational Models Defining a New Era in Vision: A Survey and Outlook. Available online: https://arxiv.org/abs/2307.13721v1 (accessed on 21 July 2024).
- What Are Foundation Models?—Foundation Models in Generative AI Explained—AWS. Available online: https://aws.amazon.com/what-is/foundation-models/ (accessed on 21 July 2024).
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-Supervised Learning in Medicine and Healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, M.; Leskovec, J.; Liang, P. LinkBERT: Pretraining Language Models with Document Links. arXiv 2022, arXiv:2203.15827. [Google Scholar]
- Vaid, A.; Jiang, J.; Sawant, A.; Lerakis, S.; Argulian, E.; Ahuja, Y.; Lampert, J.; Charney, A.; Greenspan, H.; Narula, J.; et al. A Foundational Vision Transformer Improves Diagnostic Performance for Electrocardiograms. Npj Digit. Med. 2023, 6, 108. [Google Scholar] [CrossRef]
- Hao, M.; Gong, J.; Zeng, X.; Liu, C.; Guo, Y.; Cheng, X.; Wang, T.; Ma, J.; Zhang, X.; Song, L. Large-Scale Foundation Model on Single-Cell Transcriptomics. Nat. Methods 2024, 21, 1481–1491. [Google Scholar] [CrossRef]
- Moor, M.; Banerjee, O.; Abad, Z.S.H.; Krumholz, H.M.; Leskovec, J.; Topol, E.J.; Rajpurkar, P. Foundation Models for Generalist Medical Artificial Intelligence. Nature 2023, 616, 259–265. [Google Scholar] [CrossRef]
- Ferrara, E. Fairness and Bias in Artificial Intelligence: A Brief Survey of Sources, Impacts, and Mitigation Strategies. Sci 2024, 6, 3. [Google Scholar] [CrossRef]
- Jacobides, M.G.; Brusoni, S.; Candelon, F. The Evolutionary Dynamics of the Artificial Intelligence Ecosystem. Strategy Sci. 2021, 6, 412–435. [Google Scholar] [CrossRef]
- Yoo, S. A Study on AI Business Ecosystem. J. Inst. Internet Broadcast. Commun. 2020, 20, 21–27. [Google Scholar] [CrossRef]
- Winter, N.R.; Cearns, M.; Clark, S.R.; Leenings, R.; Dannlowski, U.; Baune, B.T.; Hahn, T. From Multivariate Methods to an AI Ecosystem. Mol. Psychiatry 2021, 26, 6116–6120. [Google Scholar] [CrossRef]
- Penman-Aguilar, A.; Talih, M.; Huang, D.; Moonesinghe, R.; Bouye, K.; Beckles, G. Measurement of Health Disparities, Health Inequities, and Social Determinants of Health to Support the Advancement of Health Equity. J. Public Health Manag. Pract. JPHMP 2016, 22, S33–S42. [Google Scholar] [CrossRef]
- Rajotte, J.-F.; Bergen, R.; Buckeridge, D.L.; Emam, K.E.; Ng, R.; Strome, E. Synthetic Data as an Enabler for Machine Learning Applications in Medicine. iScience 2022, 25, 105331. [Google Scholar] [CrossRef] [PubMed]
- Synthetic Data in AI: Challenges, Applications, and Ethical Implications. Available online: https://arxiv.org/html/2401.01629v1 (accessed on 23 July 2024).
- Fang, T.; Lu, N.; Niu, G.; Sugiyama, M. Rethinking Importance Weighting for Deep Learning under Distribution Shift. Adv. Neural Inf. Process. Syst. 2020, 33, 11996–12007. [Google Scholar]
- Vaidya, A.; Chen, R.J.; Williamson, D.F.K.; Song, A.H.; Jaume, G.; Yang, Y.; Hartvigsen, T.; Dyer, E.C.; Lu, M.Y.; Lipkova, J.; et al. Demographic Bias in Misdiagnosis by Computational Pathology Models. Nat. Med. 2024, 30, 1174–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cui, Z.; Wu, Y.; Gu, L.; Harada, T. Estimating and Improving Fairness with Adversarial Learning. arXiv 2021, arXiv:2103.04243. [Google Scholar]
- Yang, J.; Soltan, A.A.S.; Eyre, D.W.; Yang, Y.; Clifton, D.A. An Adversarial Training Framework for Mitigating Algorithmic Biases in Clinical Machine Learning. Npj Digit. Med. 2023, 6, 55. [Google Scholar] [CrossRef]
- Kamishima, T.; Akaho, S.; Sakuma, J. Fairness-Aware Learning through Regularization Approach. In Proceedings of the 2011 IEEE 11th International Conference on Data Mining Workshops, Vancouver, BC, Canada, 11 December 2011; pp. 643–650. [Google Scholar]
- Olfat, M.; Mintz, Y. Flexible Regularization Approaches for Fairness in Deep Learning. In Proceedings of the 2020 59th IEEE Conference on Decision and Control (CDC), Jeju, Republic of Korea, 14–18 December 2020; pp. 3389–3394. [Google Scholar]
- Webster, K.; Wang, X.; Tenney, I.; Beutel, A.; Pitler, E.; Pavlick, E.; Chen, J.; Chi, E.; Petrov, S. Measuring and Reducing Gendered Correlations in Pre-Trained Models. arXiv 2021, arXiv:2010.06032. [Google Scholar]
- Zafar, M.B.; Valera, I.; Rodriguez, M.G.; Gummadi, K.P. Fairness Beyond Disparate Treatment & Disparate Impact: Learning Classification without Disparate Mistreatment. In Proceedings of the 26th International Conference on World Wide Web, Perth, Australia, 3 April 2017; pp. 1171–1180. [Google Scholar]
- Machine Learning Glossary: Fairness. Available online: https://developers.google.com/machine-learning/glossary/fairness (accessed on 24 July 2024).
- Gallegos, I.O.; Rossi, R.A.; Barrow, J.; Tanjim, M.M.; Kim, S.; Dernoncourt, F.; Yu, T.; Zhang, R.; Ahmed, N.K. Bias and Fairness in Large Language Models: A Survey. Comput. Linguist. 2024, 50, 1097–1179. [Google Scholar] [CrossRef]
- Chouldechova, A. Fair Prediction with Disparate Impact: A Study of Bias in Recidivism Prediction Instruments. Big Data 2017, 5, 153–163. [Google Scholar] [CrossRef]
- Li, X.; Xiong, H.; Li, X.; Wu, X.; Zhang, X.; Liu, J.; Bian, J.; Dou, D. Interpretable Deep Learning: Interpretation, Interpretability, Trustworthiness, and Beyond. Knowl. Inf. Syst. 2022, 64, 3197–3234. [Google Scholar] [CrossRef]
- Henriques, J.; Rocha, T.; de Carvalho, P.; Silva, C.; Paredes, S. Interpretability and Explainability of Machine Learning Models: Achievements and Challenges. In Proceedings of the International Conference on Biomedical and Health Informatics 2022, Concepción, Chile, 24–26 November 2022; Pino, E., Magjarević, R., de Carvalho, P., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 81–94. [Google Scholar]
- Lu, K.; Mardziel, P.; Wu, F.; Amancharla, P.; Datta, A. Gender Bias in Neural Natural Language Processing. In Logic, Language, and Security; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Zhao, J.; Wang, T.; Yatskar, M.; Ordonez, V.; Chang, K.-W. Gender Bias in Coreference Resolution: Evaluation and Debiasing Methods. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, New Orleans, LA, USA, 1–6 June 2018; Short Papers. Walker, M., Ji, H., Stent, A., Eds.; Association for Computational Linguistics: New Orleans, LA, USA, June 2018; Volume 2, pp. 15–20. [Google Scholar]
- Dinan, E.; Fan, A.; Williams, A.; Urbanek, J.; Kiela, D.; Weston, J. Queens Are Powerful Too: Mitigating Gender Bias in Dialogue Generation. In Proceedings of the Conference on Empirical Methods in Natural Language Processing (EMNLP); Webber, B., Cohn, T., He, Y., Liu, Y., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 8173–8188. [Google Scholar]
- Bolukbasi, T.; Chang, K.-W.; Zou, J.Y.; Saligrama, V.; Kalai, A.T. Man Is to Computer Programmer as Woman Is to Homemaker? Debiasing Word Embeddings. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: New York, NY, USA, 2016; Volume 29. [Google Scholar]
- Cheng, P.; Hao, W.; Yuan, S.; Si, S.; Carin, L. Fairfil: Contrastive neural debiasing method for pretrained text encoders. arXiv 2021, arXiv:2103.06413. [Google Scholar]
- Ma, W.; Scheible, H.; Wang, B.; Veeramachaneni, G.; Chowdhary, P.; Sun, A.; Koulogeorge, A.; Wang, L.; Yang, D.; Vosoughi, S. Deciphering Stereotypes in Pre-Trained Language Models. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing, Singapore, 6–10 December 2023; Bouamor, H., Pino, J., Bali, K., Eds.; Association for Computational Linguistics: Singapore, 2023; pp. 11328–11345. [Google Scholar]
- Seth, A.; Hemani, M.; Agarwal, C. DeAR: Debiasing Vision-Language Models With Additive Residuals. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 6820–6829. [Google Scholar]
- Shen, T.; Jin, R.; Huang, Y.; Liu, C.; Dong, W.; Guo, Z.; Wu, X.; Liu, Y.; Xiong, D. Large Language Model Alignment: A Survey. arXiv 2023, arXiv:2309.15025. [Google Scholar]
- Kaufmann, T.; Weng, P.; Bengs, V.; Hüllermeier, E. A Survey of Reinforcement Learning from Human Feedback. arXiv 2024, arXiv:2312.14925. [Google Scholar]
- Lee, H.; Phatale, S.; Mansoor, H.; Mesnard, T.; Ferret, J.; Lu, K.; Bishop, C.; Hall, E.; Carbune, V.; Rastogi, A.; et al. RLAIF: Scaling Reinforcement Learning from Human Feedback with AI Feedback. arXiv 2023, arXiv:2309.00267. [Google Scholar]
- Rafailov, R.; Sharma, A.; Mitchell, E.; Ermon, S.; Manning, C.D.; Finn, C. Direct Preference Optimization: Your Language Model Is Secretly a Reward Model. arXiv 2024, arXiv:2305.18290. [Google Scholar]
- Ntoutsi, E.; Fafalios, P.; Gadiraju, U.; Iosifidis, V.; Nejdl, W.; Vidal, M.-E.; Ruggieri, S.; Turini, F.; Papadopoulos, S.; Krasanakis, E.; et al. Bias in Data-Driven Artificial Intelligence Systems—An Introductory Survey. WIREs Data Min. Knowl. Discov. 2020, 10, e1356. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine; Policy and Global Affairs; Committee on Women in Science, Engineering, and Medicine. Committee on Improving the Representation of Women and Underrepresented Minorities in Clinical Trials and Research. In Improving Representation in Clinical Trials and Research: Building Research Equity for Women and Underrepresented Groups; Bibbins-Domingo, K., Helman, A., Eds.; The National Academies Collection: Reports Funded by National Institutes of Health; National Academies Press (US): Washington, DC, USA, 2022; ISBN 978-0-309-27820-1. [Google Scholar]
- Aldrighetti, C.M.; Niemierko, A.; Van Allen, E.; Willers, H.; Kamran, S.C. Racial and Ethnic Disparities Among Participants in Precision Oncology Clinical Studies. JAMA Netw. Open 2021, 4, e2133205. [Google Scholar] [CrossRef]
- Yang, G.; Mishra, M.; Perera, M.A. Multi-Omics Studies in Historically Excluded Populations: The Road to Equity. Clin. Pharmacol. Ther. 2023, 113, 541–556. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting Racial Bias in an Algorithm Used to Manage the Health of Populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Zhang, B.H.; Lemoine, B.; Mitchell, M. Mitigating Unwanted Biases with Adversarial Learning. In Proceedings of the 2018 AAAI/ACM Conference on AI, Ethics, and Society, New Orleans, LA, USA, 2–3 February 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 335–340. [Google Scholar]
- Thomasian, N.M.; Eickhoff, C.; Adashi, E.Y. Advancing Health Equity with Artificial Intelligence. J. Public Health Policy 2021, 42, 602–611. [Google Scholar] [CrossRef]
- Navigli, R.; Conia, S.; Ross, B. Biases in Large Language Models: Origins, Inventory, and Discussion. J. Data Inf. Qual. 2023, 15, 1–21. [Google Scholar] [CrossRef]
- Park, Y.-J.; Pillai, A.; Deng, J.; Guo, E.; Gupta, M.; Paget, M.; Naugler, C. Assessing the Research Landscape and Clinical Utility of Large Language Models: A Scoping Review. BMC Med. Inform. Decis. Mak. 2024, 24, 72. [Google Scholar] [CrossRef] [PubMed]
- Omiye, J.A.; Lester, J.C.; Spichak, S.; Rotemberg, V.; Daneshjou, R. Large Language Models Propagate Race-Based Medicine. Npj Digit. Med. 2023, 6, 195. [Google Scholar] [CrossRef] [PubMed]
- Challenging Systematic Prejudices: An Investigation into Bias against Women and Girls in Large Language Models—UNESCO Digital Library. Available online: https://unesdoc.unesco.org/ark:/48223/pf0000388971 (accessed on 23 July 2024).
- Petreski, D.; Hashim, I.C. Word Embeddings Are Biased. But Whose Bias Are They Reflecting? AI Soc. 2023, 38, 975–982. [Google Scholar] [CrossRef]
- Hart, S. Shapley Value. In Game Theory; Eatwell, J., Milgate, M., Newman, P., Eds.; Palgrave Macmillan: London, UK, 1989; pp. 210–216. ISBN 978-1-349-20181-5. [Google Scholar]
- Zhou, K.; Lai, E.; Jiang, J. VLStereoSet: A Study of Stereotypical Bias in Pre-Trained Vision-Language Models. In Proceedings of the 2nd Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics and the 12th International Joint Conference on Natural Language Processing (Volume 1: Long Papers); He, Y., Ji, H., Li, S., Liu, Y., Chang, C.-H., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2022; pp. 527–538. [Google Scholar]
- Saravanan, A.P.; Kocielnik, R.; Jiang, R.; Han, P.; Anandkumar, A. Exploring Social Bias in Downstream Applications of Text-to-Image Foundation Models. arXiv 2023, arXiv:2312.10065. [Google Scholar]
- Hartsock, I.; Rasool, G. Vision-Language Models for Medical Report Generation and Visual Question Answering: A Review. arXiv 2024, arXiv:2403.02469. [Google Scholar]
- Kirk, H.R.; Vidgen, B.; Röttger, P.; Hale, S.A. The Benefits, Risks and Bounds of Personalizing the Alignment of Large Language Models to Individuals. Nat. Mach. Intell. 2024, 6, 383–392. [Google Scholar] [CrossRef]
- Sun, H. Supervised Fine-Tuning as Inverse Reinforcement Learning. arXiv 2024, arXiv:2403.12017. [Google Scholar]
- Lee, B.W.; Cho, H.; Yoo, K.M. Instruction Tuning with Human Curriculum. In Proceedings of the Findings of the Association for Computational Linguistics: NAACL 2024; Duh, K., Gomez, H., Bethard, S., Eds.; Association for Computational Linguistics: Mexico City, Mexico, 2024; pp. 1281–1309. [Google Scholar]
- Chang, C.T.; Farah, H.; Gui, H.; Rezaei, S.J.; Bou-Khalil, C.; Park, Y.-J.; Swaminathan, A.; Omiye, J.A.; Kolluri, A.; Chaurasia, A.; et al. Red Teaming Large Language Models in Medicine: Real-World Insights on Model Behavior. medRxiv, 2024. [Google Scholar] [CrossRef]
- Wang, H.; Xiong, W.; Xie, T.; Zhao, H.; Zhang, T. Interpretable Preferences via Multi-Objective Reward Modeling and Mixture-of-Experts. arXiv 2024, arXiv:2406.12845. [Google Scholar]
- Bai, Y.; Kadavath, S.; Kundu, S.; Askell, A.; Kernion, J.; Jones, A.; Chen, A.; Goldie, A.; Mirhoseini, A.; McKinnon, C.; et al. Constitutional AI: Harmlessness from AI Feedback. arXiv 2022, arXiv:2212.08073. [Google Scholar]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Jagodnik, K.M.; Koplev, S.; Jenkins, S.L.; Ohno-Machado, L.; Paten, B.; Schurer, S.C.; Dumontier, M.; Verborgh, R.; Bui, A.; Ping, P.; et al. Developing a Framework for Digital Objects in the Big Data to Knowledge (BD2K) Commons: Report from the Commons Framework Pilots Workshop. J. Biomed. Inform. 2017, 71, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Hermjakob, H.; Kleemola, M.; Moilanen, K.; Tuominen, M.; Sansone, S.-A.; Lister, A.; David, R.; Panagiotopoulou, M.; Ohmann, C.; Belien, J.; et al. BY-COVID D3.2: Implementation of Cloud-Based, High Performance, Scalable Indexing System; 2022. Available online: https://zenodo.org/records/7129553 (accessed on 12 August 2024).
- Longpre, S.; Mahari, R.; Obeng-Marnu, N.; Brannon, W.; South, T.; Gero, K.; Pentland, S.; Kabbara, J. Data Authenticity, Consent, & Provenance for AI Are All Broken: What Will It Take to Fix Them? arXiv 2024, arXiv:2404.12691. [Google Scholar]
- Frasca, M.; La Torre, D.; Pravettoni, G.; Cutica, I. Explainable and Interpretable Artificial Intelligence in Medicine: A Systematic Bibliometric Review. Discov. Artif. Intell. 2024, 4, 15. [Google Scholar] [CrossRef]
- Gosiewska, A.; Kozak, A.; Biecek, P. Simpler Is Better: Lifting Interpretability-Performance Trade-off via Automated Feature Engineering. Decis. Support Syst. 2021, 150, 113556. [Google Scholar] [CrossRef]
- Sevillano-García, I.; Luengo, J.; Herrera, F. SHIELD: A Regularization Technique for eXplainable Artificial Intelligence. arXiv 2024, arXiv:2404.02611. [Google Scholar]
- Ivanovs, M.; Kadikis, R.; Ozols, K. Perturbation-Based Methods for Explaining Deep Neural Networks: A Survey. Pattern Recognit. Lett. 2021, 150, 228–234. [Google Scholar] [CrossRef]
- Lee, S.-I.; Topol, E.J. The Clinical Potential of Counterfactual AI Models. Lancet 2024, 403, 717. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Sabirsh, A.; Wählby, C.; Sintorn, I.-M. SimSearch: A Human-in-The-Loop Learning Framework for Fast Detection of Regions of Interest in Microscopy Images. IEEE J. Biomed. Health Inform. 2022, 26, 4079–4089. [Google Scholar] [CrossRef]
- Holzinger, A. Interactive Machine Learning for Health Informatics: When Do We Need the Human-in-the-Loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xiao, L.; Sun, Y.; Zhang, J.; Ma, T.; He, L. A Survey of Human-in-the-Loop for Machine Learning. Future Gener. Comput. Syst. 2022, 135, 364–381. [Google Scholar] [CrossRef]
- Jobin, A.; Ienca, M.; Vayena, E. The Global Landscape of AI Ethics Guidelines. Nat. Mach. Intell. 2019, 1, 389–399. [Google Scholar] [CrossRef]
- Felzmann, H.; Fosch-Villaronga, E.; Lutz, C.; Tamò-Larrieux, A. Towards Transparency by Design for Artificial Intelligence. Sci. Eng. Ethics 2020, 26, 3333–3361. [Google Scholar] [CrossRef]
- Balasubramaniam, N.; Kauppinen, M.; Hiekkanen, K.; Kujala, S. Transparency and Explainability of AI Systems: Ethical Guidelines in Practice. In Proceedings of the Requirements Engineering: Foundation for Software Quality; Gervasi, V., Vogelsang, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 3–18. [Google Scholar]
- Bommasani, R.; Klyman, K.; Longpre, S.; Xiong, B.; Kapoor, S.; Maslej, N.; Narayanan, A.; Liang, P. Foundation Model Transparency Reports. arXiv 2024, arXiv:2402.16268. [Google Scholar]
- Interpretability versus Explainability—Model Explainability with AWS Artificial Intelligence and Machine Learning Solutions. Available online: https://docs.aws.amazon.com/whitepapers/latest/model-explainability-aws-ai-ml/interpretability-versus-explainability.html (accessed on 24 July 2024).
- Gundersen, O.E.; Kjensmo, S. State of the Art: Reproducibility in Artificial Intelligence. Proc. AAAI Conf. Artif. Intell. 2018, 32, 1644–1651. [Google Scholar] [CrossRef]
- Mohamed, A.K.Y.S.; Auer, D.; Hofer, D.; Küng, J. A Systematic Literature Review for Authorization and Access Control: Definitions, Strategies and Models. Int. J. Web Inf. Syst. 2022, 18, 156–180. [Google Scholar] [CrossRef]
- Kotsenas, A.L.; Balthazar, P.; Andrews, D.; Geis, J.R.; Cook, T.S. Rethinking Patient Consent in the Era of Artificial Intelligence and Big Data. J. Am. Coll. Radiol. 2021, 18, 180–184. [Google Scholar] [CrossRef]
- Murdoch, B. Privacy and Artificial Intelligence: Challenges for Protecting Health Information in a New Era. BMC Med. Ethics 2021, 22, 122. [Google Scholar] [CrossRef]
- Pan, B.; Stakhanova, N.; Ray, S. Data Provenance in Security and Privacy. ACM Comput. Surv. 2023, 55, 1–35. [Google Scholar] [CrossRef]
- Mani, V.; Manickam, P.; Alotaibi, Y.; Alghamdi, S.; Khalaf, O.I. Hyperledger Healthchain: Patient-Centric IPFS-Based Storage of Health Records. Electronics 2021, 10, 3003. [Google Scholar] [CrossRef]
- Acar, A.; Aksu, H.; Uluagac, A.S.; Conti, M. A Survey on Homomorphic Encryption Schemes: Theory and Implementation. ACM Comput. Surv. 2018, 51, 1–35. [Google Scholar] [CrossRef]
- Ko, S.; Jeon, K.; Morales, R. The Hybrex Model for Confidentiality and Privacy in Cloud Computing. In Proceedings of the 3rd USENIX Conference on Hot Topics in Cloud Computing, Portland, OR, USA, 14–15 June 2011; p. 8. [Google Scholar]
- Ghadi, Y.Y.; Shah, S.F.A.; Mazhar, T.; Shahzad, T.; Ouahada, K.; Hamam, H. Enhancing Patient Healthcare with Mobile Edge Computing and 5G: Challenges and Solutions for Secure Online Health Tools. J. Cloud Comput. 2024, 13, 93. [Google Scholar] [CrossRef]
- Raith, P.; Nastic, S.; Dustdar, S. Serverless Edge Computing—Where We Are and What Lies Ahead. IEEE Internet Comput. 2023, 27, 50–64. [Google Scholar] [CrossRef]
- Characterizing Browser-Based Medical Imaging AI with Serverless Edge Computing: Towards Addressing Clinical Data Security Constraints. Available online: https://edrn.nci.nih.gov/data-and-resources/publications/37063644-3301-characterizing-browser-based-medical-imaging-ai-with-serverless-edge-computing-towards-addressing-clinical-data-security-constraints/ (accessed on 27 July 2024).
- Sadilek, A.; Liu, L.; Nguyen, D.; Kamruzzaman, M.; Serghiou, S.; Rader, B.; Ingerman, A.; Mellem, S.; Kairouz, P.; Nsoesie, E.O.; et al. Privacy-First Health Research with Federated Learning. Npj Digit. Med. 2021, 4, 132. [Google Scholar] [CrossRef] [PubMed]
- Rights (OCR), O. for C. Guidance Regarding Methods for De-Identification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. Available online: https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html (accessed on 27 July 2024).
- Dernoncourt, F.; Lee, J.Y.; Uzuner, O.; Szolovits, P. De-Identification of Patient Notes with Recurrent Neural Networks. J. Am. Med. Inform. Assoc. 2017, 24, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Aziz, M.M.A.; Mohammed, N. De-Identification of Electronic Health Record Using Neural Network. Sci. Rep. 2020, 10, 18600. [Google Scholar] [CrossRef]
- Ko, M.; Jin, M.; Wang, C.; Jia, R. Practical Membership Inference Attacks Against Large-Scale Multi-Modal Models: A Pilot Study. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 1–6 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 4848–4858. [Google Scholar]
- Ben Hamida, S.; Mrabet, H.; Chaieb, F.; Jemai, A. Assessment of Data Augmentation, Dropout with L2 Regularization and Differential Privacy against Membership Inference Attacks. Multimed. Tools Appl. 2024, 83, 44455–44484. [Google Scholar] [CrossRef]
- Hu, H.; Salcic, Z.; Sun, L.; Dobbie, G.; Yu, P.S.; Zhang, X. Membership Inference Attacks on Machine Learning: A Survey 2022.
- Song, L.; Mittal, P. Systematic Evaluation of Privacy Risks of Machine Learning Models. In Proceedings of the 30th USENIX Security Symposium, Vancouver, BC, Canada, 11–13 August 2021; pp. 2615–2632. [Google Scholar]
- Dar, S.U.H.; Seyfarth, M.; Kahmann, J.; Ayx, I.; Papavassiliu, T.; Schoenberg, S.O.; Frey, N.; Baeßler, B.; Foersch, S.; Truhn, D.; et al. Unconditional Latent Diffusion Models Memorize Patient Imaging Data: Implications for Openly Sharing Synthetic Data. arXiv 2024, arXiv:2402.01054. [Google Scholar]
- El-Mhamdi, E.-M.; Farhadkhani, S.; Guerraoui, R.; Gupta, N.; Hoang, L.-N.; Pinot, R.; Rouault, S.; Stephan, J. On the Impossible Safety of Large AI Models. arXiv 2023, arXiv:2209.15259. [Google Scholar]
- Feldman, V. Does Learning Require Memorization? A Short Tale about a Long Tail. In Proceedings of the 52nd Annual ACM SIGACT Symposium on Theory of Computing, Chicago, IL, USA, 22–26 June 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 954–959. [Google Scholar]
- Carlini, N.; Ippolito, D.; Jagielski, M.; Lee, K.; Tramer, F.; Zhang, C. Quantifying Memorization Across Neural Language Models. arXiv 2023, arXiv:2202.07646. [Google Scholar]
- Lee, K.; Ippolito, D.; Nystrom, A.; Zhang, C.; Eck, D.; Callison-Burch, C.; Carlini, N. Deduplicating Training Data Makes Language Models Better. arXiv 2022, arXiv:2107.06499. [Google Scholar]
- König, K.; Pechmann, A.; Thiele, S.; Walter, M.C.; Schorling, D.; Tassoni, A.; Lochmüller, H.; Müller-Reible, C.; Kirschner, J. De-Duplicating Patient Records from Three Independent Data Sources Reveals the Incidence of Rare Neuromuscular Disorders in Germany. Orphanet J. Rare Dis. 2019, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Aponte-Novoa, F.A.; Orozco, A.L.S.; Villanueva-Polanco, R.; Wightman, P. The 51% Attack on Blockchains: A Mining Behavior Study. IEEE Access 2021, 9, 140549–140564. [Google Scholar] [CrossRef]
- Carvalho, G.; Cabral, B.; Pereira, V.; Bernardino, J. Edge Computing: Current Trends, Research Challenges and Future Directions. Computing 2021, 103, 993–1023. [Google Scholar] [CrossRef]
- Humayun, M.; Alsirhani, A.; Alserhani, F.; Shaheen, M.; Alwakid, G. Transformative Synergy: SSEHCET—Bridging Mobile Edge Computing and AI for Enhanced eHealth Security and Efficiency. J. Cloud Comput. 2024, 13, 37. [Google Scholar] [CrossRef]
- Meng, L.; Li, D. Novel Edge Computing-Based Privacy-Preserving Approach for Smart Healthcare Systems in the Internet of Medical Things. J. Grid Comput. 2023, 21, 66. [Google Scholar] [CrossRef]
- Neamatullah, I.; Douglass, M.M.; Lehman, L.H.; Reisner, A.; Villarroel, M.; Long, W.J.; Szolovits, P.; Moody, G.B.; Mark, R.G.; Clifford, G.D. Automated De-Identification of Free-Text Medical Records. BMC Med. Inform. Decis. Mak. 2008, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Sucholutsky, I.; Griffiths, T.L. Alignment with Human Representations Supports Robust Few-Shot Learning. arXiv 2023, arXiv:2301.11990. [Google Scholar]
- Packhäuser, K.; Gündel, S.; Münster, N.; Syben, C.; Christlein, V.; Maier, A. Deep Learning-Based Patient Re-Identification Is Able to Exploit the Biometric Nature of Medical Chest X-Ray Data. Sci. Rep. 2022, 12, 14851. [Google Scholar] [CrossRef]
- Narayanan, A.; Shmatikov, V. Robust De-Anonymization of Large Sparse Datasets. In Proceedings of the 2008 IEEE Symposium on Security and Privacy (sp 2008), Oakland, CA, USA, 18–22 May 2008; pp. 111–125. [Google Scholar]
- Johnson, A.; Pollard, T.; Mark, R. MIMIC-III Clinical Database. 2015. Available online: https://physionet.org/content/mimiciii/1.4/ (accessed on 10 August 2024).
- Nasr, M.; Song, S.; Thakurta, A.; Papernot, N.; Carlini, N. Adversary Instantiation: Lower Bounds for Differentially Private Machine Learning. In Proceedings of the 2021 IEEE Symposium on security and privacy (SP), San Francisco, CA, USA, 24–27 May 2021. [Google Scholar]
- Song, C.; Shmatikov, V. Auditing Data Provenance in Text-Generation Models. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 25 July 2019. [Google Scholar]
- Xu, T.; Liu, C.; Zhang, K.; Zhang, J. Membership Inference Attacks Against Medical Databases. In Proceedings of the Neural Information Processing; Luo, B., Cheng, L., Wu, Z.-G., Li, H., Li, C., Eds.; Springer Nature: Singapore, 2024; pp. 15–25. [Google Scholar]
- Kaya, Y.; Dumitras, T. When Does Data Augmentation Help With Membership Inference Attacks? In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR. 2021; pp. 5345–5355. Available online: https://proceedings.mlr.press/v139/kaya21a.html (accessed on 10 August 2024).
- Zhang, Z.; Yan, C.; Malin, B.A. Membership Inference Attacks against Synthetic Health Data. J. Biomed. Inform. 2022, 125, 103977. [Google Scholar] [CrossRef]
- Liu, Z.; Miao, Z.; Zhan, X.; Wang, J.; Gong, B.; Yu, S.X. Large-Scale Long-Tailed Recognition in an Open World. arXiv 2019, arXiv:1904.05160. [Google Scholar]
- Wu, T.; Liu, Z.; Huang, Q.; Wang, Y.; Lin, D. Adversarial Robustness Under Long-Tailed Distribution. arXiv 2021, arXiv:2104.02703. [Google Scholar]
- Carlini, N.; Jagielski, M.; Zhang, C.; Papernot, N.; Terzis, A.; Tramer, F. The Privacy Onion Effect: Memorization Is Relative 2022.
- Manber, U.; Myers, G. Suffix Arrays: A New Method for On-Line String Searches. SIAM J. Comput. 1993, 22, 935–948. [Google Scholar] [CrossRef]
- Broder, A.Z. On the Resemblance and Containment of Documents. In Proceedings of the Compression and Complexity of SEQUENCES 1997 (Cat. No.97TB100171), Salerno, Italy, 13 June 1997; pp. 21–29. [Google Scholar]
- Baker, D.B.; Knoppers, B.M.; Phillips, M.; van Enckevort, D.; Kaufmann, P.; Lochmuller, H.; Taruscio, D. Privacy-Preserving Linkage of Genomic and Clinical Data Sets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Bouderhem, R. Shaping the Future of AI in Healthcare through Ethics and Governance. Humanit. Soc. Sci. Commun. 2024, 11, 1–12. [Google Scholar] [CrossRef]
- High-Level Expert Group on Artificial Intelligence|Shaping Europe’s Digital Future. Available online: https://digital-strategy.ec.europa.eu/en/policies/expert-group-ai (accessed on 28 July 2024).
- CAHAI—Ad Hoc Committee on Artificial Intelligence—Artificial Intelligence—www.Coe.Int. Available online: https://www.coe.int/en/web/artificial-intelligence/cahai (accessed on 28 July 2024).
- Ethics of Artificial Intelligence|UNESCO. Available online: https://www.unesco.org/en/artificial-intelligence/recommendation-ethics (accessed on 28 July 2024).
- Ethics and Governance of Artificial Intelligence for Health. Available online: https://www.who.int/publications/i/item/9789240029200 (accessed on 28 July 2024).
- World Health Organization. Regulatory Considerations on Artificial Intelligence for Health; World Health Organization: Geneva, Switzerland, 2023; ISBN 978-92-4-007887-1. [Google Scholar]
- Using Artificial Intelligence and Algorithms. Available online: https://www.ftc.gov/business-guidance/blog/2020/04/using-artificial-intelligence-algorithms (accessed on 29 July 2024).
- Tabassi, E. Artificial Intelligence Risk Management Framework (AI RMF 1.0); National Institute of Standards and Technology (U.S.): Gaithersburg, MD, USA, 2023; p. NIST AI 100-1.
- Rep. McNerney, J. [D-C.-9 H.R.2575—116th Congress (2019–2020): AI in Government Act of 2020. Available online: https://www.congress.gov/bill/116th-congress/house-bill/2575 (accessed on 29 July 2024).
- House, T.W. Executive Order on the Safe, Secure, and Trustworthy Development and Use of Artificial Intelligence. Available online: https://www.whitehouse.gov/briefing-room/presidential-actions/2023/10/30/executive-order-on-the-safe-secure-and-trustworthy-development-and-use-of-artificial-intelligence/ (accessed on 24 July 2024).
- Deshpande, A.; Sharp, H. Responsible AI Systems: Who Are the Stakeholders? In Proceedings of the 2022 AAAI/ACM Conference on AI, Ethics, and Society, Oxford, UK, 1–3 August 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 227–236. [Google Scholar]
- Whittaker, R.; Dobson, R.; Jin, C.K.; Style, R.; Jayathissa, P.; Hiini, K.; Ross, K.; Kawamura, K.; Muir, P. An Example of Governance for AI in Health Services from Aotearoa New Zealand. Npj Digit. Med. 2023, 6, 1–7. [Google Scholar] [CrossRef]
- PCAST Releases Report on Supercharging Research: Harnessing Artificial Intelligence to Meet Global Challenges|PCAST. Available online: https://www.whitehouse.gov/pcast/briefing-room/2024/04/29/pcast-releases-report-on-supercharging-research-harnessing-artificial-intelligence-to-meet-global-challenges/ (accessed on 29 July 2024).
- Scott, I.A.; Carter, S.M.; Coiera, E. Exploring Stakeholder Attitudes towards AI in Clinical Practice. BMJ Health Care Inform. 2021, 28, e100450. [Google Scholar] [CrossRef]
- Dwivedi, R.; Dave, D.; Naik, H.; Singhal, S.; Omer, R.; Patel, P.; Qian, B.; Wen, Z.; Shah, T.; Morgan, G.; et al. Explainable AI (XAI): Core Ideas, Techniques, and Solutions. ACM Comput. Surv. 2023, 55, 1–33. [Google Scholar] [CrossRef]
- Li, L. How to Co-Design Software/Hardware Architecture for AI/ML in a New Era? Available online: https://towardsdatascience.com/how-to-co-design-software-hardware-architecture-for-ai-ml-in-a-new-era-b296f2842fe2 (accessed on 29 July 2024).
- Co-Design and Ethical Artificial Intelligence for Health: An Agenda for Critical Research and Practice—Joseph Donia, James A. Shaw. 2021. Available online: https://journals.sagepub.com/doi/full/10.1177/20539517211065248 (accessed on 29 July 2024).
- Olczak, J.; Pavlopoulos, J.; Prijs, J.; Ijpma, F.F.A.; Doornberg, J.N.; Lundström, C.; Hedlund, J.; Gordon, M. Presenting Artificial Intelligence, Deep Learning, and Machine Learning Studies to Clinicians and Healthcare Stakeholders: An Introductory Reference with a Guideline and a Clinical AI Research (CAIR) Checklist Proposal. Acta Orthop. 2021, 92, 513–525. [Google Scholar] [CrossRef]
- Zicari, R.V.; Ahmed, S.; Amann, J.; Braun, S.A.; Brodersen, J.; Bruneault, F.; Brusseau, J.; Campano, E.; Coffee, M.; Dengel, A.; et al. Co-Design of a Trustworthy AI System in Healthcare: Deep Learning Based Skin Lesion Classifier. Front. Hum. Dyn. 2021, 3, 688152. [Google Scholar] [CrossRef]
- Zicari, R.V.; Brodersen, J.; Brusseau, J.; Düdder, B.; Eichhorn, T.; Ivanov, T.; Kararigas, G.; Kringen, P.; McCullough, M.; Möslein, F.; et al. Z-Inspection®: A Process to Assess Trustworthy AI. IEEE Trans. Technol. Soc. 2021, 2, 83–97. [Google Scholar] [CrossRef]
- De Silva, D.; Alahakoon, D. An Artificial Intelligence Life Cycle: From Conception to Production. Patterns 2022, 3, 100489. [Google Scholar] [CrossRef] [PubMed]
- Vyhmeister, E.; Castane, G.; Östberg, P.-O.; Thevenin, S. A Responsible AI Framework: Pipeline Contextualisation. AI Ethics 2023, 3, 175–197. [Google Scholar] [CrossRef]

| Category | Strategy | Limitation/Challenge |
|---|---|---|
| Challenge: Underrepresentation of certain demographic groups (e.g., African Americans or women) in biomedical data can result in biased and unfair model decisions. | ||
| Data Level (all dataset types) |
|
|
| Training Level (datasets with labeled demographic metadata) |
|
|
| Evaluation Level (datasets with labeled demographic metadata) |
| 1–3. Relying on a single fairness metric might give an incomplete or misleading picture of a model’s bias. 4. Application of techniques can be resource intensive. Effectiveness of explainability methods often depends on human interpretation. |
| Challenge: Human stereotypical biases can contaminate natural language data and affect the fairness of large language and vision-language biomedical models. | ||
| LLM |
|
|
| VLM |
|
|
| Versatile (LLM and VLM) |
|
|
| Category | Strategy | Limitation/Challenge |
|---|---|---|
| Challenge: The absence of standardized data management protocols throughout its lifecycle obstructs collaboration in biomedical AI, particularly in integrating diverse datasets for comprehensive model development. | ||
| Biomedical Data |
|
|
| Challenge: The decision-making process and output of a biomedical AI model must be transparent, understandable, and verifiable by human experts to build the trust required for integration into clinical and translational settings. | ||
| Biomedical AI systems |
|
|
| Category | Strategy | Limitation/Challenge |
|---|---|---|
| Challenge: Unauthorized access to sensitive biomedical data jeopardizes individual privacy, undermines stakeholder trust, and threatens compliance with regulatory guidelines. | ||
| Data Life Cycle |
|
|
| Challenge: Developing biomedical AI in the cloud offers flexible, ready-to-use, and scalable infrastructure. However, ensuring the security and privacy of sensitive data during storage and computation is challenging in this environment. | ||
| Data Storage |
|
|
| Computation |
|
|
| Challenge: Sensitive biomedical data are vulnerable to adversarial attacks, including the risk of re-identifying individuals or inferring whether an individual’s data were used to train a model. | ||
| Patient Re-identification |
|
|
| Membership Inference Attacks |
|
|
| Challenge: Large-scale biomedical models can memorize data, making them vulnerable to unique security risks such as data leaks, manipulation to produce misleading outputs, and the exposure of sensitive patient information. | ||
| Over-Parameterized Models and “Long-Tailed” Data |
|
|
| Bias Mitigation Strategies | Data Privacy & Security | Fairness | Stakeholders and Governance | AI Lifecycle Ecosystem | Clinical Integration | Standardization of Ethical FMs | Biomedical Contexts/Applications | |
|---|---|---|---|---|---|---|---|---|
| Fairness and Bias in AI: A Brief Survey of Sources, Impact, and Mitigation Strategies [9] | X | X | X | |||||
| The Evolutionary Dynamics of the AI Ecosystem [10] | X | X | ||||||
| Bias and Fairness in LLMs: A Survey [25] | X | X | ||||||
| Assessing the Research Landscape and Clinical Utility of LLMs: A Scoping Review [48] | X | X | X | |||||
| An AI Life Cycle: From Conception to Production [144] | X | X | X | |||||
| Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundational Models | X | X | X | X | X | X | X | X |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankar, B.S.; Gilliland, D.; Rincon, J.; Hermjakob, H.; Yan, Y.; Adam, I.; Lemaster, G.; Wang, D.; Watson, K.; Bui, A.; et al. Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models. Bioengineering 2024, 11, 984. https://doi.org/10.3390/bioengineering11100984
Sankar BS, Gilliland D, Rincon J, Hermjakob H, Yan Y, Adam I, Lemaster G, Wang D, Watson K, Bui A, et al. Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models. Bioengineering. 2024; 11(10):984. https://doi.org/10.3390/bioengineering11100984
Chicago/Turabian StyleSankar, Baradwaj Simha, Destiny Gilliland, Jack Rincon, Henning Hermjakob, Yu Yan, Irsyad Adam, Gwyneth Lemaster, Dean Wang, Karol Watson, Alex Bui, and et al. 2024. "Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models" Bioengineering 11, no. 10: 984. https://doi.org/10.3390/bioengineering11100984
APA StyleSankar, B. S., Gilliland, D., Rincon, J., Hermjakob, H., Yan, Y., Adam, I., Lemaster, G., Wang, D., Watson, K., Bui, A., Wang, W., & Ping, P. (2024). Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models. Bioengineering, 11(10), 984. https://doi.org/10.3390/bioengineering11100984






