Abstract
Background: Magnetic Resonance Imaging (MRI) data collected from multiple centres can be heterogeneous due to factors such as the scanner used and the site location. To reduce this heterogeneity, the data needs to be harmonised. In recent years, machine learning (ML) has been used to solve different types of problems related to MRI data, showing great promise. Objective: This study explores how well various ML algorithms perform in harmonising MRI data, both implicitly and explicitly, by summarising the findings in relevant peer-reviewed articles. Furthermore, it provides guidelines for the use of current methods and identifies potential future research directions. Method: This review covers articles published through PubMed, Web of Science, and IEEE databases through June 2022. Data from studies were analysed based on the criteria of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). Quality assessment questions were derived to assess the quality of the included publications. Results: a total of 41 articles published between 2015 and 2022 were identified and analysed. In the review, MRI data has been found to be harmonised either in an implicit (n = 21) or an explicit (n = 20) way. Three MRI modalities were identified: structural MRI (n = 28), diffusion MRI (n = 7) and functional MRI (n = 6). Conclusion: Various ML techniques have been employed to harmonise different types of MRI data. There is currently a lack of consistent evaluation methods and metrics used across studies, and it is recommended that the issue be addressed in future studies. Harmonisation of MRI data using ML shows promises in improving performance for ML downstream tasks, while caution should be exercised when using ML-harmonised data for direct interpretation.
1. Introduction
Neuroimaging technologies have rapidly developed in recent years. They are now widely used in various neurological research centres to study various clinical syndromes [1,2,3]. To both reduce logistics costs and increase the diversity of clinical exemplars, large-scale neuroimaging studies often combine datasets from multiple centres [4,5]. The presence of heterogeneity in Magnetic Resonance Imaging (MRI) machine characteristics, such as variations in software, MRI equipment, reconstruction algorithms and scan protocols, can lead to inter- and intra-site variations. Such variations can affect the signal intensity and derived metrics, which may obscure the effect of interest [6,7,8,9,10,11] and lead to the failure of downstream and machine learning (ML) analyses [12,13].
To address these issues, harmonisation can be applied to remove non-biological factors that can cause variations in MRI data across different centres and institutions. This can be achieved through applying protocol or data harmonisation. An acquisition protocol is harmonised by specifying the type of MRI machine, as well as the imaging parameters, e.g., the field of view and the analysis methods [14]. However, using standardised acquisition protocols is not sufficient to achieve the desired level of standardisation and cannot be applied to existing datasets [15,16,17]. In contrast, data harmonisation aims to improve the comparability of MRI data so that scan results are comparable across different settings/domains (e.g., sites, scanners, and institutions) [18]. The underlying objective is to remove non-biological factors while preserving biological factors among participants from different domains [18]. This differs from data normalisation, which compares and integrates MRI data across subjects within the same domain by transforming the data into a standardised system. Harmonisation is performed across different domains.
Data harmonisation can be challenging due to the presence of confounding variables and unknown factors that cause cross-site variations. Direct mapping to a constant value may not be sufficient because it could eliminate important variables of interest. The use of ML, especially deep learning, to address the scanner effect has grown significantly due to its ability to model complex relationships within messy data sets. Using ML techniques, data can be harmonised either explicitly or implicitly. Explicit approaches aim to predict the target image or its derived metrics given the reference image. ML models are trained to harmonise data which can then be directly inspected by a radiologist. Models used in an implicit manner, on the other hand, are trained on a downstream task (e.g., classification and segmentation), and harmonisation is performed implicitly through the optimisation process during training. Therefore, the harmonised data are not visible. A representation of explicit and implicit methods for data can be seen in Figure 1.
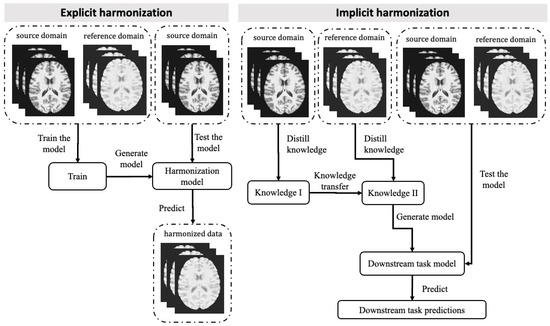
Figure 1.
Workflow of Implicit and Explicit Computational Harmonisation Approaches for MRI Data. The explicit approach (left panel) involves training data from both source and reference domains to generate the harmonisation model. The source domain refers to the dataset that requires harmonisation, while the reference domain refers to the dataset that serves as a standard. The model is then tested using additional source domain data, resulting in the generation of harmonised data. In the implicit approach (right panel), data from the source and reference domains are processed separately, and the distilled knowledge is transferred from one to the other. The downstream task model is generated based on the transferred knowledge, and both source and reference domain data can be fed into the model to generate task-related predictions.
This paper presents a systematic review of harmonisation methods for brain MRI data, with a specific focus on ML-based approaches applied in both explicit and implicit ways. Our primary objective is to provide an overview of the latest techniques and advancements in MRI harmonisation that will be valuable for radiologists, neuroimaging researchers, and other professionals working with large-scale, multicentre MRI datasets. While several reviews have been conducted to summarise the harmonisation methods used in medical imaging [18,19,20], none have focused on ML-based approaches. The field also lacks a summary of implicit methods. Therefore, this review aims to fill these gaps and offer valuable insights into available resources when developing harmonisation methods.
2. Materials and Methods
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were followed in conducting this review. Our main interest is: What ML algorithms are available, and how are they being applied by researchers to harmonise brain MRI data.
2.1. Inclusion Criteria and Search Terms
To focus on our main question of interest, we reviewed works that meet the following inclusion criteria: (1) address the multi-site/scanner/centre issues by using data harmonisation methods; (2) apply ML techniques; and (3) are applied to MRI images. The search terms were generated by considering each inclusion criterion individually. For the first criterion, we used terms such as “harmonisation”, “normalisation”, and “standardisation”, as well as those associated with multicentre, such as “multi-site”, “inter-site”, and “multi-scanner”. For the second criterion, we used a comprehensive range of search terms encompassing ML algorithms that had used or could potentially be used for harmonisation. The third criterion involved keywords related to MRI, such as “DTI”, “MRI”, “DWI”, and “functional”. All keywords used for searching is shown in Table 1. To form the sophisticated search terms, the Boolean expression ‘OR’ was used to combine within-group keywords, while ‘AND’ was used to combine three groups. Parenthetically, articles from both journals and conference proceedings have been included in this review since they both make significant contributions to the fields of ML and bioengineering.

Table 1.
Search terms for electronic databases used.
2.2. Screening and Selection Process
Articles published in journal articles or conference proceedings between Jan 2010 and June 2022 were searched using three search engines: PubMed, IEEE, and Web of Science. Following the PRISMA, the first phase of screening removed duplicate articles collected from multiple sources. In the second phase of screening, we discarded all papers that conclusively did not belong in the scope of this review, such as those with abstracts and titles which contained no keywords related to “harmonisation” or “machine learning”. Afterwards, the articles that passed the screening phase were assessed for eligibility by reviewing the full texts. In addition to the usual search engines, papers identified from the reference list of included papers were also evaluated for eligibility using the same criteria. Searching and screening were conducted independently by two researchers (G.W and A.W).
2.3. Exclusion Criteria
Articles that were excluded from the review include those that (a) were not peer-reviewed; (b) were books, letters, notes, graduate theses, and patents; (c) were not able to be accessed; (d) were not written in English; (e) were surveys or literature reviews; (f) did not use multi-site or multi-scanner datasets; (g) were not applied on human neuro MRI; (h) did not sufficiently describe an ML approach or an experiment or validation methods; (i) did not contain any quantitative results; or (j) did not describe an experiment or validation study.
2.4. Data Extraction
From the data, we extracted: the author name, publishing details, data set details, harmonised domain, training inputs and validation strategies, evaluation methods, computational libraries, ML techniques and study design. More details about the data characteristics are presented in Table 2. Extracted data were evaluated and resolved (if necessary) by another reviewer (V.S.).

Table 2.
Data characters of the Included studies.
2.5. Quality Assessment
The modified QualSyst Assessment Tool for quantitative studies was used to assess the methodological quality, credibility and relevance of the included studies [21]. The assessment involved the use of questions that were derived from the scale, focusing on aspects such as research questions, study design, data collection, data analysis, and results reporting. Additionally, questions related to the quality of ML-based models, such as the ML algorithms, training procedure and comparative analysis, were incorporated, as suggested by [22]. These questions are listed in Table 3. Each of them had three optional answers: “Yes”, “Partly”, or “No”, with scores of 1, 0.5, or 0, respectively. For each study, the final score was calculated by summing up the scores given to each question, ranging from 0 to 10. A cut-off score of 5 (50% of the maximum total) was considered acceptable when assessing the reliability of studies. Final scoring was agreed upon by two reviewers (G.W. and A.W.).

Table 3.
Quality assessment questions.
3. Results
Figure 2 shows the PRISMA flowchart. We identified a total of 1112 studies from three different bibliographic databases, as well as six additional studies from other sources. After removing duplicates, 1053 studies were screened based on their titles and abstracts, resulting in 97 studies for full-text evaluation. Eventually, 41 articles published between 2015 and June 2022 remained for quality assessment. The quality scores for the included studies, ranging from 5.5 to 10 with a mean score of 8.16, are provided in Supplementary Materials. These findings suggest that the studies included in this review are of high quality. Scores for questions 8 and 9 were relatively low, however, indicating that several studies did not conduct adequate comparative analyses. Specifically, 24% of the studies did not perform any comparative analysis, and 48.8% of the studies compared their proposed method to either statistical or ML-based approaches. The results also indicated a slight advantage in scoring for studies utilising implicit approaches compared to those utilising explicit approaches. However, the sample size was relatively small, and subject characteristics were unclear, contributing to bias. A table with extensive details for individual key aspects can be seen in Supplementary Materials.
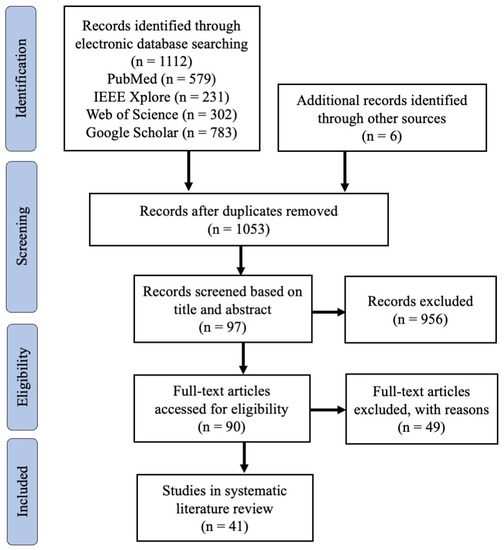
Figure 2.
The PRISMA flow diagram for the systematic review detailing the database searches, the number of abstracts screened, and the full texts retrieved.
3.1. Article Distribution among Journals and Conference Proceedings
The eligible articles chosen for the study were published across sixteen different journals and three conference proceedings. In total, 29 papers were published in journals, and 12 papers were from conference proceedings. Among these, the Medical Image Computing and Computer Assisted Intervention Society conference proceedings contained the most publications used, comprising 9 out of 41 publications. Following this, six publications from NeuroImage and five from Medical Image Analysis. The publications covered a diverse range of journals, including those in the fields of biomedical engineering, neuroscience, signal processing, and computer science. Figure 3 depicts all the included publishing sources, journals, and conference proceedings.
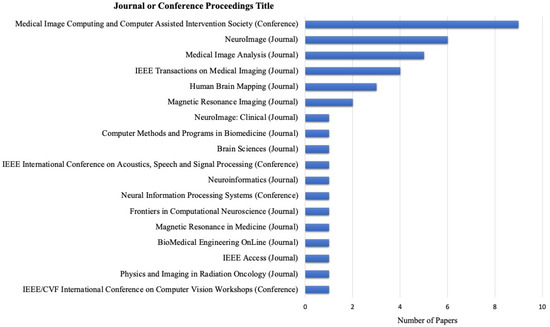
Figure 3.
The number of publications per publisher.
3.2. Application of ML Methods
A total of 41 studies were conducted that describe ML being applied to harmonise MRI data in either an explicit (n = 20) or implicit (n = 21) way. The applied ML algorithms were plotted by year, shown in Figure 4. Implicit approaches first appeared in the included studies in 2015, while explicit harmonisation first appeared in 2019. Generally, the number of papers has grown gradually since 2019. Explicit approaches included generative adversarial networks (GANs) (n = 7), autoencoders (n = 7), convolutional neural networks (CNNs) (n = 3), regression (n = 1), transformer networks (n = 1) and sparse dictionary learning (n = 1). Regarding implicit approaches, most of the applications were fine-tuning (n = 10), followed by adversarial transfer learning (n = 7), feature extraction network-based transfer learning (n = 3) and multi-task learning (n = 1). These ML methods are presented in Table 4, along with a brief description and the number of articles for each method.
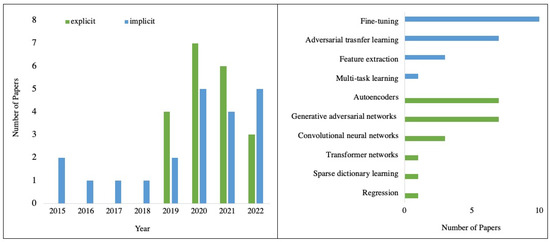
Figure 4.
Breakdown of the papers included in this review in the year of publication and harmonisation methods. The number of papers for 2022 has been extrapolated from those published before June.

Table 4.
The ML methods used in the publications as well as the number of papers in each category and a brief description of each method.
3.3. MRI Modality and Harmonised Features
Three different MRI modalities were identified: structural MRI (sMRI) (n = 28), diffusion MRI (dMRI) (n = 7), and functional MRI (fMRI) (n = 6). Papers were also categorised according to the harmonisation features: raw signals (n = 29) and image-derived features (n = 12). Of the 28 studies on sMRI, 24 harmonised raw intensity signals, while four studies focused on derived measures, including brain volumes [25,46], cortical thickness [36] and mixed image-based features [66]. Four studies harmonised the raw diffusion signals of dMRI through dictionary representation [31] and spherical harmonics representation [29,39,48], while three studies focused on derived features [28,41,65]. For fMRI data, only one study harmonised raw signals [61], whereas five harmonised derived functional connectome-based features [57,60,62,70,73]. Results showed that raw signals of sMRI and dMRI data were more frequently used than fMRI raw signals. One possible explanation for this is that fMRI data are time-series signals, which have a higher degree of dimensionality and are, therefore, more complex and difficult to process and analyse.
3.4. Evaluation of Methodology
The reliability of harmonised data can be evaluated directly (n = 20) and indirectly (n = 25), depending on whether travelling subject datasets or downstream ML models were available. Direct evaluation methods were more widely used in explicit approaches. One method was to evaluate the appearance of the harmonisation results. The contrast of harmonised images was found to be more similar to that of reference scans, as documented in studies such as [27]. Studies also performed subject-wise comparisons by measuring similarities between individual scans from different domains. This was accomplished using various metrics such as mean error (in studies, e.g., [27,28,36]), structural similarity index (in studies, e.g., [27,35,49]) and coefficient of variation (in studies, e.g., [28,48]).
Alternatively, a group-wise analysis can be performed to determine whether subjects between different groups, such as different age groups [25], become more distinguishable after harmonisation. Studies that used explicit methods commonly employed statistical tests to compute the probabilities of differences. The most commonly used metrics were Cohen’s d value (e.g., [36,41]), Pearson’s r value (e.g., [41]), and Kullback–Leibler divergence (e.g., [31,46]). Implicit methods, on the other hand, used the t-SNE algorithm to directly visualise the data distributions based on the features extracted from the trained network. Three studies have used this method, which revealed a significant change in domain clustering after harmonisation [51,52,71].
Using indirect evaluation methods, researchers set up the downstream task and measured the harmonisation effect by comparing task performance before and after harmonisation. Implicit methods relied solely on indirect evaluation methods, while only four studies demonstrated the harmonised results using downstream tasks in explicit methods [33,35,37,40].
There were a variety of approaches taken to validate the models developed. Validation by splitting into a testing and validation dataset was performed in all studies. In total, 22 studies used k-fold cross-validation.
3.5. Data Characteristics and Reproducibility
MRI data is typically stored in a 3D or 4D format. Three categorises were identified: 1D (n = 6), 2D (n = 19), pseudo 3D (n = 3) which is constructed by 2D data, and 3D (n = 13). We also documented the accessibility of the datasets, the number of subjects, and the availability of travel subjects. Thirty papers in our review used public datasets that are available for download, eight studies used private datasets (n = 8), and three studies used both. Thirteen studies utilised travelling subject datasets. In total, nine datasets containing travelling subjects were found, as shown in Table 5. Five of them are publicly available.

Table 5.
Datasets containing multiple scans of the same subject were used in the reviewed papers.
We divided the articles into five categories based on the size of the datasets used. The number of studies in each category is shown in Figure 5. Most of the studies used datasets with sizes of 101–1000 subjects (n = 20), followed by 1–20 subjects (n = 7), 1001–10,000 subjects (n = 8), 21–100 subjects (n = 5) and more than 100,000 subjects (n = 1). In terms of reproducibility, less than 30% of the studies (n = 11) reported their implemented codes. Of the 60% of studies (n = 23) that described the libraries used for implementation, PyTorch (n = 11) was the most widely used library, followed by Keras and TensorFlow (n = 6).

Figure 5.
Size of the dataset used in the reviewed studies.
4. Discussion
This systematic review evaluated the use of ML-based algorithms for the purpose of MRI data harmonisation. We summarise the main findings in methodology, datasets, and evaluation methods, highlighting their limitations and offering suggestions for future directions.
4.1. ML-Based Methods for Harmonisation
ML methods have provided efficient solutions for explicitly generating harmonised MRI data across different domains. The most widely used models are GANs and autoecoders, which have shown promising results in reducing multi-site variation through image-to-image synthesis [40,46,49]. GANs perform domain translation by learning domain-invariant features [34,80]. One issue is how they are limited to mapping between two specific scanners for most studies [36,37,38,39,40,41]. When mapping between multiple sites, multiple generative models are required, increasing the difficulty of training [35]. Another issue is that unpaired image-to-image translations may not preserve heterogeneity and individual quantitative information [81,82].
Autoencoder-based methods aim to harmonise data in terms of disentangled representations [83]. All related work in this review attempted to extract scanner-related features for harmonisation [43,44,45,46,47,48,49]. Similar to GANs, data from multiple sources and target domains are required for training. However, when an additional dataset from a new scanner is given, re-training only needs to be conducted on the new dataset. It is worth noting that none of the studies examined the minimum number of subjects required to disentangle the representation during training. Future study is needed to address this gap in knowledge.
Implicit approaches focus on finding task-specific, scanner-related confounds using either feature- or model-based techniques. Feature-based techniques, including feature extractors and adversarial networks, perform distribution alignment. This is, however, challenging, especially when optimising adversarial objectives due to the complex feature space of 3D or 4D MRI data. While most studies focused on alignment between two specific domains, one study attempted to create a global shared domain, resulting in a more generalisable and transferable feature representation that can be applied to new domains [61].
Fine-tuning and multi-task learning are model-based transfer learning techniques [74,84]. One future direction for these methods is to investigate the optimal number of subjects required for model transferring [85]. Research should also explore the adaptability of models to unseen domains.
4.2. Dataset Used for Harmonisation
Travelling subject datasets are essential for explicit harmonisation, as they can ensure that the ML models do not learn the population bias [10,46,86]. Current travelling subject datasets, however, have limitations due to their small size, the long inter-scan interval between consecutive scans of the same individual and scan-rescan reliability issues. Most datasets contain no more than 20 travelling subjects, and while a few are relatively large, they are primarily designed for longitudinal study for capturing brain changes over time. Addressing scanner-related problems can be difficult when the biological features of the brain differ significantly between the paired scans. Scan-rescan reliability can be influenced by factors such as positioning, shim setting and some unknown factors [87,88]. These variabilities are usually neglected in a study, but they can be significant, particularly given the small size of the travelling subject dataset.
Implicit approaches or unsupervised learning do not require a travelling-subject dataset. Instead, researchers need to define the source and reference domains. Large-scale public datasets may not, however, provide enough information, such as site information regarding where scans were acquired from, making harmonisation challenging. One solution is to simplify the problem by performing mapping between broader domains, for example, between different field strengths (e.g., 1.5 T, 3.0 T). Studies have shown that harmonising data within broader domains can reduce non-biological variations [33,47]; however, whether this is sufficient has yet to be explored.
As the number of public datasets grows, ML models can be validated more effectively against more diverse datasets. While there are shared travelling subject dataset collections, many have not been recently updated. One example is the multiBrain collection (https://github.com/Conxz/multiBrain, accessed on 15 February 2023), which has not been updated since 2019. Additionally, we identified the RMP Rumination fMRI Dataset, which is a travelling subject dataset that has not been used in any of our included studies [89]. It is plausible that this dataset has been used in low-quality studies that were not included in this review. Further evaluation is necessary to determine the efficacy of this dataset for harmonisation. To address the growing problem of scanner-related issues, an open and maintained platform for sharing travelling subject datasets should be developed and established in the future. Public data sources should include site and scanner information, being as specific as possible.
4.3. Data Representation for Model Training
Most ML models in this review only used 2D inputs. In general, 3D models are more powerful for exploring medical image features and yield better learning performance [90]. Developing models that capture 3D and 4D structural information is, however, challenging due to subtle scanner differences. Current 3D models primarily use patch-based architectures due to limited computational resources. Nevertheless, how to improve the state-of-the-art patch-based networks to integrate location information for harmonisation is still an open research question. Possible solutions could be to incorporate brain location information into a patch-based model, as suggested in [91], or to use a multi-scale patch-based network, as demonstrated in [92]. In addition, 4D models have been introduced in the field of medical images [93], but their applications in harmonisation are lacking. Developing 4D models to harmonise fMRI data or introduce domain adaption techniques in 4D models would be a promising future direction.
4.4. Performance Evaluation for Harmonisation Models
Ideally, harmonisation should remove all non-biological factors while preserving biological factors. Since there is no reference for harmonisation, methods evaluation can be difficult due to unknown sources of variability. Evaluation methods for explicit harmonisation methods have been performed using various quantitative methods, but heterogeneity in evaluation methods across studies makes it difficult to synthesise results quantitatively or conduct meta-analyses. Implicit approaches are simpler to evaluate by comparing prediction accuracy before and after harmonisation.
Image quality assessment (IQA) is also important for the evaluation of image-to-image tasks. Studies utilising explicit harmonisation methods have shown that applying ML-based harmonisation can enhance image quality, as demonstrated using full-reference IQA metrics such as signal-to-noise ratio and the contrast-to-noise ratio [35,40,43,44,49]. No-reference IQA methods, e.g., blind/referenceless image spatial quality evaluator [94], are, however, lacking.
It is recommended that researchers use more advanced IQA methods, such as in [95] or ML-based methods, such as in [96,97]. A reliable and robust reference-free IQA framework for harmonisation should be developed and used in future studies. Such a framework should be capable of evaluating different MRI modalities and different sequential stages (e.g., slice-wise, volume-wise and subject-wise) [98]. In addition to evaluating whether the quality of the harmonised image has been improved, it is also important to assess whether the criteria have been met, as well as the trade-off between the computational cost and the improved quality. A thorough quality check is essential before utilising harmonised images for any analysis.
4.5. Comparative Analysis
For explicit approaches, studies often compare their proposed methods to either statistical methods or other state-of-the-art ML methods. In statistical comparative analysis experiments, studies consistently suggest that their proposed ML-based methods outperform the statistical features alignment methods [28,41,44,44,45,46,47,49]. When comparing different ML methods, some studies suggest that autoencoders outperform GANs [44,49], while others find that supervised CNNs models outperform Encoder-based methods [27] and transformer networks outperform GANs [33]. Some studies also show improvements over previous, similar studies [31,48].
For implicit approaches, studies have consistently demonstrated that incorporating harmonisation leads to better performance compared to baseline experiments which do not apply harmonisation. Transfer learning approaches tend to outperform statistical covariate techniques [50,52,55,56,59,60,61,63,64,65,69,71,74], and some studies suggest that adversarial transfer learning methods outperform non-adversarial methods [51,53,55]. Multi-task learning is superior to single-task learning in classification tasks [75]. Some studies also demonstrate improvements over various feature-based methods when using their proposed methods [57,60,61,62,69,72]. It is important to note that studies that perform different regularisation techniques are not counted as comparative analyses in this study.
Although several studies have attempted to compare different models using the same dataset [99,100], no conclusions regarding whether high-performing ML models are necessary for MR data harmonisation can be drawn. This review does not include these studies as they do not fall within the scope of developing new methods. These studies, however, highlight the importance of carefully evaluating and comparing different methods to determine their effectiveness for a given task and dataset.
4.6. Toward Clinical Application of AI
The application of AI in clinical settings is an exciting prospect. Harmonisation can improve the accuracy and generalisability of AI models, allowing for more accurate results and better generalising to new datasets. To further enhance personalised, predictive and explainable computational models, new ML methods, such as brain-inspired computation [101], are worth exploring, along with traditional methods. An example of using a brain-inspired approach on longitudinal sMRI data is presented in [102]. In [103], fMRI and DTI neuroimaging data are integrated with a spiking neural network model based on the NeuCube [104,105] architecture for a better personalised and explainable prediction of response to treatment of Schizophrenia patients.
The ideal harmonisation method that can be applied in a clinical setting should be capable of removing variable data caused by using multiple scanners without requiring the testing data to be drawn from the same group as the training data. This, however, remains an ongoing challenge.
4.7. Recommendation for Future Studies
To harmonise several MRI datasets, the choice of ML methods should depend on the specific research goals and constraints of the datasets. Therefore, we have the following suggestions for researchers:
- Understanding the purpose of harmonisation. Use explicit approaches for harmonising intensity values and image-derived metrics or correcting known sources, whereas using implicit approaches to improve the performance of a downstream task or to correct unknown sources of variability.
- Consider the nature and properties of the dataset, including the size, dimensionality, and variability in the data. Generally, the more extensive and diverse the dataset, the more complex the ML model can be. For example, when using a travelling-subject dataset for training, researchers may consider using simpler ML models to reduce the risks of overfitting.
- When defining the target and reference domains, ensure that there is enough training data in each domain so that the model can learn the relevant information.
- Conduct experiments using different ML approaches, varying feature extraction techniques, and adjusting hyperparameters. Evaluate the performance using the consistent metrics and interpret the results of image analysis carefully.
- Consider the trade-off between accuracy and interpretability when choosing an ML method. ML methods, such as GANs, are more complex, so although they may produce highly accurate results, they can be difficult to interpret. In contrast, other methods, such as regression models, may be more interpretable despite giving less accurate results.
In general, we suggest using ML-based harmonisation methods to help improve the performance of the downstream task but to be careful when using ML-harmonised data for direct interpretation. Future studies should focus on developing specific evaluation and IQA methods for ML-harmonised data. In addition, this review focuses on brain MRI, but we believe that the methodologies can be generalised to MRI data of other body parts, such as lungs and breasts [106,107], in the field of harmonisation.
4.8. Limitation of Systematic Review
This systematic review has several limitations. First, this review did not evaluate the performance of ML models or compare ML models with other models due to a lack of consistent evaluation metrics. This limitation may hopefully be alleviated by summarising the findings in the comparative experiments conducted in all included studies, as well as the significant usage of the ML models and a broad range of relevant characteristics, such as the evaluation methods. We recommend that future researchers define consistent evaluation methods and metrics for MRI data harmonisation to facilitate comparative analysis for different ML-based applications. Additionally, it is possible that a study may have been overlooked, such as for non-English articles, unpublished or internal studies, or a study that was published after the search procedure was conducted.
5. Conclusions
The rapid development of ML in the current big data era necessitates the use of large-scale datasets. It is essential to apply harmonisation when a dataset is heterogeneous with respect to scanner, site, and acquisition parameters. Data harmonisation helps minimise the non-biological variations within a dataset. It can be applied as an essential pre-processing step to minimise scanner-related errors and improve the medical image quality, and potentially improve the performance and generalisability of downstream ML-based prediction models. Harmonisation is therefore important in the medical field, where accurate and consistent diagnoses can have a significant impact on patient outcomes,
This paper performed a systematic review of studies that used ML on MRI data harmonisation for inter-scanner variability removal. We identified and summarised different aspects found in the harmonisation literature, including data sources, modality, features, and ML methods. Our main contribution lies in identifying the current gaps and limitations in the literature and providing recommendations for future research directions. Specifically, we argue in the paper that:
- -
- There is a large amount of diverse imaging data (sMRI, dMRI, fMRI, DTI, longitudinal, static etc.) related to the same problem, but collected at different sites under different conditions, that need to be harmonised.
- -
- New methods for neuroimage data harmonisation are needed for better results.
- -
- Following harmonisation, methods for the integration of the multimodal harmonised data are needed for the development of better (1) personalised, (2) predictive and (3) explainable computational models.
- -
- The use of harmonisation as a strategy for improving downstream tasks is recommended; however, consistent evaluation methods are needed.
A growing interest in harmonisation will lead to adopting more advanced ML techniques when developing harmonisation methods. There should be a particular focus on establishing a shared space and creating a universal model for harmonising MRI data, regardless of the scanner, site characteristics, and type of neuroimaging data. The research space should also include some other data modalities.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bioengineering10040397/s1, Table S1: Papers used explicit approach; Table S2: Papers used implicit approach; Table S3: Quality assessment of all included articles; Sub-Table for S3 (S3-1): Quality assessment of all articles that used explicit approaches; Sub-Table for S3 (S3-2): Quality assessment of all articles that used implicit approaches. Reference [108] is cited in the Supplementary Materials.
Author Contributions
Conceptualisation, fomal analysis, methodology, G.W. and A.W.; validation, investigation, G.W., V.S. and A.W.; visualisation, data curation, writing—original draft preparation, G.W.; writing—review and editing, G.W., A.W., V.S., S.J.H., J.F., M.Q. and N.K.; project administration, A.W., M.Q., V.S. and S.J.H.; supervision, A.W., M.Q., V.S. and S.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Ministry of Business, Innovation and Employment (MBIE) of New Zealand [grant number PROP70240-CNZSDS-UOA] and the Health Research Council of New Zealand [grant number 21/144].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We extend our gratitude to the MBIE Catalyst: Strategic Fund NZ-Singapore Data Science Research Programme UOAX2001 for the financial support, which enabled the contributions of Miao Qiao, Vickie Shim, Samantha Holdsworth, Alan Wang, and Grace Wen to this research project. We would also like to thank the University of Auckland and the Auckland Bioengineering Institute for their generous support.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationship that could have influenced the work reported in this paper.
References
- Oba, H.; Yagishita, A.; Terada, H.; Barkovich, A.J.; Kutomi, K.; Yamauchi, T.; Furui, S.; Shimizu, T.; Uchigata, M.; Matsumura, K.; et al. New and reliable MRI diagnosis for progressive supranuclear palsy. Neurology 2005, 64, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Traboulsee, A.L.; Li, D.K.B. The role of MRI in the diagnosis of multiple sclerosis. Adv. Neurol. 2006, 98, 125–146. [Google Scholar] [PubMed]
- Yu-Feng, Z.; Yong, H.; Chao-Zhe, Z.; Qing-Jiu, C.; Man-Qiu, S.; Meng, L.; Li-Xia, T.; Tian-Zi, J. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007, 29, 83–91. [Google Scholar] [CrossRef]
- Swanton, J.K.; Rovira, A.; Tintore, M.; Altmann, D.R.; Barkhof, F.; Filippi, M.; Huerga, E.; Miszkiel, K.A.; Plant, G.T.; Polman, C.; et al. MRI criteria for multiple sclerosis in patients presenting with clinically isolated syndromes: A multicentre retrospective study. Lancet Neurol. 2007, 6, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.A.; Zielinski, B.A.; Fletcher, P.T.; Alexander, A.L.; Lange, N.; Bigler, E.D.; Lainhart, J.E.; Anderson, J.S. Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 2013, 7, 599. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, B. Rapid gradient-echo imaging. J. Magn. Reson. Imaging 2012, 36, 1300–1313. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.; Sakaie, K.; Lee, J.-C.; Debbins, J.; Liu, Y.; Arnold, D.; Melhem, E.; Smith, C.; Philips, M.; Lowe, M.; et al. A Validation Study of Multicenter Diffusion Tensor Imaging: Reliability of Fractional Anisotropy and Diffusivity Values. Am. J. Neuroradiol. 2011, 33, 695–700. [Google Scholar] [CrossRef]
- Seo, Y.; Wang, Z.J.; Morriss, M.C.; Rollins, N.K. Minimum SNR and acquisition for bias-free estimation of fractional anisotropy in diffusion tensor imaging—A comparison of two analytical techniques and field strengths. Magn. Reson. Imaging 2012, 30, 1123–1133. [Google Scholar] [CrossRef]
- Milidonis, X.; Lennen, R.J.; Jansen, M.A.; Mueller, S.; Boehm-Sturm, P.; Holmes, W.M.; Sena, E.S.; Macleod, M.R.; Marshall, I. Multicenter Evaluation of Geometric Accuracy of MRI Protocols Used in Experimental Stroke. PLoS ONE 2016, 11, e0162545. [Google Scholar] [CrossRef]
- Palacios, E.; Martin, A.; Boss, M.; Ezekiel, F.; Chang, Y.; Yuh, E.; Vassar, M.; Schnyer, D.; MacDonald, C.; Crawford, K.; et al. Toward Precision and Reproducibility of Diffusion Tensor Imaging: A Multicenter Diffusion Phantom and Traveling Volunteer Study. Am. J. Neuroradiol. 2016, 38, 537–545. [Google Scholar] [CrossRef]
- Schwartz, D.L.; Tagge, I.; Powers, K.; Ahn, S.; Bakshi, R.; Calabresi, P.A.; Constable, R.T.; Grinstead, J.; Henry, R.G.; Nair, G.; et al. Multisite reliability and repeatability of an advanced brain MRI protocol. J. Magn. Reson. Imaging 2019, 50, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Mårtensson, G.; Ferreira, D.; Granberg, T.; Cavallin, L.; Oppedal, K.; Padovani, A.; Rektorova, I.; Bonanni, L.; Pardini, M.; Kramberger, M.G.; et al. The reliability of a deep learning model in clinical out-of-distribution MRI data: A multicohort study. Med. Image Anal. 2020, 66, 101714. [Google Scholar] [CrossRef] [PubMed]
- Knoll, F.; Hammernik, K.; Kobler, E.; Pock, T.; Recht, M.P.; Sodickson, D. Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn. Reson. Med. 2019, 81, 116–128. [Google Scholar] [CrossRef]
- Clarke, W.; Mougin, O.; Driver, I.D.; Rua, C.; Morgan, A.; Asghar, M.; Clare, S.; Francis, S.; Wise, R.G.; Rodgers, C.T.; et al. Multi-site harmonization of 7 tesla MRI neuroimaging protocols. Neuroimage 2019, 206, 116335. [Google Scholar] [CrossRef] [PubMed]
- Jovicich, J.; Marizzoni, M.; Bosch, B.; Bartrés-Faz, D.; Arnold, J.; Benninghoff, J.; Wiltfang, J.; Roccatagliata, L.; Picco, A.; Nobili, F.; et al. Multisite longitudinal reliability of tract-based spatial statistics in diffusion tensor imaging of healthy elderly subjects. Neuroimage 2014, 101, 390–403. [Google Scholar] [CrossRef]
- Shinohara, R.; Oh, J.; Nair, G.; Calabresi, P.; Davatzikos, C.; Doshi, J.; Henry, R.; Kim, G.; Linn, K.; Papinutto, N.; et al. Volumetric Analysis from a Harmonized Multisite Brain MRI Study of a Single Subject with Multiple Sclerosis. Am. J. Neuroradiol. 2017, 38, 1501–1509. [Google Scholar] [CrossRef]
- Biberacher, V.; Schmidt, P.; Keshavan, A.; Boucard, C.C.; Righart, R.; Sämann, P.; Preibisch, C.; Fröbel, D.; Aly, L.; Hemmer, B.; et al. Intra-and interscanner variability of magnetic resonance imaging based volumetry in multiple sclerosis. Neuroimage 2016, 142, 188–197. [Google Scholar] [CrossRef]
- Pinto, M.S.; Paolella, R.; Billiet, T.; Van Dyck, P.; Guns, P.-J.; Jeurissen, B.; Ribbens, A.; Dekker, A.J.D.; Sijbers, J. Harmonization of Brain Diffusion MRI: Concepts and Methods. Front. Neurosci. 2020, 14, 396. [Google Scholar] [CrossRef]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef]
- Stamoulou, E.; Spanakis, C.; Manikis, G.C.; Karanasiou, G.; Grigoriadis, G.; Foukakis, T.; Tsiknakis, M.; Fotiadis, D.I.; Marias, K. Harmonization Strategies in Multicenter MRI-Based Radiomics. J. Imaging 2022, 8, 303. [Google Scholar] [CrossRef]
- Kmet, L.M.; Cook, L.S.; Lee, R.C. Standard Quality Assessment Criteria for Evaluating Primary Research Papers from a Variety of Fields; Alberta Heritage Foundation for Medical Research: Edmonton, AB, Canada, 2004. [Google Scholar] [CrossRef]
- Wen, J.; Li, S.; Lin, Z.; Hu, Y.; Huang, C. Systematic literature review of machine learning based software development effort estimation models. Inf. Softw. Technol. 2012, 54, 41–59. [Google Scholar] [CrossRef]
- Ho, T.K. Random Decision Forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995. [Google Scholar] [CrossRef]
- Fortin, J.-P.; Cullen, N.; Sheline, Y.I.; Taylor, W.D.; Aselcioglu, T.; Cook, P.A.; Adams, P.; Cooper, C.; Fava, M.; McGrath, P.J.; et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 2018, 167, pp. 104–120. [Google Scholar] [CrossRef]
- Garcia-Dias, R.; Scarpazza, C.; Baecker, L.; Vieira, S.; Pinaya, W.H.; Corvin, A.; Redolfi, A.; Nelson, B.; Crespo-Facorro, B.; McDonald, C.; et al. Neuroharmony: A new tool for harmonizing volumetric MRI data from unseen scanners. Neuroimage 2020, 220, 117127. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, L.T.; Zhang, Q.; Armstrong, D.; Deen, M.J. Convolutional neural networks for medical image analysis: State-of-the-art, comparisons, improvement and perspectives. Neurocomputing 2021, 444, 92–110. [Google Scholar] [CrossRef]
- Dewey, B.E.; Zhao, C.; Reinhold, J.C.; Carass, A.; Fitzgerald, K.C.; Sotirchos, E.S.; Saidha, S.; Oh, J.; Pham, D.L.; Calabresi, P.A.; et al. DeepHarmony: A deep learning approach to contrast harmonization across scanner changes. Magn. Reson. Imaging 2019, 64, 160–170. [Google Scholar] [CrossRef]
- Tong, Q.; Gong, T.; He, H.; Wang, Z.; Yu, W.; Zhang, J.; Zhai, L.; Cui, H.; Meng, X.; Tax, C.W.; et al. A deep learning–based method for improving reliability of multicenter diffusion kurtosis imaging with varied acquisition protocols. Magn. Reson. Imaging 2020, 73, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Koppers, S.; Bloy, L.; Berman, J.I.; Tax, C.M.W.; Edgar, J.C.; Merhof, D. Spherical Harmonic Residual Network for Diffusion Signal Harmonization. In Proceedings of the International MICCAI Workshop, Granada, Spain, 20 September 2018; pp. 173–182. [Google Scholar] [CrossRef]
- Kreutz-Delgado, K.; Murray, J.F.; Rao, B.D.; Engan, K.; Lee, T.-W.; Sejnowski, T.J. Dictionary Learning Algorithms for Sparse Representation. Neural Comput. 2003, 15, 349–396. [Google Scholar] [CrossRef]
- St-Jean, S.; Viergever, M.A.; Leemans, A. Harmonization of diffusion MRI data sets with adaptive dictionary learning. Hum. Brain Mapp. 2020, 41, 4478–4499. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2022, 3, 59–78. [Google Scholar] [CrossRef]
- Robinson, R.; Dou, Q.; de Castro, D.C.; Kamnitsas, K.; de Groot, M.; Summers, R.M.; Rueckert, D.; Glocker, B. Image-Level Harmonization of Multi-site Data Using Image-and-Spatial Transformer Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 710–719. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2014, 63, 139–144. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Y.; Wang, Y.; Shi, Z.; Yu, J. A Universal Intensity Standardization Method Based on a Many-to-One Weak-Paired Cycle Generative Adversarial Network for Magnetic Resonance Images. IEEE Trans. Med. Imaging 2019, 38, 2059–2069. [Google Scholar] [CrossRef]
- Zhao, F.; Wu, Z.; Wang, L.; Lin, W.; Xia, S.; Shen, D.; Li, G.; The UNC/UMN Baby Connectome Project Consortium. Harmonization of Infant Cortical Thickness Using Surface-to-Surface Cycle-Consistent Adversarial Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019 22nd International Conference, Shenzhen, China, 13–17 October 2019; Volume 11767, pp. 475–483. [Google Scholar] [CrossRef]
- Ren, M.; Dey, N.; Fishbaugh, J.; Gerig, G. Segmentation-Renormalized Deep Feature Modulation for Unpaired Image Harmonization. IEEE Trans. Med. Imaging 2021, 40, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Maiti, P.; Thomopoulos, S.; Zhu, A.; Chai, Y.; Kim, H.; Jahanshad, N. Style Transfer Using Generative Adversarial Networks for Multi-site MRI Harmonization. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021 24th International Conference, Strasbourg, France, 27 September–1 October 2021; pp. 313–322. [Google Scholar] [CrossRef]
- Weninger, L.; Ahmad, M.; Merhof, D. From Supervised to Unsupervised Harmonization of Diffusion Mri Acquisitions. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Arai, H.; Onga, Y.; Ikuta, K.; Chayama, Y.; Iyatomi, H.; Oishi, K. Disease-Oriented Image Embedding With Pseudo-Scanner Standardization for Content-Based Image Retrieval on 3D Brain MRI. IEEE Access 2021, 9, 165326–165340. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, Y.; Li, J.; Xue, X.; Liu, S.; Wang, M.; Gao, X.; Wang, Q.; Yang, J.; Li, X. Inter-site harmonization based on dual generative adversarial networks for diffusion tensor imaging: Application to neonatal white matter development. Biomed. Eng. Online 2020, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. In Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014, Banff, AB, Canada, 14–16 April 2014. [Google Scholar] [CrossRef]
- Dewey, B.E.; Zuo, L.; Carass, A.; He, Y.; Liu, Y.; Mowry, E.M.; Newsome, S.; Oh, J.; Calabresi, P.A.; Prince, J.L. A Disentangled Latent Space for Cross-Site MRI Harmonization. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 720–729. [Google Scholar] [CrossRef]
- Zuo, L.; Dewey, B.E.; Liu, Y.; He, Y.; Newsome, S.D.; Mowry, E.M.; Resnick, S.M.; Prince, J.L.; Carass, A. Unsupervised MR harmonization by learning disentangled representations using information bottleneck theory. Neuroimage 2021, 243, 118569. [Google Scholar] [CrossRef] [PubMed]
- Torbati, M.E.; Tudorascu, D.L.; Minhas, D.S.; Maillard, P.; DeCarli, C.S.; Hwang, S.J. Multi-scanner Harmonization of Paired Neuroimaging Data via Structure Preserving Embedding Learning. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW), Montreal, BC, Canada, 11–17 October 2021; pp. 3277–3286. [Google Scholar] [CrossRef]
- Tian, D.; Zeng, Z.; Sun, X.; Tong, Q.; Li, H.; He, H.; Gao, J.-H.; He, Y.; Xia, M. A deep learning-based multisite neuroimage harmonization framework established with a traveling-subject dataset. Neuroimage 2022, 257, 119297. [Google Scholar] [CrossRef] [PubMed]
- Fatania, K.; Clark, A.; Frood, R.; Scarsbrook, A.; Al-Qaisieh, B.; Currie, S.; Nix, M. Harmonisation of scanner-dependent contrast variations in magnetic resonance imaging for radiation oncology, using style-blind auto-encoders. Phys. Imaging Radiat. Oncol. 2022, 22, 115–122. [Google Scholar] [CrossRef]
- Moyer, D.; Steeg, G.V.; Tax, C.M.W.; Thompson, P.M. Scanner invariant representations for diffusion MRI harmonization. Magn. Reson. Med. 2020, 84, 2174–2189. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Dewey, B.E.; Carass, A.; Liu, Y.; He, Y.; Calabresi, P.A.; Prince, J.L. Information-Based Disentangled Representation Learning for Unsupervised MR Harmonization. In Proceedings of the Information Processing in Medical Imaging 27th International Conference, IPMI 2021, Virtual, 28–30 June 2021; pp. 346–359. [Google Scholar] [CrossRef]
- Tzeng, E.; Hoffman, J.; Saenko, K.; Darrell, T. Adversarial Discriminative Domain Adaptation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2962–2971. [Google Scholar]
- Guan, H.; Liu, Y.; Yang, E.; Yap, P.-T.; Shen, D.; Liu, M. Multi-site MRI harmonization via attention-guided deep domain adaptation for brain disorder identification. Med. Image Anal. 2021, 71, 102076. [Google Scholar] [CrossRef]
- Dinsdale, N.K.; Jenkinson, M.; Namburete, A.I. Deep learning-based unlearning of dataset bias for MRI harmonisation and confound removal. Neuroimage 2020, 228, 117689. [Google Scholar] [CrossRef] [PubMed]
- Orbes-Arteaga, M.; Varsavsky, T.; Sudre, C.H.; Eaton-Rosen, Z.; Haddow, L.J.; Sørensen, L.; Nielsen, M.; Pai, A.; Ourselin, S.; Modat, M.; et al. Multi-domain Adaptation in Brain MRI Through Paired Consistency and Adversarial Learning. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Springer: Cham, Switzerland, 2019; pp. 54–62. [Google Scholar] [CrossRef]
- Ackaouy, A.; Courty, N.; Vallée, E.; Commowick, O.; Barillot, C.; Galassi, F. Unsupervised Domain Adaptation With Optimal Transport in Multi-Site Segmentation of Multiple Sclerosis Lesions From MRI Data. Front. Comput. Neurosci. 2020, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, M.; Pan, Y.; Shen, D. Unsupervised Conditional Consensus Adversarial Network for Brain Disease Identification with Structural MRI. In Proceedings of the 10th International Workshop, MLMI 2019, Shenzhen, China, 13 October 2019; pp. 391–399. [Google Scholar] [CrossRef]
- Delisle, P.-L.; Anctil-Robitaille, B.; Desrosiers, C.; Lombaert, H. Realistic image normalization for multi-domain segmentation. Med. Image Anal. 2021, 74, 102191. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-L.; Hsieh, W.-T.; Yang, H.-C.; Lee, C.-C. Conditional Domain Adversarial Transfer for Robust Cross-Site ADHD Classification Using Functional MRI. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1190–1194. [Google Scholar] [CrossRef]
- Delisle, P.-L.; Anctil-Robitaille, B.; Desrosiers, C.; Lombaert, H. Adversarial Normalization for Multi Domain Image Segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 849–853. [Google Scholar] [CrossRef]
- Murez, Z.; Kolouri, S.; Kriegman, D.; Ramamoorthi, R.; Kim, K. Image to Image Translation for Domain Adaptation. arXiv 2017. [Google Scholar] [CrossRef]
- Wang, N.; Yao, D.; Ma, L.; Liu, M. Multi-site clustering and nested feature extraction for identifying autism spectrum disorder with resting-state fMRI. Med. Image Anal. 2022, 75, 102279. [Google Scholar] [CrossRef] [PubMed]
- Yousefnezhad, M.; Selvitella, A.; Zhang, D.; Greenshaw, A.J.; Greiner, R. Shared space transfer learning for analyzing multi-site fMRI data. In Proceedings of the 34th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 6–12 December 2020; pp. 15990–16000. [Google Scholar]
- Wang, M.; Zhang, D.; Huang, J.; Yap, P.-T.; Shen, D.; Liu, M. Identifying Autism Spectrum Disorder With Multi-Site fMRI via Low-Rank Domain Adaptation. IEEE Trans. Med. Imaging 2019, 39, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, J.; Hong, Q.-Q.; Teku, R.; Wang, S.-H.; Zhang, Y.-D. Transfer learning for medical images analyses: A survey. Neurocomputing 2022, 489, 230–254. [Google Scholar] [CrossRef]
- Monte-Rubio, G.C.; Segura, B.; Strafella, A.P.; van Eimeren, T.; Ibarretxe-Bilbao, N.; Diez-Cirarda, M.; Eggers, C.; Lucas-Jiménez, O.; Ojeda, N.; Peña, J.; et al. Parameters from site classification to harmonize MRI clinical studies: Application to a multi-site Parkinson’s disease dataset. Hum. Brain Mapp. 2022, 43, 3130–3142. [Google Scholar] [CrossRef]
- Chen, C.-L.; Hsu, Y.-C.; Yang, L.-Y.; Tung, Y.-H.; Luo, W.-B.; Liu, C.-M.; Hwang, T.-J.; Hwu, H.-G.; Tseng, W.-Y.I. Generalization of diffusion magnetic resonance imaging–based brain age prediction model through transfer learning. Neuroimage 2020, 217, 116831. [Google Scholar] [CrossRef]
- Wachinger, C.; Reuter, M. Domain adaptation for Alzheimer’s disease diagnostics. Neuroimage 2016, 139, 470–479. [Google Scholar] [CrossRef]
- Ghafoorian, M.; Mehrtash, A.; Kapur, T.; Karssemeijer, N.; Marchiori, E.; Pesteie, M.; Guttmann, C.R.; de Leeuw, F.-E.; Tempany, C.M.; Van Ginneken, B. Transfer learning for domain adaptation in mri: Application in brain lesion segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; pp. 516–524. [Google Scholar]
- Balboni, E.; Nocetti, L.; Carbone, C.; Dinsdale, N.; Genovese, M.; Guidi, G.; Malagoli, M.; Chiari, A.; Namburete, A.I.L.; Jenkinson, M.; et al. The impact of transfer learning on 3D deep learning convolutional neural network segmentation of the hippocampus in mild cognitive impairment and Alzheimer disease subjects. Hum. Brain Mapp. 2022, 43, 3427–3438. [Google Scholar] [CrossRef]
- Shi, C.; Xin, X.; Zhang, J. Domain Adaptation Using a Three-Way Decision Improves the Identification of Autism Patients from Multisite fMRI Data. Brain Sci. 2021, 11, 603. [Google Scholar] [CrossRef]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; de Bruijne, M. Transfer Learning Improves Supervised Image Segmentation Across Imaging Protocols. IEEE Trans. Med. Imaging 2014, 34, 1018–1030. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chaudhari, P.; Davatzikos, C. Embracing the disharmony in medical imaging: A Simple and effective framework for domain adaptation. Med. Image Anal. 2021, 76, 102309. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-L.; Xin, X.-W.; Zhang, J.-C. Domain adaptation based on rough adjoint inconsistency and optimal transport for identifying autistic patients. Comput. Methods Programs Biomed. 2022, 215, 106615. [Google Scholar] [CrossRef] [PubMed]
- Van Opbroek, A.; Vernooij, M.W.; Ikram, M.A.; de Bruijne, M. Weighting training images by maximizing distribution similarity for supervised segmentation across scanners. Med. Image Anal. 2015, 24, 245–254. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q. An overview of multi-task learning. Natl. Sci. Rev. 2017, 5, 30–43. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, T.; Zanetti, M.V.; Shen, H.; Satterthwaite, T.D.; Wolf, D.H.; Gur, R.E.; Fan, Y.; Hu, D.; Busatto, G.F.; et al. Classification of multi-site MR images in the presence of heterogeneity using multi-task learning. NeuroImage Clin. 2018, 19, 476–486. [Google Scholar] [CrossRef]
- Ning, L.; Bonet-Carne, E.; Grussu, F.; Sepehrband, F.; Kaden, E.; Veraart, J.; Blumberg, S.B.; Khoo, C.S.; Palombo, M.; Coll-Font, J.; et al. Muti-shell Diffusion MRI Harmonisation and Enhancement Challenge (MUSHAC): Progress and Results. In Proceedings of the Computational Diffusion MRI International MICCAI Workshop, Granada, Spain, 20 September 2018; pp. 217–224. [Google Scholar] [CrossRef]
- Van Essen, D.C.; Ugurbil, K.; Auerbach, E.; Barch, D.; Behrens, T.E.J.; Bucholz, R.; Chang, A.; Chen, L.; Corbetta, M.; Curtiss, S.W.; et al. The Human Connectome Project: A data acquisition perspective. NeuroImage 2012, 62, 2222–2231. [Google Scholar] [CrossRef]
- Tanaka, S.C.; Yamashita, A.; Yahata, N.; Itahashi, T.; Lisi, G.; Yamada, T.; Ichikawa, N.; Takamura, M.; Yoshihara, Y.; Kunimatsu, A.; et al. A multi-site, multi-disorder resting-state magnetic resonance image database. Sci. Data 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Marcus, D.S.; Fotenos, A.F.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies: Longitudinal MRI Data in Nondemented and Demented Older Adults. J. Cogn. Neurosci. 2010, 22, 2677–2684. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Sundar, L.K.S.; Muzik, O.; Buvat, I.; Bidaut, L.; Beyer, T. Potentials and caveats of AI in hybrid imaging. Methods 2020, 188, 4–19. [Google Scholar] [CrossRef]
- Sundar, L.K.S.; Iommi, D.; Muzik, O.; Chalampalakis, Z.; Klebermass, E.-M.; Hienert, M.; Rischka, L.; Lanzenberger, R.; Hahn, A.; Pataraia, E.; et al. Conditional Generative Adversarial Networks Aided Motion Correction of Dynamic 18F-FDG PET Brain Studies. J. Nucl. Med. 2020, 62, 871–879. [Google Scholar] [CrossRef]
- Mathieu, E.; Rainforth, T.; Siddharth, N.; Teh, Y.W. Disentangling Disentanglement in Variational Autoencoders. ICML 2019, 97, 4402–4412. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Shin, J.; Zhang, L.; Gurudu, S.; Gotway, M.; Liang, J. Fine-tuning Convolutional Neural Networks for Biomedical Image Analysis: Actively and Incrementally. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017), Las Vegas, NV, USA, 27–30 June 2017; pp. 4761–4772. [Google Scholar] [CrossRef]
- Maikusa, N.; Zhu, Y.; Uematsu, A.; Yamashita, A.; Saotome, K.; Okada, N.; Kasai, K.; Okanoya, K.; Yamashita, O.; Tanaka, S.C.; et al. Comparison of traveling-subject and ComBat harmonization methods for assessing structural brain characteristics. Hum. Brain Mapp. 2021, 42, 5278–5287. [Google Scholar] [CrossRef] [PubMed]
- Bartzokis, G.; Mintz, J.; Marx, P.; Osborn, D.; Gutkind, D.; Chiang, F.; Phelan, C.; Marder, S.R. Reliability of in vivo volume measures of hippocampus and other brain structures using MRI. Magn. Reson. Imaging 1993, 11, 993–1006. [Google Scholar] [CrossRef]
- Morey, R.A.; Selgrade, E.S.; Ii, H.R.W.; Huettel, S.A.; Wang, L.; McCarthy, G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum. Brain Mapp. 2010, 31, 1751–1762. [Google Scholar] [CrossRef]
- Chen, X.; Chen, N.-X.; Shen, Y.-Q.; Li, H.-X.; Li, L.; Lu, B.; Zhu, Z.-C.; Fan, Z.; Yan, C.-G. The subsystem mechanism of default mode network underlying rumination: A reproducible neuroimaging study. Neuroimage 2020, 221, 117185. [Google Scholar] [CrossRef]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D Deep Learning on Medical Images: A Review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef]
- Kao, P.-Y.; Shailja, S.; Jiang, J.; Zhang, A.; Khan, A.; Chen, J.W.; Manjunath, B.S. Improving Patch-Based Convolutional Neural Networks for MRI Brain Tumor Segmentation by Leveraging Location Information. Front. Neurosci. 2020, 13, 1449. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Wu, T.-H.; Lai, S.-H. Multi-Scale Patch-Based Representation Learning for Image Anomaly Detection and Segmentation. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 3065–3073. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Huang, H.; Zhang, W.; Zhao, S.; Makkie, M.; Zhang, M.; Li, Q.; Liu, T. Four-Dimensional Modeling of fMRI Data via Spatio–Temporal Convolutional Neural Networks (ST-CNNs). IEEE Trans. Cogn. Dev. Syst. 2019, 12, 451–460. [Google Scholar] [CrossRef]
- Mittal, A.; Moorthy, A.K.; Bovik, A.C. Blind/Referenceless Image Spatial Quality Evaluator. In Proceedings of the 2011 Conference Record of the Forty Fifth Asilomar Conference on Signals, Systems and Computers (ASILOMAR), Pacific Grove, CA, USA, 6–9 November 2011; pp. 723–727. [Google Scholar] [CrossRef]
- Obuchowicz, R.; Oszust, M.; Bielecka, M.; Bielecki, A.; Piórkowski, A. Magnetic Resonance Image Quality Assessment by Using Non-Maximum Suppression and Entropy Analysis. Entropy 2020, 22, 220. [Google Scholar] [CrossRef] [PubMed]
- Küstner, T.; Gatidis, S.; Liebgott, A.; Schwartz, M.; Mauch, L.; Martirosian, P.; Schmidt, H.; Schwenzer, N.; Nikolaou, K.; Bamberg, F.; et al. A machine-learning framework for automatic reference-free quality assessment in MRI. Magn. Reson. Imaging 2018, 53, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fu, J.; Li, X.; Zhou, W.; Liu, S.; Zhang, X.; Wu, W.; Jia, C.; Liu, Y.; Chen, Z. RTN: Reinforced Transformer Network for Coronary CT Angiography Vessel-level Image Quality Assessment. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022 25th International Conference, Singapore, 18–22 September 2022; pp. 644–653. [Google Scholar] [CrossRef]
- Liu, S.; Thung, K.-H.; Lin, W.; Yap, P.-T.; Shen, D.; The UNC/UMN Baby Connectome Project Consortium. Multi-stage Image Quality Assessment of Diffusion MRI via Semi-supervised Nonlocal Residual Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019 22nd International Conference, Shenzhen, China, 13–17 October 2019; pp. 521–528. [Google Scholar] [CrossRef]
- Tax, C.M.; Grussu, F.; Kaden, E.; Ning, L.; Rudrapatna, U.; Evans, C.J.; St-Jean, S.; Leemans, A.; Koppers, S.; Merhof, D.; et al. Cross-scanner and cross-protocol diffusion MRI data harmonisation: A benchmark database and evaluation of algorithms. Neuroimage 2019, 195, 285–299. [Google Scholar] [CrossRef]
- Ning, L.; Bonet-Carne, E.; Grussu, F.; Sepehrband, F.; Kaden, E.; Veraart, J.; Blumberg, S.B.; Khoo, C.S.; Palombo, M.; Kokkinos, I.; et al. Cross-scanner and cross-protocol multi-shell diffusion MRI data harmonization: Algorithms and results. Neuroimage 2020, 221, 117128. [Google Scholar] [CrossRef]
- Kasabov, N.K. Time-Space, Spiking Neural Networks and Brain-Inspired Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Doborjeh, M.; Doborjeh, Z.; Merkin, A.; Bahrami, H.; Sumich, A.; Krishnamurthi, R.; Medvedev, O.N.; Crook-Rumsey, M.; Morgan, C.; Kirk, I.; et al. Personalised predictive modelling with brain-inspired spiking neural networks of longitudinal MRI neuroimaging data and the case study of dementia. Neural Netw. 2021, 144, 522–539. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, N.; McNabb, C.B.; Kasabov, N.; Russell, B.R. Integrating Space, Time, and Orientation in Spiking Neural Networks: A Case Study on Multimodal Brain Data Modeling. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5249–5263. [Google Scholar] [CrossRef]
- Kasabov, N.K. NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Netw. 2014, 52, 62–76. [Google Scholar] [CrossRef]
- Kasabov, N.K.; Doborjeh, M.G.; Doborjeh, Z.G. Mapping, Learning, Visualization, Classification, and Understanding of fMRI Data in the NeuCube Evolving Spatiotemporal Data Machine of Spiking Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2016, 28, 887–899. [Google Scholar] [CrossRef]
- Whitney, H.M.; Li, H.; Ji, Y.; Liu, P.; Giger, M.L. Harmonization of radiomic features of breast lesions across international DCE-MRI datasets. J. Med. Imaging 2020, 7, 012707. [Google Scholar] [CrossRef]
- Crombé, A.; Kind, M.; Fadli, D.; Le Loarer, F.; Italiano, A.; Buy, X.; Saut, O. Intensity harmonization techniques influence radiomics features and radiomics-based predictions in sarcoma patients. Sci. Rep. 2020, 10, 15496. [Google Scholar] [CrossRef]
- Mirzaalian, H.; Ning, L.; Savadjiev, P.; Pasternak, O.; Bouix, S.; Michailovich, O.; Karmacharya, S.; Grant, G.; Marx, C.E.; Morey, R.A.; et al. Multi-site harmonization of diffusion MRI data in a registration framework. Brain Imaging Behav. 2018, 12, 284–295. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).