Image Translation of Breast Ultrasound to Pseudo Anatomical Display by CycleGAN
Abstract
1. Introduction
2. Materials and Methods
2.1. Cycle Generative Adversarial Network (CycleGAN)
2.2. BUSI Dataset
2.3. Optic/Anatomic Dataset
2.4. Training Parameters
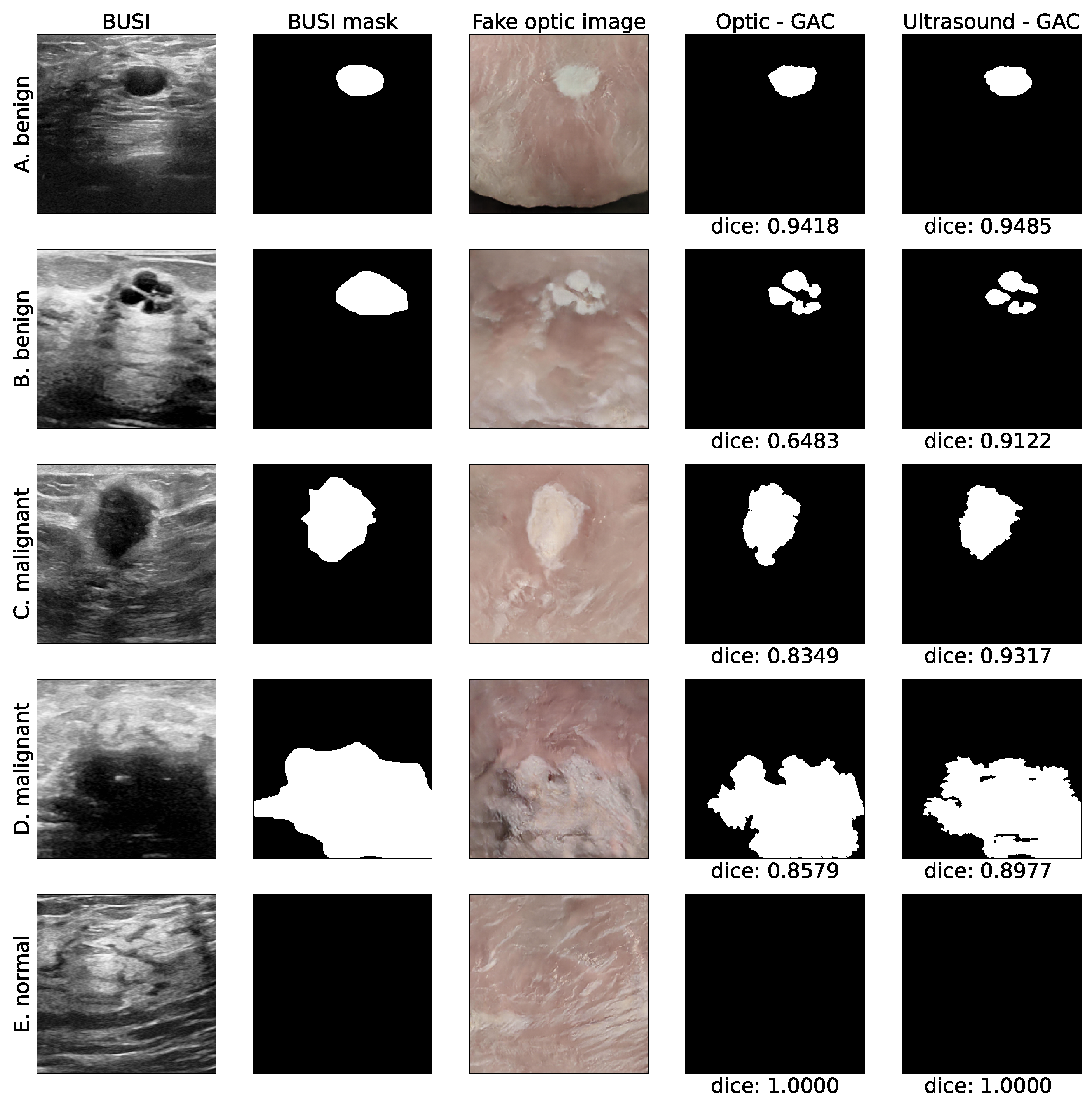
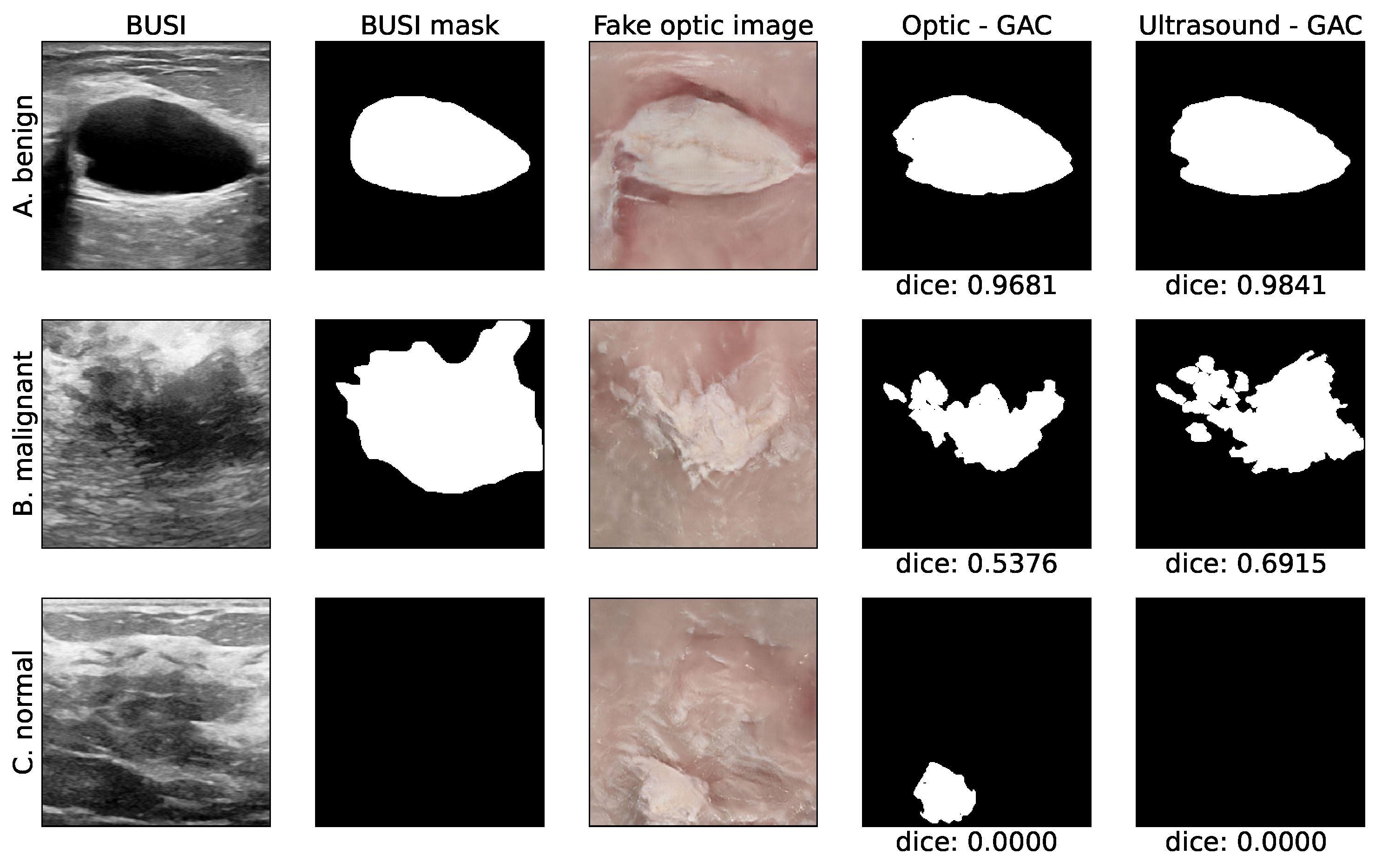
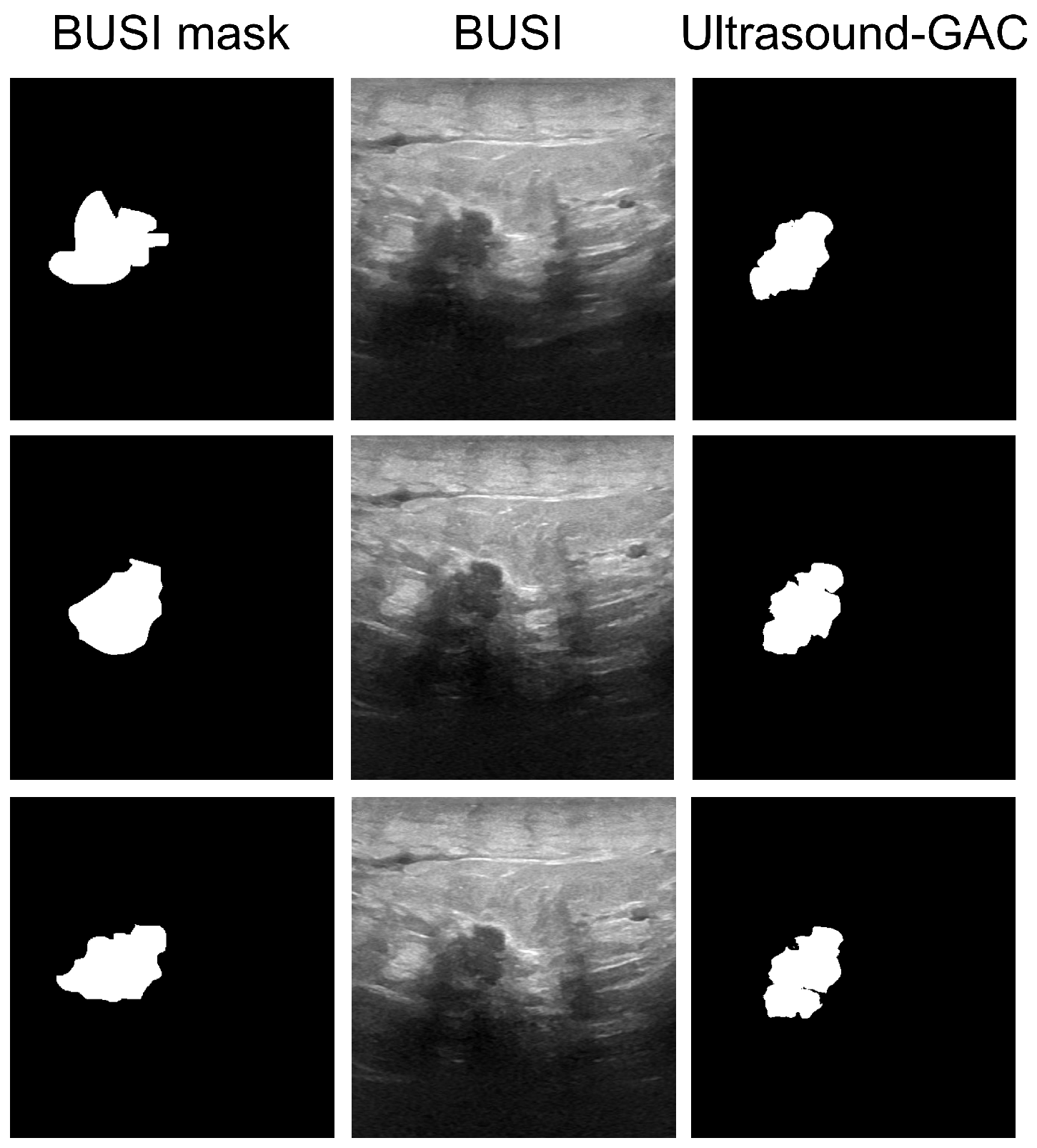
2.5. Automated Segmentation
2.6. Evaluation Protocol
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Gilbert, F.J.; Pinker-Domenig, K. Diagnosis and staging of breast cancer: When and how to use mammography, tomosynthesis, ultrasound, contrast-enhanced mammography, and magnetic resonance imaging. In Diseases of the Chest, Breast, Heart and Vessels 2019–2022: Diagnostic and Interventional Imaging; Springer: Cham, Switzerland, 2019; pp. 155–166. [Google Scholar]
- Na, S.P.; Houserkovaa, D. The role of various modalities in breast imaging. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2007, 151, 209–218. [Google Scholar]
- Smith, N.B.; Webb, A. Introduction to Medical Imaging: Physics, Engineering and Clinical Applications; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Azhari, H.; Kennedy, J.A.; Weiss, N.; Volokh, L. From Signals to Image; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Ngô, C.; Pollet, A.G.; Laperrelle, J.; Ackerman, G.; Gomme, S.; Thibault, F.; Fourchotte, V.; Salmon, R.J. Intraoperative ultrasound localization of nonpalpable breast cancers. Ann. Surg. Oncol. 2007, 14, 2485–2489. [Google Scholar] [CrossRef] [PubMed]
- Colakovic, N.; Zdravkovic, D.; Skuric, Z.; Mrda, D.; Gacic, J.; Ivanovic, N. Intraoperative ultrasound in breast cancer surgery—From localization of non-palpable tumors to objectively measurable excision. World J. Surg. Oncol. 2018, 16, 1–7. [Google Scholar] [CrossRef]
- Van Velthoven, V. Intraoperative Ultrasound Imaging: Comparison of pAthomorphological Findings in US versus CT, MRI and Intraoperative Findings. Acta Neurochir. Suppl. 2003, 85, 95–99. [Google Scholar]
- Balog, J.; Sasi-Szabó, L.; Kinross, J.; Lewis, M.R.; Muirhead, L.J.; Veselkov, K.; Mirnezami, R.; Dezső, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 2013, 5, 194ra93. [Google Scholar] [CrossRef]
- Haka, A.S.; Volynskaya, Z.; Gardecki, J.A.; Nazemi, J.; Shenk, R.; Wang, N.; Dasari, R.R.; Fitzmaurice, M.; Feld, M.S. Diagnosing breast cancer using Raman spectroscopy: Prospective analysis. J. Biomed. Opt. 2009, 14, 054023. [Google Scholar] [CrossRef]
- Dixon, J.M.; Renshaw, L.; Young, O.; Kulkarni, D.; Saleem, T.; Sarfaty, M.; Sreenivasan, R.; Kusnick, C.; Thomas, J.; Williams, L. Intra-operative assessment of excised breast tumour margins using ClearEdge imaging device. Eur. J. Surg. Oncol. (EJSO) 2016, 42, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.B.; Gao, S.; Zhu, N.; Habimana-Griffin, L.; Akers, W.J.; Liang, R.; Gruev, V.; Margenthaler, J.; Achilefu, S. Optical see-through cancer vision goggles enable direct patient visualization and real-time fluorescence-guided oncologic surgery. Ann. Surg. Oncol. 2017, 24, 1897–1903. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.L.; Gadd, M.A.; Lanahan, C.R.; Rai, U.; Tang, R.; Rice-Stitt, T.; Merrill, A.L.; Strasfeld, D.B.; Ferrer, J.M.; Brachtel, E.F.; et al. Real-time, intraoperative detection of residual breast cancer in lumpectomy cavity walls using a novel cathepsin-activated fluorescent imaging system. Breast Cancer Res. Treat. 2018, 171, 413–420. [Google Scholar] [CrossRef]
- Scimone, M.T.; Krishnamurthy, S.; Maguluri, G.; Preda, D.; Park, J.; Grimble, J.; Song, M.; Ban, K.; Iftimia, N. Assessment of breast cancer surgical margins with multimodal optical microscopy: A feasibility clinical study. PLoS ONE 2021, 16, e0245334. [Google Scholar] [CrossRef]
- Schwarz, J.; Schmidt, H. Technology for intraoperative margin assessment in breast cancer. Ann. Surg. Oncol. 2020, 27, 2278–2287. [Google Scholar] [CrossRef]
- Jafari, M.H.; Girgis, H.; Van Woudenberg, N.; Moulson, N.; Luong, C.; Fung, A.; Balthazaar, S.; Jue, J.; Tsang, M.; Nair, P.; et al. Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, N.; Zhao, Z.; Fan, X.; Chang, E.I.; Xu, Y. MRI cross-modality image-to-image translation. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Namburete, A.I.; Papageorghiou, A.T.; Noble, J.A. Self-Supervised Ultrasound to MRI Fetal Brain Image Synthesis. IEEE Trans. Med. Imaging 2020, 39, 4413–4424. [Google Scholar] [CrossRef] [PubMed]
- Kearney, V.; Ziemer, B.P.; Perry, A.; Wang, T.; Chan, J.W.; Ma, L.; Morin, O.; Yom, S.S.; Solberg, T.D. Attention-aware discrimination for MR-to-CT image translation using cycle-consistent generative adversarial networks. Radiology. Artif. Intell. 2020, 2, e190027. [Google Scholar] [CrossRef]
- Vedula, S.; Senouf, O.; Bronstein, A.M.; Michailovich, O.V.; Zibulevsky, M. Towards CT-quality ultrasound imaging using deep learning. arXiv 2017, arXiv:1710.06304. [Google Scholar]
- Pang, Y.; Lin, J.; Qin, T.; Chen, Z. Image-to-image translation: Methods and applications. IEEE Trans. Multimed. 2021, 24, 3859–3881. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Rezende, D.J.; Mohamed, S.; Wierstra, D. Stochastic backpropagation and approximate inference in deep generative models. In Proceedings of the 31st International Conference on International Conference on Machine Learning, Beijing, China, 21–26 June 2014; JMLR: Brookline, MA, USA, 2014; pp. 1278–1286. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Hiasa, Y.; Otake, Y.; Takao, M.; Matsuoka, T.; Takashima, K.; Carass, A.; Prince, J.L.; Sugano, N.; Sato, Y. Cross-modality image synthesis from unpaired data using CycleGAN. In Simulation and Synthesis in Medical Imaging: Third International Workshop, SASHIMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 31–41. [Google Scholar]
- Modanwal, G.; Vellal, A.; Buda, M.; Mazurowski, M.A. MRI image harmonization using cycle-consistent generative adversarial network. In Proceedings of the SPIE Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 15–20 February 2020; Volume 11314, p. 1131413. [Google Scholar]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef]
- Glassman, L.; Hazewinkel, M. The Radiology Assistant: MRI of the Breast. 2013. Available online: https://radiologyassistant.nl/breast/mri/mri-of-the-breast, (accessed on 5 October 2021).
- Samardar, P.; de Paredes, E.S.; Grimes, M.M.; Wilson, J.D. Focal asymmetric densities seen at mammography: US and pathologic correlation. Radiographics 2002, 22, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Franquet, T.; De Miguel, C.; Cozcolluela, R.; Donoso, L. Spiculated lesions of the breast: Mammographic-pathologic correlation. Radiographics 1993, 13, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.G.; Whitman, G.J.; Fornage, B.D.; Stelling, C.B.; Huynh, P.T.; Sahin, A.A. Tubular carcinoma of the breast: Mammographic and sonographic features. Am. J. Roentgenol. 2000, 174, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Neila, P.; Baumela, L.; Alvarez, L. A morphological approach to curvature-based evolution of curves and surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 36, 2–17. [Google Scholar] [CrossRef]
- Lee, G. Morphsnakes (Version 0.19.3). 2021. Available online: https://github.com/scikit-image/scikit-image/blob/v0.19.3/skimage/segmentation/morphsnakes.py, (accessed on 21 March 2023).
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]


| Pros | Cons |
|---|---|
| Radiation-free | Low SNR |
| Real time imaging | Operator dependent |
| Radiation-free | Low SNR |
| Real time imaging | Operator dependent |
| Enables arbitrary for cross-section imaging | Non intuitive black and white image |
| Tumor detection pre and intra-operative | |
| Margin assessment | |
| Cost effective |
| Tumor Type | Median BUSI | Median MorphGAC | Mean ± Std BUSI | Mean ± Std MorphGAC | |
|---|---|---|---|---|---|
| Dice | Benign | 0.85 | 0.91 | 0.67 ± 0.36 | 0.70 ± 0.38 |
| Malignant | 0.58 | 0.70 | 0.53 ± 0.30 | 0.60 ± 0.32 | |
| all | 0.77 | 0.83 | 0.62 ± 0.35 | 0.67 ± 0.36 | |
| Center error [%] | Benign | 0.56 | 0.58 | 5.09 ± 11.23 | 4.22 ± 10.78 |
| Malignant | 4.13 | 3.27 | 7.21 ± 10.29 | 7.21 ± 10.65 | |
| all | 1.17 | 0.73 | 5.76 ± 10.95 | 5.14 ± 10.79 | |
| Area index [%] | Benign | 0.74 | 0.40 | 2.84 ± 5.21 | 2.11 ± 5.08 |
| Malignant | 9.25 | 4.34 | 11.64 ± 8.79 | 6.12 ± 6.49 | |
| all | 2.31 | 0.71 | 5.56 ± 7.67 | 3.35 ± 5.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barkat, L.; Freiman, M.; Azhari, H. Image Translation of Breast Ultrasound to Pseudo Anatomical Display by CycleGAN. Bioengineering 2023, 10, 388. https://doi.org/10.3390/bioengineering10030388
Barkat L, Freiman M, Azhari H. Image Translation of Breast Ultrasound to Pseudo Anatomical Display by CycleGAN. Bioengineering. 2023; 10(3):388. https://doi.org/10.3390/bioengineering10030388
Chicago/Turabian StyleBarkat, Lilach, Moti Freiman, and Haim Azhari. 2023. "Image Translation of Breast Ultrasound to Pseudo Anatomical Display by CycleGAN" Bioengineering 10, no. 3: 388. https://doi.org/10.3390/bioengineering10030388
APA StyleBarkat, L., Freiman, M., & Azhari, H. (2023). Image Translation of Breast Ultrasound to Pseudo Anatomical Display by CycleGAN. Bioengineering, 10(3), 388. https://doi.org/10.3390/bioengineering10030388