Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explainability
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
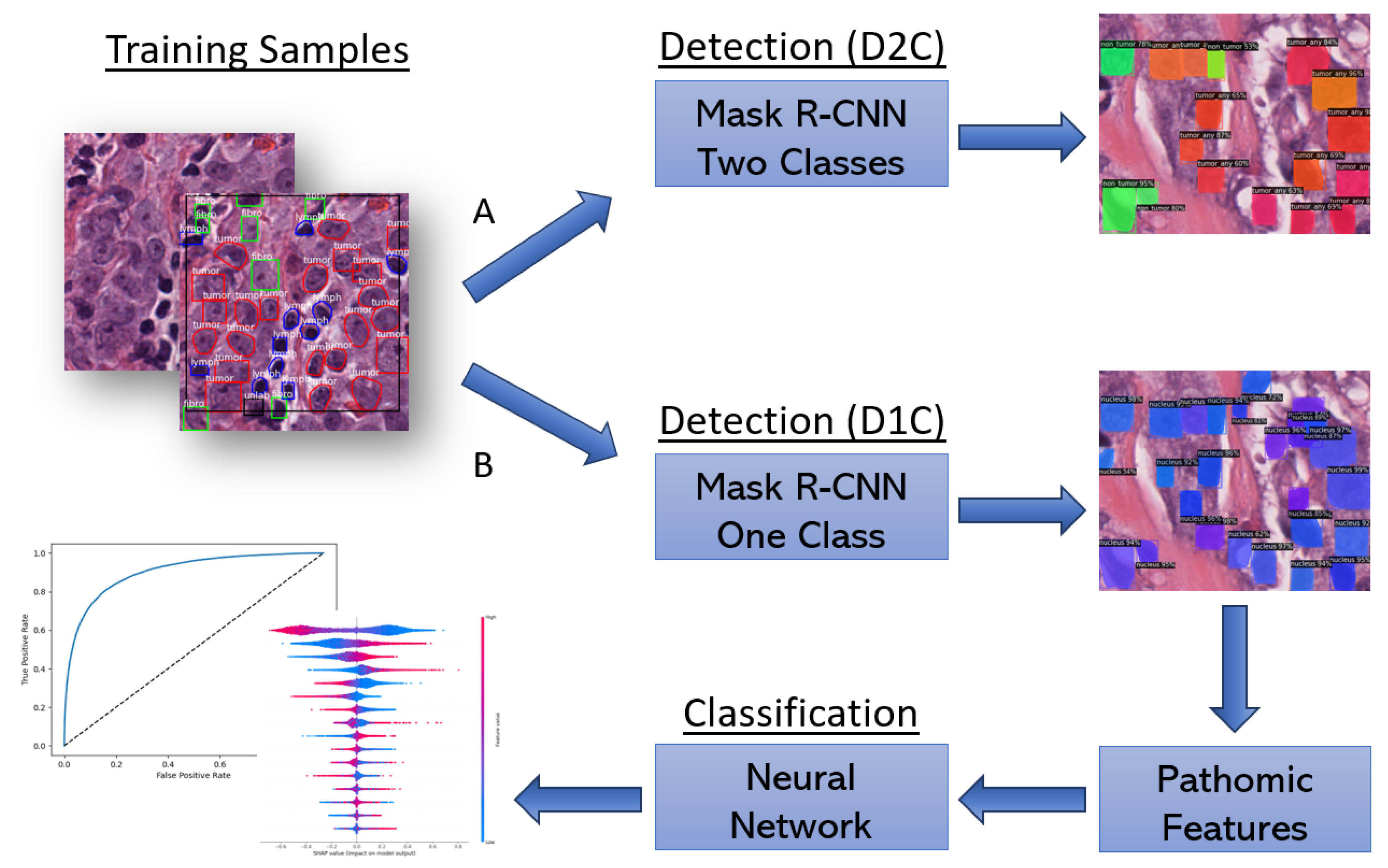
2.2. Proposed Workflow
2.2.1. End-to-End Approach
2.2.2. Two-Stage Approach
2.2.3. Evaluation Metrics
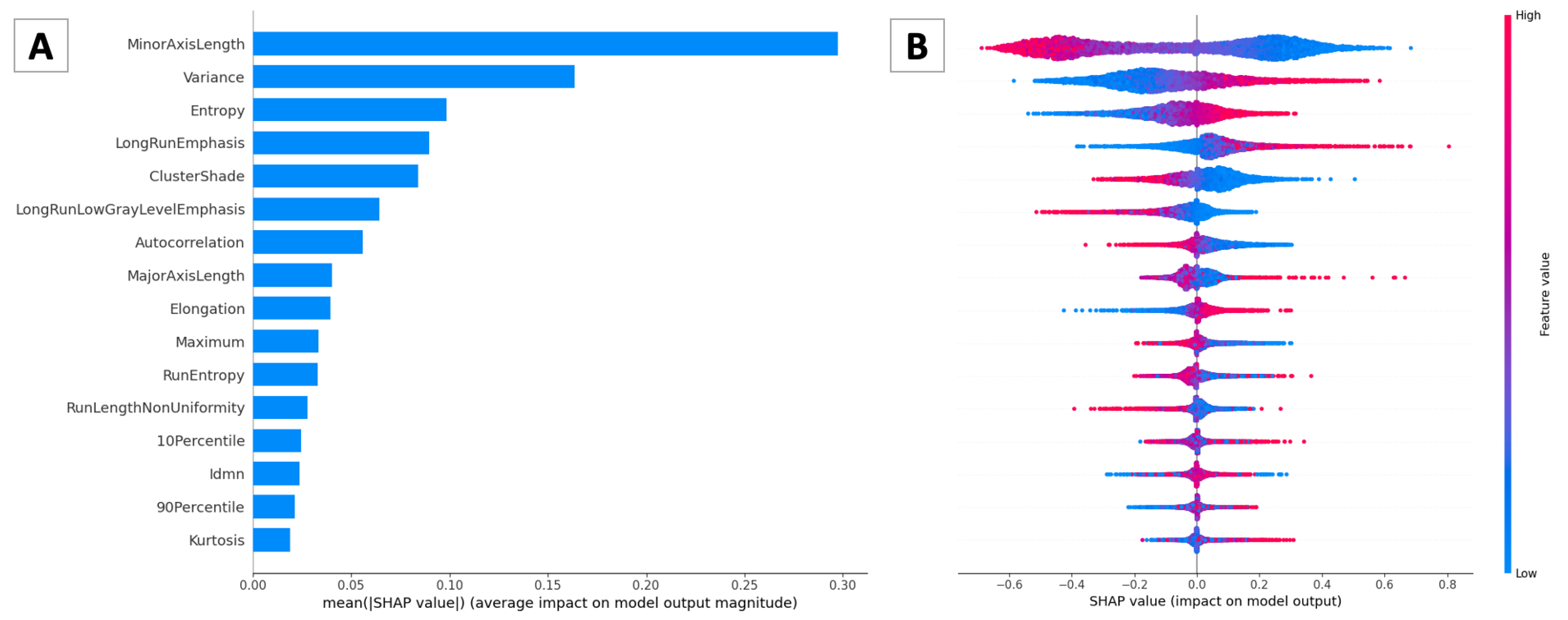
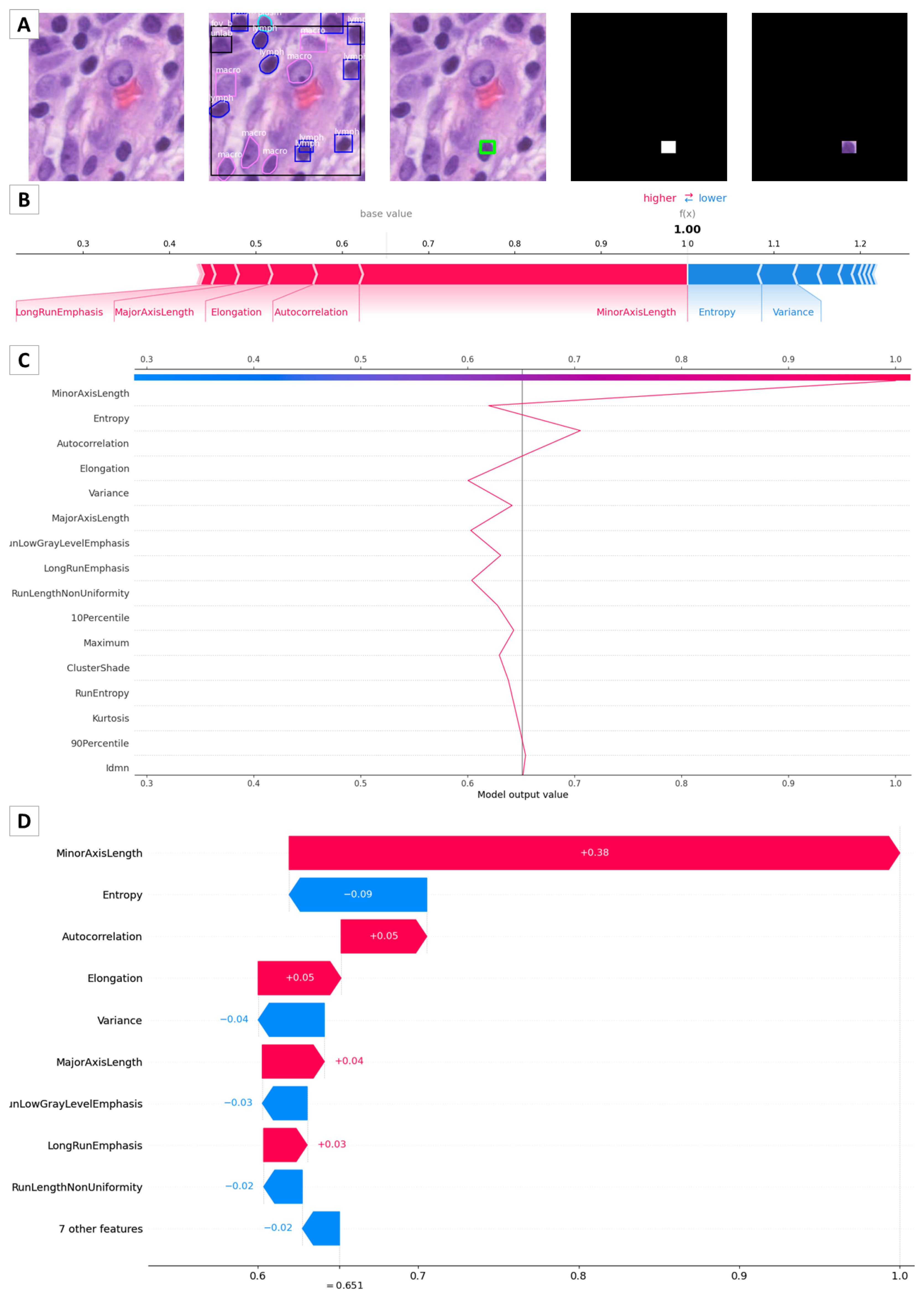
2.3. SHAP
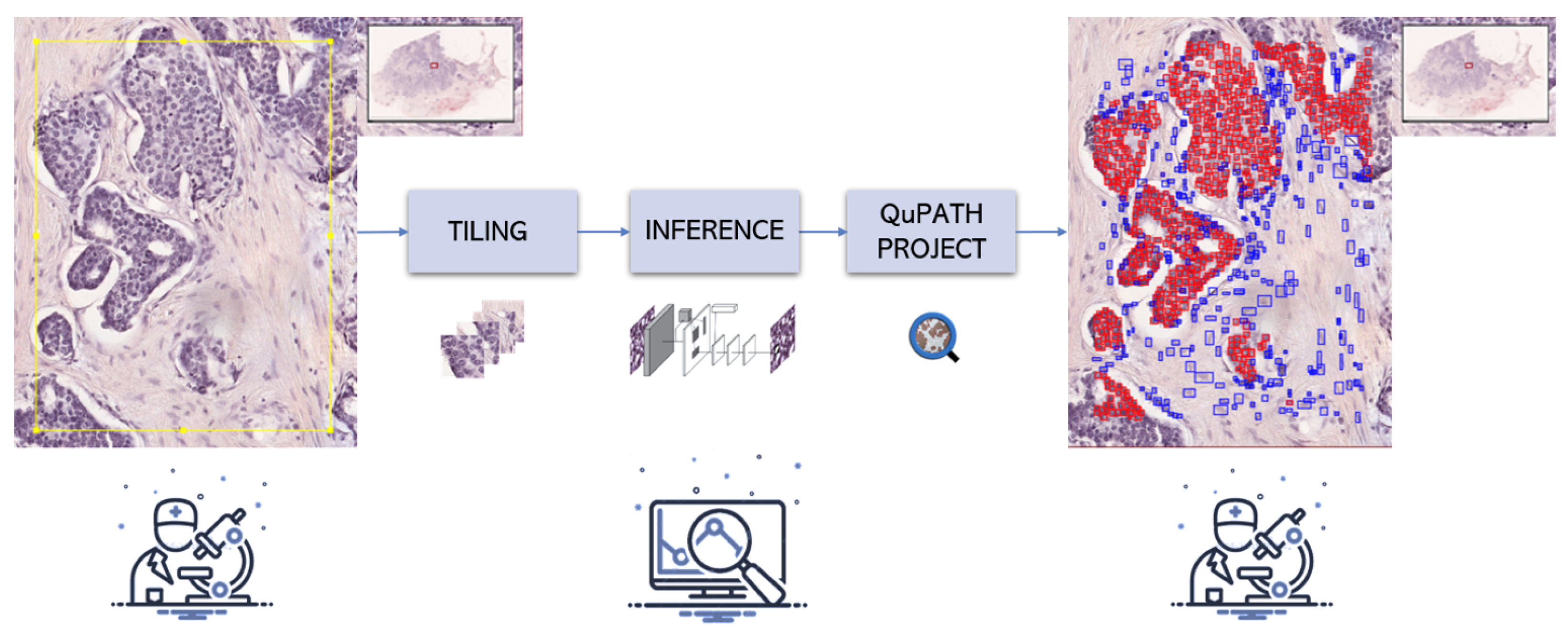
2.4. Integration with QuPath
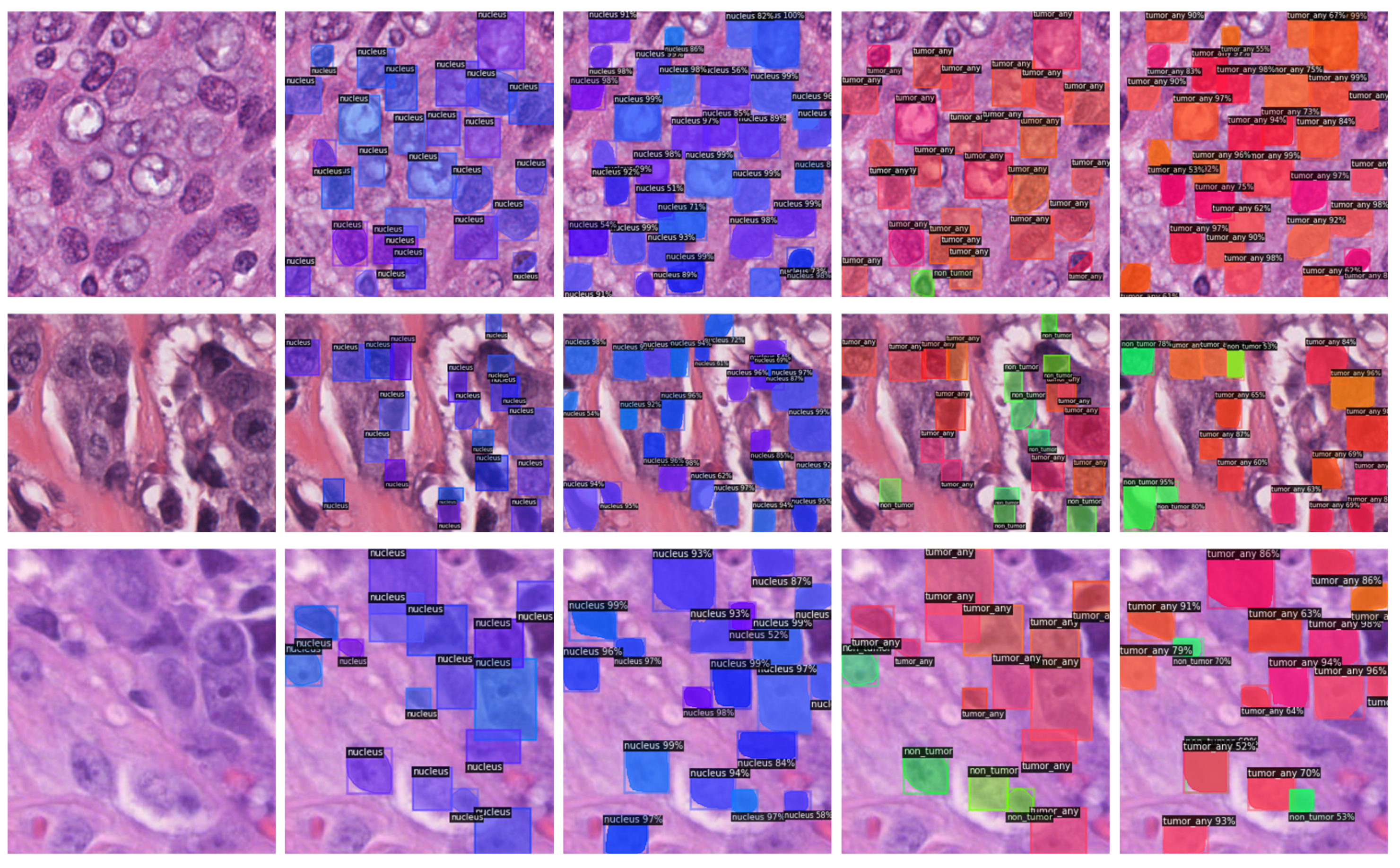
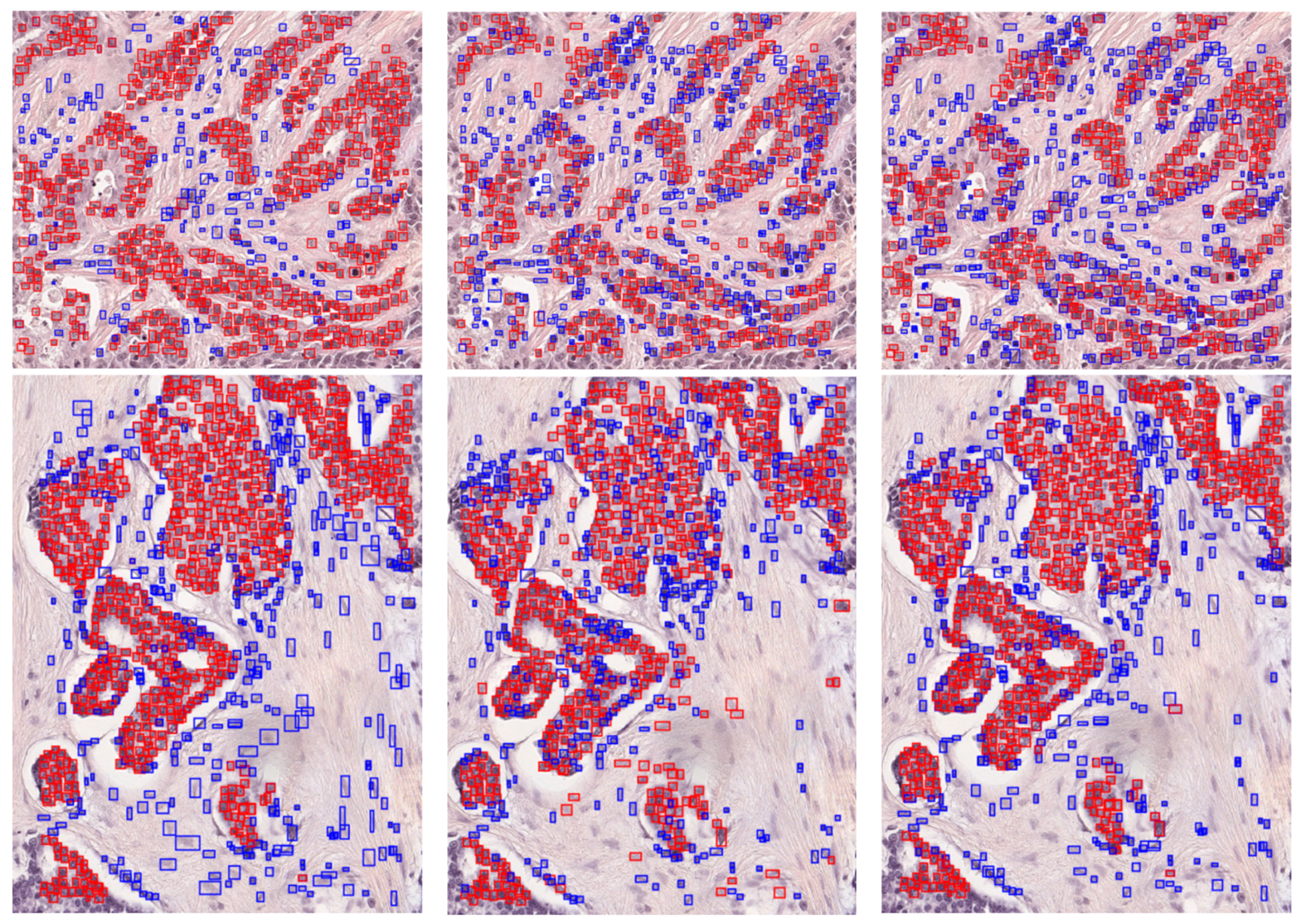
3. Results
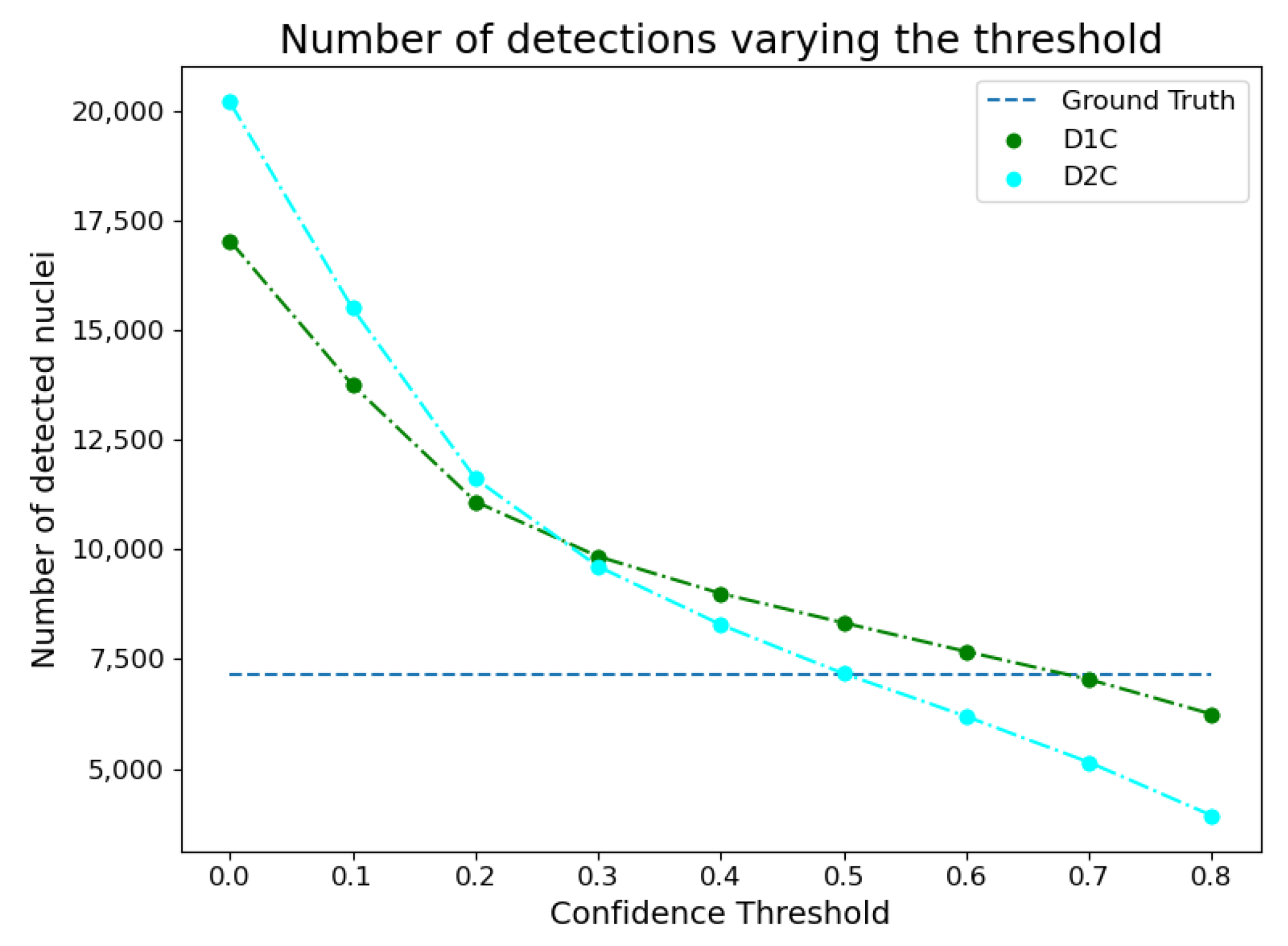
3.1. Instance Segmentation
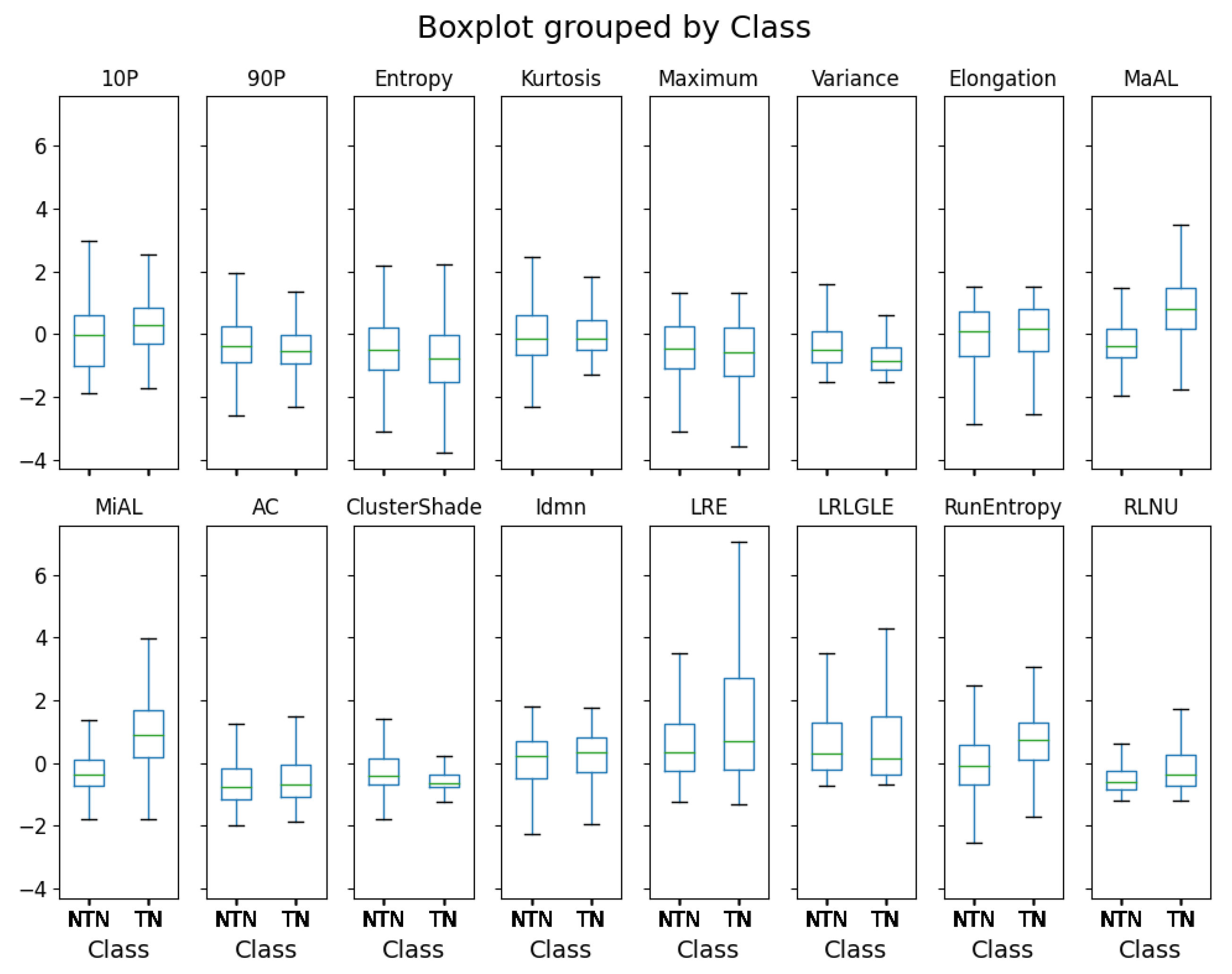
3.2. Pathomic Features and SHAP Analysis
4. Discussion
4.1. Instance Segmentation and Classification
4.2. Clinical Perspective
5. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rodriguez, J.P.M.; Rodriguez, R.; Silva, V.W.K.; Kitamura, F.C.; Corradi, G.C.A.; de Marchi, A.C.B.; Rieder, R. Artificial intelligence as a tool for diagnosis in digital pathology whole slide images: A systematic review. J. Pathol. Inform. 2022, 100138. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kurc, T.; Sharma, A.; Almeida, J.S.; Saltz, J. The emergence of pathomics. Curr. Pathobiol. Rep. 2019, 7, 73–84. [Google Scholar] [CrossRef]
- Manivannan, S.; Li, W.; Akbar, S.; Wang, R.; Zhang, J.; McKenna, S.J. An automated pattern recognition system for classifying indirect immunofluorescence images of HEp-2 cells and specimens. Pattern Recognit. 2016, 51, 12–26. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiang, Z.; Xie, F.; Zhang, H.; Ma, Y.; Shi, H.; Zhao, Y. Feature extraction from histopathological images based on nucleus-guided convolutional neural network for breast lesion classification. Pattern Recognit. 2017, 71, 14–25. [Google Scholar] [CrossRef]
- Van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 102470. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.M.; Buongiorno, D.; Altini, N.; Berloco, F.; Prencipe, B.; Moschetta, M.; Bevilacqua, V.; Brunetti, A. Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Appl. Sci. 2022, 12, 6230. [Google Scholar] [CrossRef]
- Altini, N.; Brunetti, A.; Puro, E.; Taccogna, M.G.; Saponaro, C.; Zito, F.A.; De Summa, S.; Bevilacqua, V. NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering 2022, 9, 475. [Google Scholar] [CrossRef]
- Altini, N.; Marvulli, T.M.; Caputo, M.; Mattioli, E.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Tommasi, S.; Bevilacqua, V.; Summa, S.D.; et al. Multi-class Tissue Classification in Colorectal Cancer with Handcrafted and Deep Features. In Proceedings of the International Conference on Intelligent Computing, Nanjing, China, 25–27 June 2021; pp. 512–525. [Google Scholar]
- Ploug, T.; Holm, S. The four dimensions of contestable AI diagnostics-A patient-centric approach to explainable AI. Artif. Intell. Med. 2020, 107, 101901. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4765–4774. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Altini, N.; Puro, E.; Taccogna, M.G.; Marino, F.; De Summa, S.; Saponaro, C.; Mattioli, E.; Zito, F.A.; Bevilacqua, V. A Dataset of Annotated Histopathological Images for Tumor Cellularity Assessment in Breast Cancer; Zenodo: Meyrin, Switzerland, 2023. [Google Scholar]
- Amgad, M.; Elfandy, H.; Hussein, H.; Atteya, L.A.; Elsebaie, M.A.; Abo Elnasr, L.S.; Sakr, R.A.; Salem, H.S.; Ismail, A.F.; Saad, A.M.; et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 2019, 35, 3461–3467. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network; Weinstein, J.; Collisson, E.; Mills, G.; Shaw, K.M.; Ozenberger, B.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Laukamp, K.R.; Shakirin, G.; Baeßler, B.; Thiele, F.; Zopfs, D.; Hokamp, N.G.; Timmer, M.; Kabbasch, C.; Perkuhn, M.; Borggrefe, J. Accuracy of radiomics-based feature analysis on multiparametric magnetic resonance images for noninvasive meningioma grading. World Neurosurg. 2019, 132, e366–e390. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Altini, N.; Prencipe, B.; Brunetti, A.; Villani, L.; Sacco, A.; Morelli, C.; Ciaccia, M.; Scardapane, A. Lung Segmentation and Characterization in COVID-19 Patients for Assessing Pulmonary Thromboembolism: An Approach Based on Deep Learning and Radiomics. Electronics 2021, 10, 2475. [Google Scholar] [CrossRef]
- Brunetti, A.; Altini, N.; Buongiorno, D.; Garolla, E.; Corallo, F.; Gravina, M.; Bevilacqua, V.; Prencipe, B. A Machine Learning and Radiomics Approach in Lung Cancer for Predicting Histological Subtype. Appl. Sci. 2022, 12, 5829. [Google Scholar] [CrossRef]
- Knabbe, J.; Das Gupta, A.; Kuner, T.; Asan, L.; Beretta, C.; John, J. Comprehensive monitoring of tissue composition using in vivo imaging of cell nuclei and deep learning. bioRxiv 2022. [Google Scholar] [CrossRef]
- Du, L.; Zhang, R.; Wang, X. Overview of two-stage object detection algorithms. In Proceedings of the Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2020; Volume 1544, p. 012033. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Wu, Y.; Kirillov, A.; Massa, F.; Lo, W.Y.; Girshick, R. Detectron2. 2019. Available online: https://github.com/facebookresearch/detectron2 (accessed on 14 March 2023).
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Scikit-Learn Machine Learning in Python. Available online: https://scikit-learn.org/stable/ (accessed on 19 February 2023).
- Detectron2 COCO-InstanceSegmentation. Available online: https://github.com/facebookresearch/detectron2/tree/main/configs/COCO-InstanceSegmentation (accessed on 10 February 2023).
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for nuclei detection, segmentation, and classification in digital histopathology: A review—current status and future potential. IEEE Rev. Biomed. Eng. 2013, 7, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Ra˛czkowski, Ł.; Moz˙ ejko, M.; Zambonelli, J.; Szczurek, E. ARA: Accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Sci. Rep. 2019, 9, 14347. [Google Scholar] [CrossRef] [PubMed]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Chu, A.; Sehgal, C.M.; Greenleaf, J.F. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Haralick, R.M.; Dinstein, I.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Thibault, G.; Fertil, B.; Navarro, C.; Pereira, S.; Cau, P.; Levy, N.; Sequeira, J.; Mari, J.l. Texture Indexes and Gray Level Size Zone Matrix Application to Cell Nuclei Classification. In Proceedings of the 10th International Conference on Pattern Recognition and Information Processing, PRIP 2009, Minsk, Belarus, 2009; pp. 140–145. Available online: https://www.researchgate.net/publication/255609273_Texture_Indexes_and_Gray_Level_Size_Zone_Matrix_Application_to_Cell_Nuclei_Classification (accessed on 28 November 2022).
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vision Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Hailemariam, Y.; Yazdinejad, A.; Parizi, R.M.; Srivastava, G.; Dehghantanha, A. An empirical evaluation of AI deep explainable tools. In Proceedings of the 2020 IEEE Globecom Workshops (GC Wkshps), Taipei, Taiwan, 7–11 December 2020; pp. 1–6. [Google Scholar]
- SHAP Documentation. Available online: https://shap.readthedocs.io/en/latest/index.html (accessed on 26 November 2022).
- Rozemberczki, B.; Watson, L.; Bayer, P.; Yang, H.T.; Kiss, O.; Nilsson, S.; Sarkar, R. The shapley value in machine learning. arXiv 2022, arXiv:2202.05594. [Google Scholar]
- Bagheri, R. Introduction to SHAP Values and Their Application in Machine Learning. Available online: https://towardsdatascience.com/introduction-to-shap-values-and-their-application-in-machine-learning-8003718e6827 (accessed on 28 November 2022).
- PAQUO Documentation. Available online: https://paquo.readthedocs.io/en/latest/index.html. (accessed on 26 November 2022).
- Molnar, C. Interpretable Machine Learning: A Guide for Making Black Box Models Explainable; Leanpub: Victoria, BC, Canada, 2022; Available online: https://christophm.github.io/interpretable-ml-book/ (accessed on 14 March 2023).
- Malato, G. How to Explain Neural Networks Using SHAP. Available online: https://www.yourdatateacher.com/2021/05/17/how-to-explain-neural-networks-using-shap/ (accessed on 28 November 2022).
- Altini, N.; Cascarano, G.D.; Brunetti, A.; Marino, F.; Rocchetti, M.T.; Matino, S.; Venere, U.; Rossini, M.; Pesce, F.; Gesualdo, L.; et al. Semantic segmentation framework for glomeruli detection and classification in kidney histological sections. Electronics 2020, 9, 503. [Google Scholar] [CrossRef]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; De Feudis, I.; Buongiorno, D.; Rossini, M.; Pesce, F.; Gesualdo, L.; Bevilacqua, V. A deep learning instance segmentation approach for global glomerulosclerosis assessment in donor kidney biopsies. Electronics 2020, 9, 1768. [Google Scholar] [CrossRef]
- Tripathi, S.; Singh, S.K. Ensembling handcrafted features with deep features: An analytical study for classification of routine colon cancer histopathological nuclei images. Multimed. Tools Appl. 2020, 79, 34931–34954. [Google Scholar] [CrossRef]
- Jahn, S.W.; Bösl, A.; Tsybrovskyy, O.; Gruber-Rossipal, C.; Helfgott, R.; Fitzal, F.; Knauer, M.; Balic, M.; Jasarevic, Z.; Offner, F.; et al. Clinically high-risk breast cancer displays markedly discordant molecular risk predictions between the MammaPrint and EndoPredict tests. Br. J. Cancer 2020, 122, 1744–1746. [Google Scholar] [CrossRef]
- Reza, S.M.; Iftekharuddin, K.M. Glioma grading using cell nuclei morphologic features in digital pathology images. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, San Diego, CA, USA, 27 February–3 March 2016; Volume 9785, pp. 735–740. [Google Scholar]
- Fischer, E.G. Nuclear morphology and the biology of cancer cells. Acta Cytol. 2020, 64, 511–519. [Google Scholar] [CrossRef]
- Yeom, C.; Kim, H.; Kwon, S.; Kang, S.H. Clinicopathologic Features of Pleomorphic Invasive Lobular Carcinoma: Comparison with Classic Invasive Lobular Carcinoma. J. Breast Dis. 2016, 4, 10–15. [Google Scholar] [CrossRef]
- Ishitha, G.; Manipadam, M.T.; BackIanathan, S.; Chacko, R.T.; Abraham, D.T.; Jacob, P.M. Clinicopathological study of triple negative breast cancers. J. Clin. Diagn. Res. 2016, 10, EC05. [Google Scholar] [CrossRef] [PubMed]
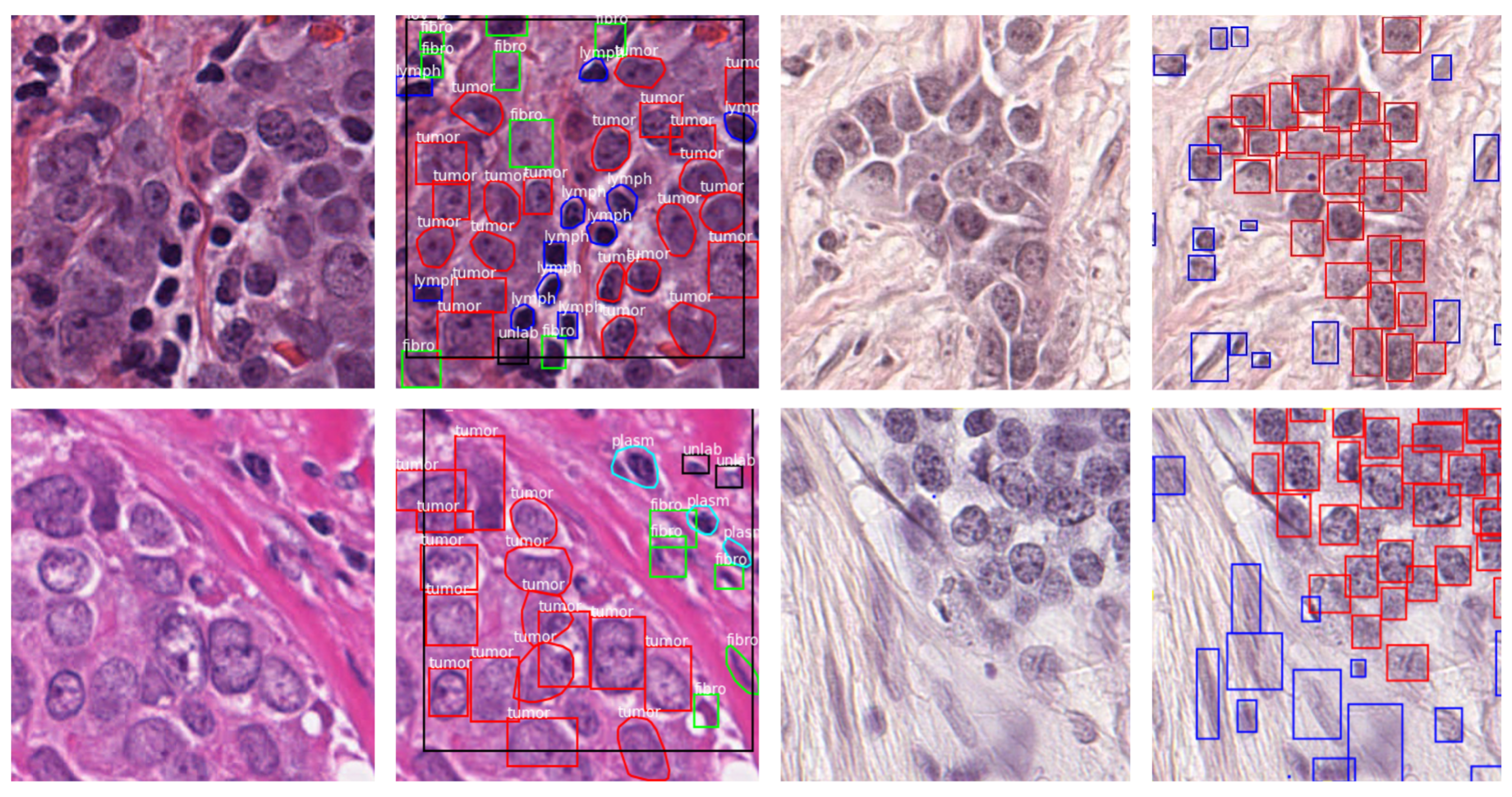
| Dataset | Publication Year | Organ | Magnification | Annotated Nuclei | Classes |
|---|---|---|---|---|---|
| NuCLS [13] | 2019 | Breast | 40× | 59,485 | Tumor, Stromal, sTILs, Other, Ambiguous |
| Local [12] | 2023 | Breast | 40× | 2024 | Tumor, Non-tumor |
| ID | C | Kernel | ||||
|---|---|---|---|---|---|---|
| SVM | 1 | 5 | rbf | |||
| 2 | 1 | rbf | ||||
| 3 | 50 | poly | ||||
| ID | Hidden Layers | Solver | Learning Rate | Activation | Max Iter | |
| ANN | 4 | (100) | adam | constant | relu | 500 |
| 5 | (150) | sgd | adaptive | relu | 500 | |
| 6 | (80, 80) | sgd | adaptive | logistic | 1000 | |
| 7 | (100, 80) | adam | constant | logistic | 1500 | |
| Approach | Architecture | ||||||
|---|---|---|---|---|---|---|---|
| D1C | R_50_DC5_1x | 0.722 | 0.891 | 0.803 | 0.711 | 0.739 | 0.527 |
| D2C | R_50_DC5_1x | 0.519 | 0.651 | 0.584 | 0.380 | 0.471 | 0.550 |
| R_50_DC5_3x | 0.533 | 0.682 | 0.603 | 0.387 | 0.482 | 0.321 | |
| R_50_FPN_1x | 0.493 | 0.628 | 0.556 | 0.374 | 0.447 | 0.320 | |
| R_50_C4_3x | 0.540 | 0.688 | 0.603 | 0.428 | 0.473 | 0.413 | |
| R_101_DC5_3x | 0.536 | 0.686 | 0.606 | 0.389 | 0.495 | 0.550 | |
| X_101_32x8d_FPN_3x | 0.483 | 0.634 | 0.543 | 0.339 | 0.433 | 0.550 |
| Approach | Architecture | ||||
|---|---|---|---|---|---|
| D1C | R_50_DC5_1x | 0.839 | 0.820 | 0.858 | 0.545 |
| D2C | R_50_DC5_1x | 0.777 | 0.699 | 0.746 | 0.545 |
| R_50_DC5_3x | 0.781 | 0.678 | 0.779 | 0.318 | |
| R_50_FPN_1x | 0.758 | 0.696 | 0.723 | 0.364 | |
| R_50_C4_3x | 0.798 | 0.718 | 0.802 | 0.409 | |
| R_101_DC5_3x | 0.783 | 0.711 | 0.768 | 0.545 | |
| X_101_32x8d_FPN_3x | 0.710 | 0.645 | 0.663 | 0.545 |
| Model | ID | Accuracy | F-Measure | AUC | Accuracy | F-Measure | AUC |
|---|---|---|---|---|---|---|---|
| (Train) | (Train) | (Train) | (Test) | (Test) | (Test) | ||
| SVM | 1 | 0.83 | 0.87 | 0.89 | 0.77 | 0.80 | 0.84 |
| 2 | 0.83 | 0.87 | 0.89 | 0.77 | 0.81 | 0.85 | |
| 3 | 0.82 | 0.87 | 0.89 | 0.77 | 0.80 | 0.82 | |
| ANN | 4 | 0.83 | 0.87 | 0.90 | 0.77 | 0.80 | 0.86 |
| 5 | 0.82 | 0.86 | 0.89 | 0.78 | 0.81 | 0.86 | |
| 6 | 0.81 | 0.86 | 0.88 | 0.78 | 0.81 | 0.84 | |
| 7 | 0.83 | 0.87 | 0.90 | 0.77 | 0.80 | 0.85 |
| ROI | Approach | Tumor Nucleus | Non-Tumor Nucleus | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F-Measure | Precision | Recall | F-Measure | ||
| 1 | D2C | 0.963 | 0.954 | 0.958 | 0.839 | 0.864 | 0.851 |
| 2 | D2C | 0.897 | 0.726 | 0.802 | 0.417 | 0.964 | 0.582 |
| 1 | D1C | 0.854 | 0.714 | 0.778 | 0.524 | 0.522 | 0.523 |
| 2 | D1C | 0.857 | 0.578 | 0.691 | 0.343 | 0.665 | 0.453 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altini, N.; Puro, E.; Taccogna, M.G.; Marino, F.; De Summa, S.; Saponaro, C.; Mattioli, E.; Zito, F.A.; Bevilacqua, V. Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explainability. Bioengineering 2023, 10, 396. https://doi.org/10.3390/bioengineering10040396
Altini N, Puro E, Taccogna MG, Marino F, De Summa S, Saponaro C, Mattioli E, Zito FA, Bevilacqua V. Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explainability. Bioengineering. 2023; 10(4):396. https://doi.org/10.3390/bioengineering10040396
Chicago/Turabian StyleAltini, Nicola, Emilia Puro, Maria Giovanna Taccogna, Francescomaria Marino, Simona De Summa, Concetta Saponaro, Eliseo Mattioli, Francesco Alfredo Zito, and Vitoantonio Bevilacqua. 2023. "Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explainability" Bioengineering 10, no. 4: 396. https://doi.org/10.3390/bioengineering10040396
APA StyleAltini, N., Puro, E., Taccogna, M. G., Marino, F., De Summa, S., Saponaro, C., Mattioli, E., Zito, F. A., & Bevilacqua, V. (2023). Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explainability. Bioengineering, 10(4), 396. https://doi.org/10.3390/bioengineering10040396









