Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework
Abstract
:1. Introduction
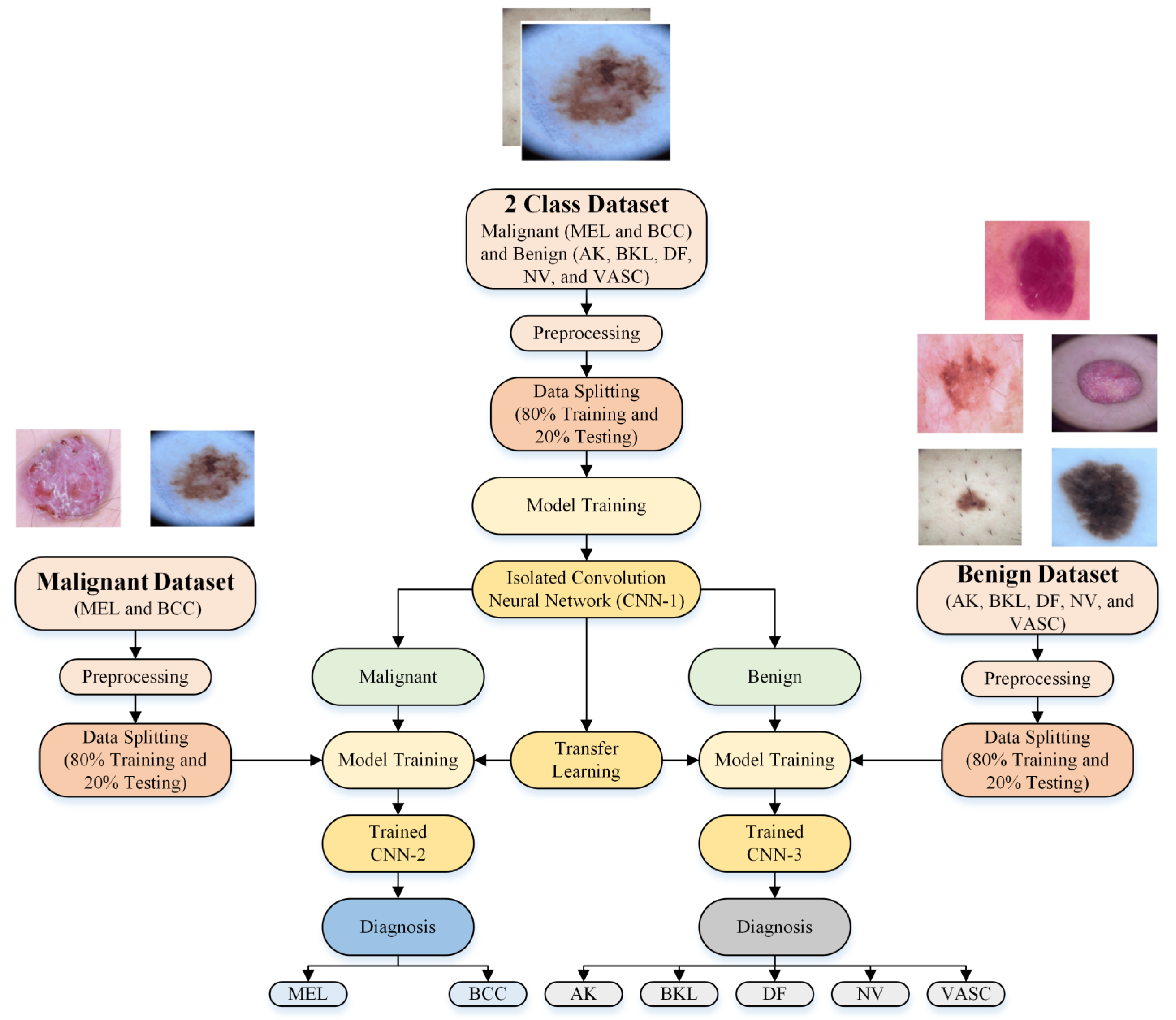
- A new multistage and multiclass identification CNN-based framework for skin lesion detection using dermoscopic images is presented;
- First, an isolated CNN was developed from scratch to classify the dermoscopic images into malignant and benign classes;
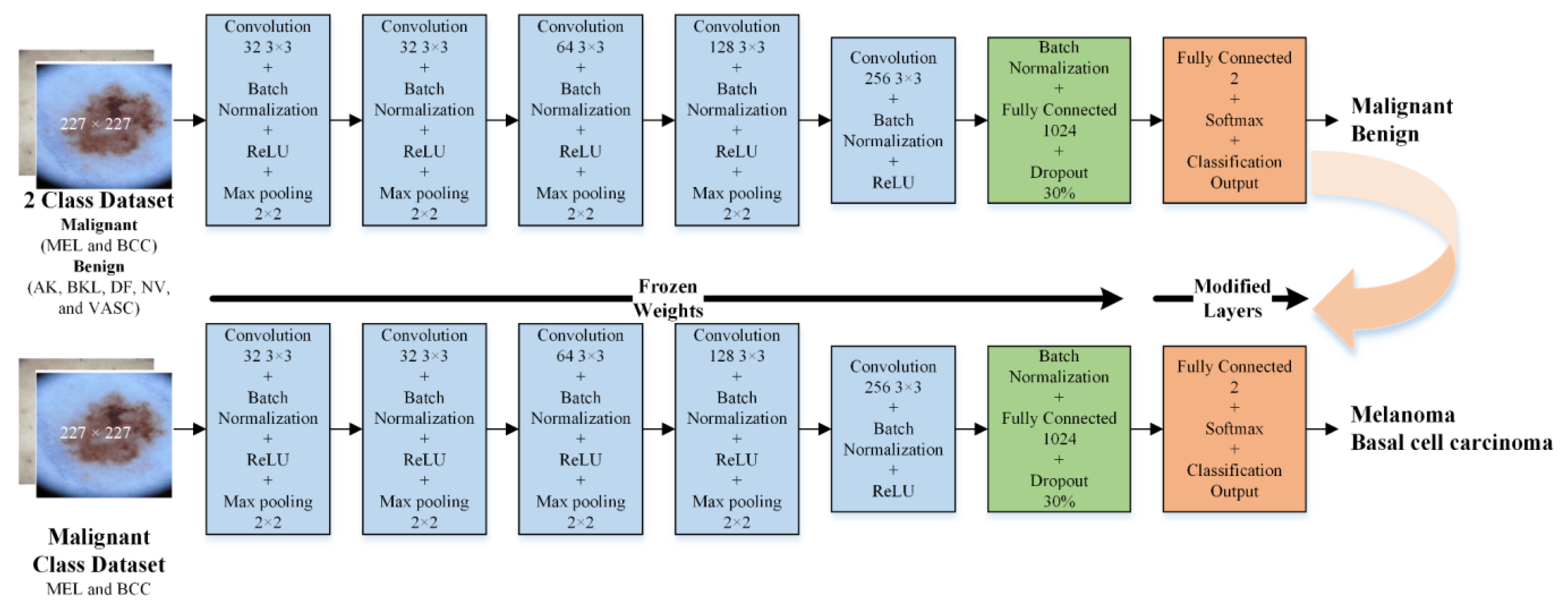
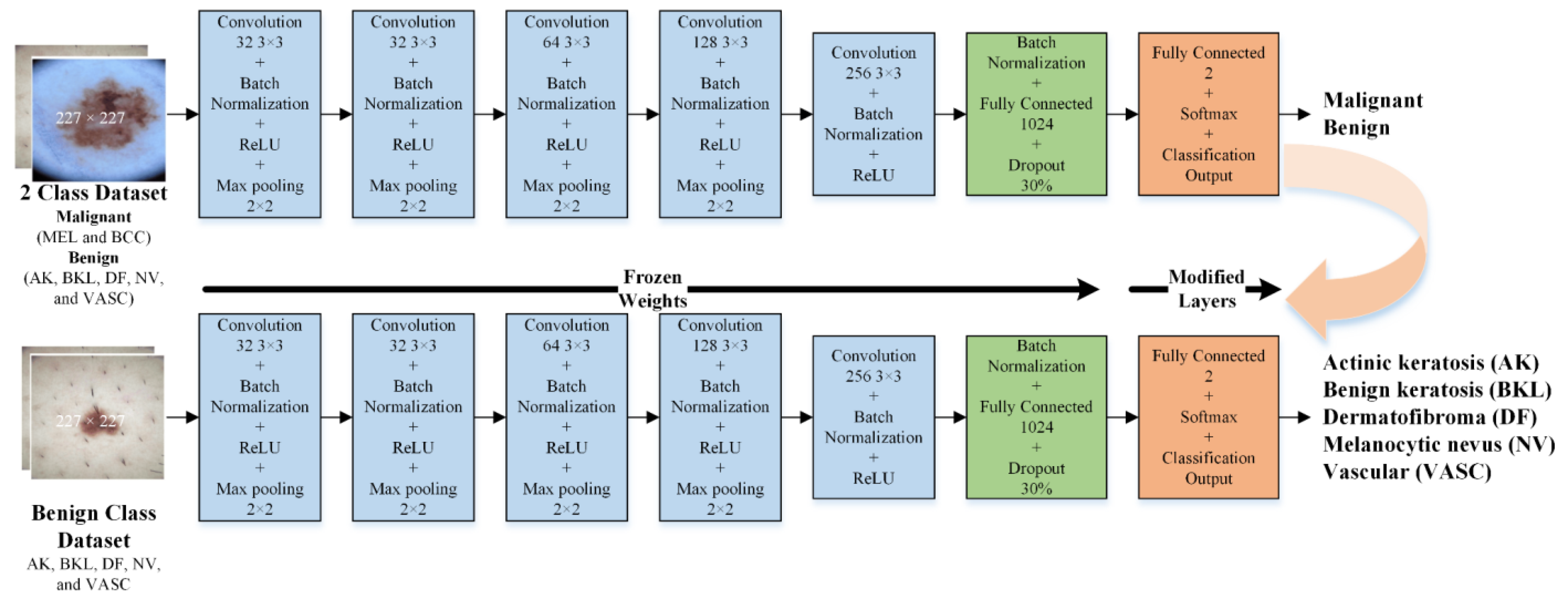
- Second, the developed isolated CNN model was used to develop two new CNN models to further classify each detected class (malignant and benign) into subcategories (MEL and BCC in the case of malignant and NV, AK, BK, DF, and VASC in the case of benign) using the idea of transfer learning. It was hypothesized that the frozen weights of the CNN developed and trained on correlated images could enhance the effectiveness of transfer learning when applied to the same type of images for subclassifying benign and malignant classes;
- The online skin lesions dataset was used to validate the proposed framework;
- The proposed multistage and multiclass framework results were also compared with the existing pretrained models and the literature.
2. Proposed Framework
2.1. Dataset Description
2.2. Preprocessing
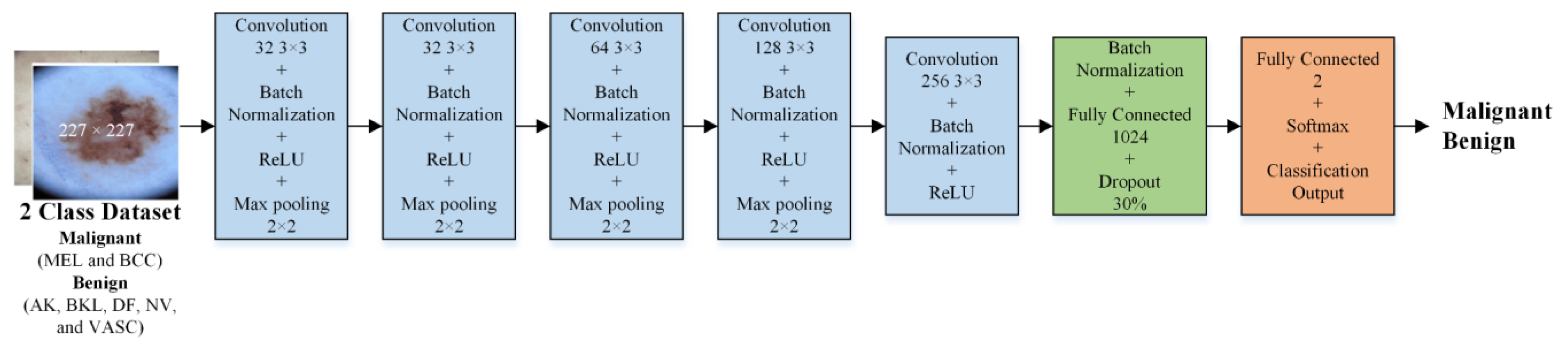
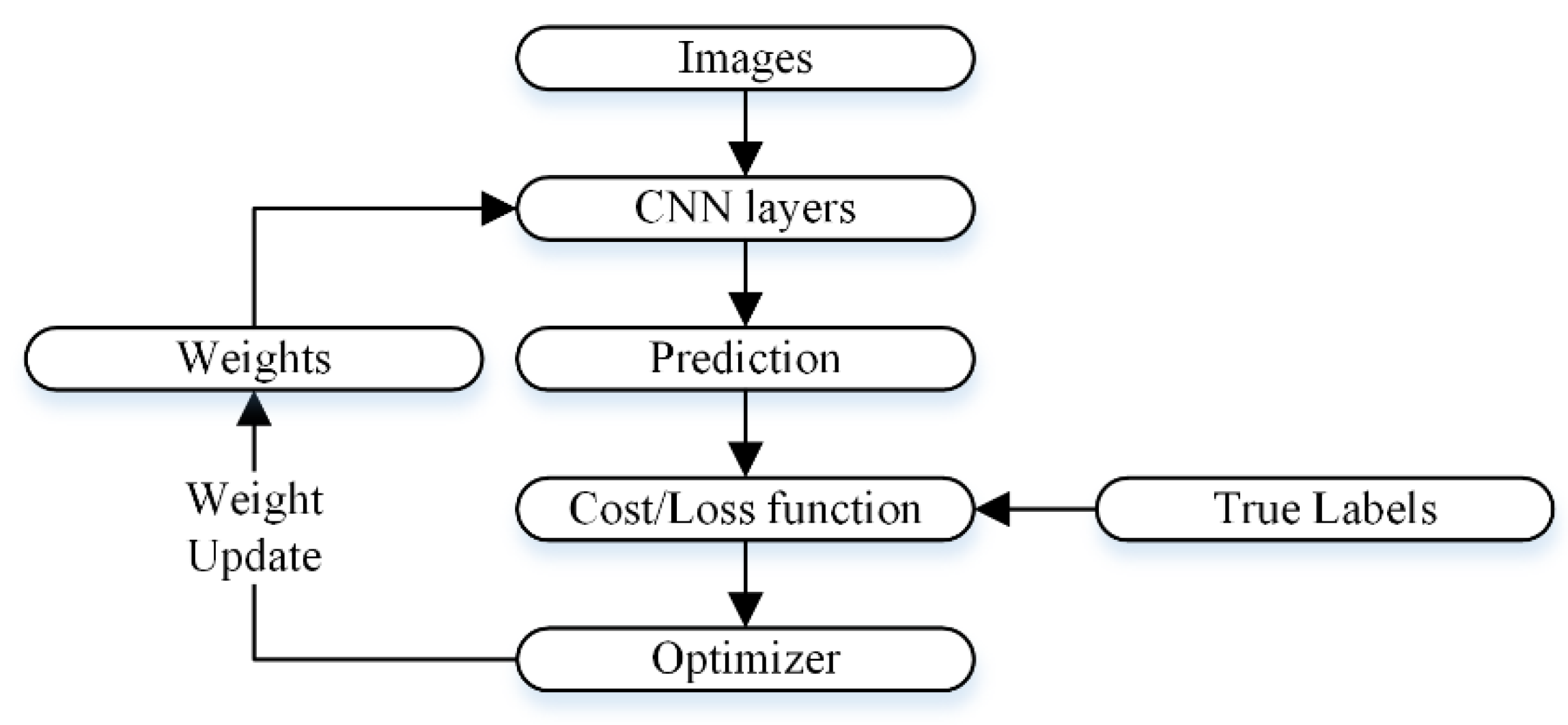
2.3. Development of CNN Models
2.3.1. Isolated CNN for Binary Class Classification
2.3.2. Developed Transfer Learned CNNs for Subcategorization
2.3.3. CNN Optimization
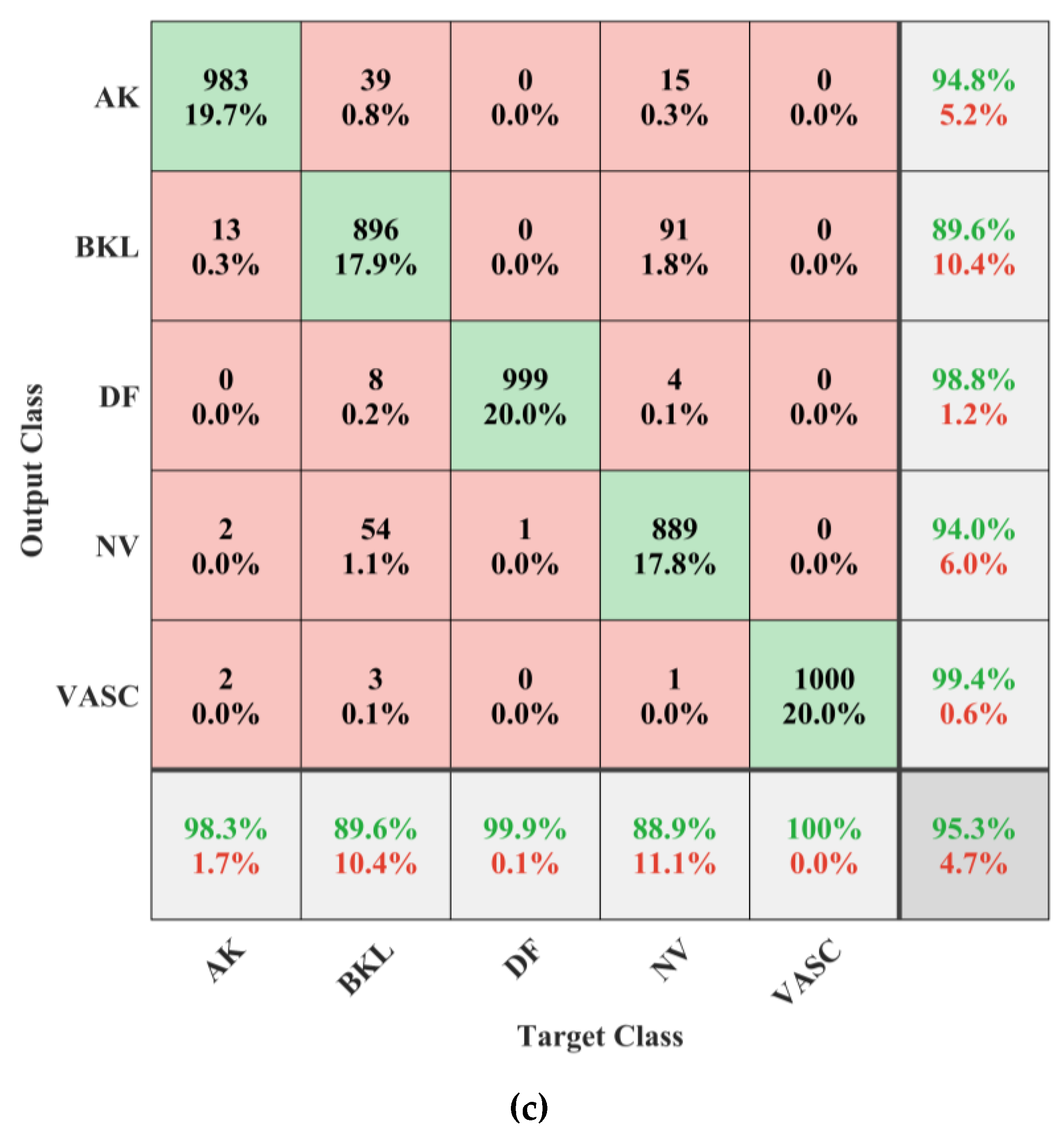
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R. Skin Cancer: An Overview of Epidemiology and Risk Factors. Semin. Oncol. Nurs. 2013, 29, 160–169. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, D.E.; Brenner, D.R.; Demers, P.A.; Villeneuve, P.J.; Friedenreich, C.M.; King, W.D. Indoor tanning and skin cancer in Canada: A meta-analysis and attributable burden estimation. Cancer Epidemiol. 2019, 59, 1–7. [Google Scholar] [CrossRef]
- Zhang, N.; Cai, Y.-X.; Wang, Y.-Y.; Tian, Y.-T.; Wang, X.-L.; Badami, B. Skin cancer diagnosis based on optimized convolutional neural network. Artif. Intell. Med. 2020, 102, 101756. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.E.; Barker, J.; Bleiker, T.O.; Chalmers, R.; Creamer, D. Rook's Textbook of Dermatology, 4 Volume Set; John Wiley & Sons: Hoboken, NJ, USA, 2016; Volume 1. [Google Scholar]
- Yaiza, J.M.; Gloria, R.A.; Belén, G.O.M.; Elena, L.-R.; Gema, J.; Antonio, M.J.; Ángel, G.C.M.; Houria, B. Melanoma cancer stem-like cells: Optimization method for culture, enrichment and maintenance. Tissue Cell 2019, 60, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Dalila, F.; Zohra, A.; Reda, K.; Hocine, C. Segmentation and classification of melanoma and benign skin lesions. Optik 2017, 140, 749–761. [Google Scholar] [CrossRef]
- Razmjooy, N.; Sheykhahmad, F.R.; Ghadimi, N. A hybrid neural network—World cup optimization algorithm for melanoma detection. Open Med. 2018, 13, 9–16. [Google Scholar] [CrossRef]
- Silveira, M.; Nascimento, J.C.; Marques, J.S.; Marcal, A.R.S.; Mendonca, T.; Yamauchi, S.; Maeda, J.; Rozeira, J. Comparison of Segmentation Methods for Melanoma Diagnosis in Dermoscopy Images. IEEE J. Sel. Top. Signal Process. 2009, 3, 35–45. [Google Scholar] [CrossRef]
- Fargnoli, M.C.; Kostaki, D.; Piccioni, A.; Micantonio, T.; Peris, K. Dermoscopy in the diagnosis and management of non-melanoma skin cancers. Eur. J. Dermatol. 2012, 22, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Argenziano, G.; Soyer, H.P.; Chimenti, S.; Talamini, R.; Corona, R.; Sera, F.; Binder, M.; Cerroni, L.; De Rosa, G.; Ferrara, G.; et al. Dermoscopy of pigmented skin lesions: Results of a consensus meeting via the Internet. J. Am. Acad. Dermatol. 2003, 48, 679–693. [Google Scholar] [CrossRef]
- Nachbar, F.; Stolz, W.; Merkle, T.; Cognetta, A.B.; Vogt, T.; Landthaler, M.; Bilek, P.; Braun-Falco, O.; Plewig, G. The ABCD rule of dermatoscopy: High prospective value in the diagnosis of doubtful melanocytic skin lesions. J. Am. Acad. Dermatol. 1994, 30, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, J.; Daneshvar, S.; Argenziano, G.; Hamarneh, G. Seven-Point Checklist and Skin Lesion Classification Using Multitask Multimodal Neural Nets. IEEE J. Biomed. Health Inform. 2019, 23, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Henning, J.S.; Dusza, S.W.; Wang, S.Q.; Marghoob, A.A.; Rabinovitz, H.S.; Polsky, D.; Kopf, A.W. The CASH (color, architecture, symmetry, and homogeneity) algorithm for dermoscopy. J. Am. Acad. Dermatol. 2007, 56, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, Z.; Akbar, A.; Kim, E.S.; Kamran, M.A.; Kim, J.H.; Jeong, M.Y. Utilizing EEG and fNIRS for the detection of sleep-deprivation-induced fatigue and its inhibition using colored light stimulation. Sci. Rep. 2023, 13, 6465. [Google Scholar] [CrossRef]
- Shoaib, Z.; Chang, W.K.; Lee, J.; Lee, S.H.; Phillips, V.Z.; Lee, S.H.; Paik, N.-J.; Hwang, H.-J.; Kim, W.-S. Investigation of neuromodulatory effect of anodal cerebellar transcranial direct current stimulation on the primary motor cortex using functional near-infrared spectroscopy. CerebellumPl 2023, 1–11. [Google Scholar] [CrossRef]
- Ali, M.U.; Kallu, K.D.; Masood, H.; Tahir, U.; Gopi, C.V.V.M.; Zafar, A.; Lee, S.W. A CNN-Based Chest Infection Diagnostic Model: A Multistage Multiclass Isolated and Developed Transfer Learning Framework. Int. J. Intell. Syst. 2023, 2023, 6850772. [Google Scholar] [CrossRef]
- Ali, M.U.; Hussain, S.J.; Zafar, A.; Bhutta, M.R.; Lee, S.W. WBM-DLNets: Wrapper-Based Metaheuristic Deep Learning Networks Feature Optimization for Enhancing Brain Tumor Detection. Bioengineering 2023, 10, 475. [Google Scholar] [CrossRef]
- Zafar, A.; Hussain, S.J.; Ali, M.U.; Lee, S.W. Metaheuristic Optimization-Based Feature Selection for Imagery and Arithmetic Tasks: An fNIRS Study. Sensors 2023, 23, 3714. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; AlRuwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain Tumor/Mass Classification Framework Using Magnetic-Resonance-Imaging-Based Isolated and Developed Transfer Deep-Learning Model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-L.; Ma, X.; Hu, F. Editorial: Machine Learning and Intelligent Communications. Mob. Netw. Appl. 2018, 23, 68–70. [Google Scholar] [CrossRef]
- Cerquitelli, T.; Meo, M.; Curado, M.; Skorin-Kapov, L.; Tsiropoulou, E.E. Machine learning empowered computer networks. Comput. Netw. 2023, 230, 109807. [Google Scholar] [CrossRef]
- Zafar, M.; Sharif, M.I.; Sharif, M.I.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Skin Lesion Analysis and Cancer Detection Based on Machine/Deep Learning Techniques: A Comprehensive Survey. Life 2023, 13, 146. [Google Scholar] [CrossRef]
- Debelee, T.G. Skin Lesion Classification and Detection Using Machine Learning Techniques: A Systematic Review. Diagnostics 2023, 13, 3147. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Part I 13. pp. 818–833. [Google Scholar]
- Thomas, S.M.; Lefevre, J.G.; Baxter, G.; Hamilton, N.A. Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer. Med. Image Anal. 2021, 68, 101915. [Google Scholar] [CrossRef] [PubMed]
- Amin, J.; Sharif, A.; Gul, N.; Anjum, M.A.; Nisar, M.W.; Azam, F.; Bukhari, S.A.C. Integrated design of deep features fusion for localization and classification of skin cancer. Pattern Recognit. Lett. 2020, 131, 63–70. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Kim, D.-H.; Kim, T.-S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef]
- Pacheco, A.G.; Ali, A.-R.; Trappenberg, T. Skin cancer detection based on deep learning and entropy to detect outlier samples. arXiv 2019, arXiv:1909.04525. [Google Scholar]
- Bibi, S.; Khan, M.A.; Shah, J.H.; Damaševičius, R.; Alasiry, A.; Marzougui, M.; Alhaisoni, M.; Masood, A. MSRNet: Multiclass Skin Lesion Recognition Using Additional Residual Block Based Fine-Tuned Deep Models Information Fusion and Best Feature Selection. Diagnostics 2023, 13, 3063. [Google Scholar] [CrossRef]
- Rosebrock, A. Finding Extreme Points in Contours with Open CV. Available online: https://pyimagesearch.com/2016/04/11/finding-extreme-points-in-contours-with-opencv/ (accessed on 2 November 2023).
- Chollet, F. Deep Learning with Python; Simon and Schuster: New York, NY, USA, 2017. [Google Scholar]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; pp. 270–279. [Google Scholar]
- Akram, M.W.; Li, G.; Jin, Y.; Chen, X.; Zhu, C.; Ahmad, A. Automatic detection of photovoltaic module defects in infrared images with isolated and develop-model transfer deep learning. Sol. Energy 2020, 198, 175–186. [Google Scholar] [CrossRef]
- Ahmed, W.; Hanif, A.; Kallu, K.D.; Kouzani, A.Z.; Ali, M.U.; Zafar, A. Photovoltaic Panels Classification Using Isolated and Transfer Learned Deep Neural Models Using Infrared Thermographic Images. Sensors 2021, 21, 5668. [Google Scholar] [CrossRef] [PubMed]
- Oyetade, I.S.; Ayeni, J.O.; Ogunde, A.O.; Oguntunde, B.O.; Olowookere, T.A. Hybridized deep convolutional neural network and fuzzy support vector machines for breast cancer detection. SN Comput. Sci. 2022, 3, 1–14. [Google Scholar] [CrossRef]
- Budhiman, A.; Suyanto, S.; Arifianto, A. Melanoma Cancer Classification Using ResNet with Data Augmentation. In Proceedings of the 2019 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), Yogyakarta, Indonesia, 5–6 December 2019; pp. 17–20. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef]
- Ali, K.; Shaikh, Z.A.; Khan, A.A.; Laghari, A.A. Multiclass skin cancer classification using EfficientNets—A first step towards preventing skin cancer. Neurosci. Inform. 2022, 2, 100034. [Google Scholar] [CrossRef]
- Carcagnì, P.; Leo, M.; Cuna, A.; Mazzeo, P.L.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of skin lesions by combining multilevel learnings in a DenseNet architecture. In Proceedings of the Image Analysis and Processing—ICIAP 2019: 20th International Conference, Trento, Italy, 9–13 September 2019; Part I 20. pp. 335–344. [Google Scholar]
- Sevli, O. A deep convolutional neural network-based pigmented skin lesion classification application and experts evaluation. Neural Comput. Appl. 2021, 33, 12039–12050. [Google Scholar] [CrossRef]
- Zafar, M.; Amin, J.; Sharif, M.; Anjum, M.A.; Mallah, G.A.; Kadry, S. DeepLabv3+-Based Segmentation and Best Features Selection Using Slime Mould Algorithm for Multi-Class Skin Lesion Classification. Mathematics 2023, 11, 364. [Google Scholar] [CrossRef]
- Bansal, P.; Garg, R.; Soni, P. Detection of melanoma in dermoscopic images by integrating features extracted using handcrafted and deep learning models. Comput. Ind. Eng. 2022, 168, 108060. [Google Scholar] [CrossRef]


| Types | Dermoscopic Images | No. of Samples |
|---|---|---|
| MEL | 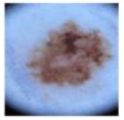 | 1113 |
| BCC | 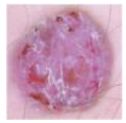 | 514 |
| AK | 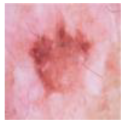 | 327 |
| BKL |  | 1099 |
| DF | 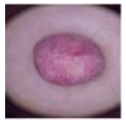 | 115 |
| NV |  | 6705 |
| VASC | 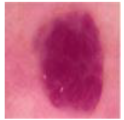 | 142 |
| Parameters | CNNs | |||||
|---|---|---|---|---|---|---|
| ResNet50 | Inception V3 | GoogleNet | DenseNet-201 | 26-Layer CNN | ||
| Training Loss | 0.0019 | 0.0011 | 0.0011 | 0.0012 | 0.0016 | |
| Training Accuracy (%) | 100 | 100 | 100 | 100 | 100 | |
| Validation Loss | 0.3425 | 0.4971 | 0.4971 | 0.3526 | 0.5216 | |
| Validation Accuracy (%) | 92.42 | 91.57 | 91.57 | 93.1 | 90.07 | |
| Training Time | 389 min 46 s | 513 min 43 s | 67 min 11 s | 1227 min 29 s | 11 min 43 s | |
| Sensitivity | AK | 0.98 | 0.96 | 0.96 | 0.985 | 0.985 |
| BCC | 0.945 | 0.935 | 0.935 | 0.935 | 0.920 | |
| BKL | 0.885 | 0.82 | 0.82 | 0.905 | 0.795 | |
| DF | 1 | 1 | 1 | 1 | 1 | |
| MEL | 0.76 | 0.865 | 0.865 | 0.855 | 0.825 | |
| NV | 0.9 | 0.83 | 0.83 | 0.84 | 0.780 | |
| VASC | 1 | 1 | 1 | 1 | 1 | |
| Specificity | AK | 0.987 | 0.991 | 0.991 | 0.993 | 0.980 |
| BCC | 0.995 | 0.985 | 0.985 | 0.993 | 0.992 | |
| BKL | 0.977 | 0.988 | 0.988 | 0.978 | 0.977 | |
| DF | 0.998 | 0.995 | 0.995 | 0.998 | 0.998 | |
| MEL | 0.981 | 0.967 | 0.967 | 0.980 | 0.964 | |
| NV | 0.975 | 0.978 | 0.978 | 0.978 | 0.978 | |
| VASC | 1 | 0.998 | 0.998 | 0.999 | 0.996 | |
| Precision | AK | 0.925 | 0.946 | 0.946 | 0.961 | 0.891 |
| BCC | 0.969 | 0.912 | 0.912 | 0.959 | 0.948 | |
| BKL | 0.863 | 0.916 | 0.916 | 0.874 | 0.850 | |
| DF | 0.985 | 0.971 | 0.971 | 0.985 | 0.990 | |
| MEL | 0.869 | 0.812 | 0.812 | 0.877 | 0.793 | |
| NV | 0.857 | 0.865 | 0.865 | 0.866 | 0.852 | |
| VASC | 1 | 0.99 | 0.99 | 0.995 | 0.976 | |
| F1 Score | AK | 0.952 | 0.953 | 0.953 | 0.973 | 0.936 |
| BCC | 0.957 | 0.923 | 0.923 | 0.947 | 0.934 | |
| BKL | 0.874 | 0.865 | 0.865 | 0.889 | 0.822 | |
| DF | 0.992 | 0.985 | 0.985 | 0.993 | 0.995 | |
| MEL | 0.811 | 0.838 | 0.838 | 0.866 | 0.809 | |
| NV | 0.878 | 0.847 | 0.847 | 0.853 | 0.815 | |
| VASC | 1 | 0.995 | 0.995 | 0.998 | 0.988 | |
| Parameters | CNN-1 | |
|---|---|---|
| Training Loss | 0.0074 | |
| Training Accuracy (%) | 100 | |
| Validation Loss | 0.4563 | |
| Validation Accuracy (%) | 93.4 | |
| Training Time | 11 min 41 s | |
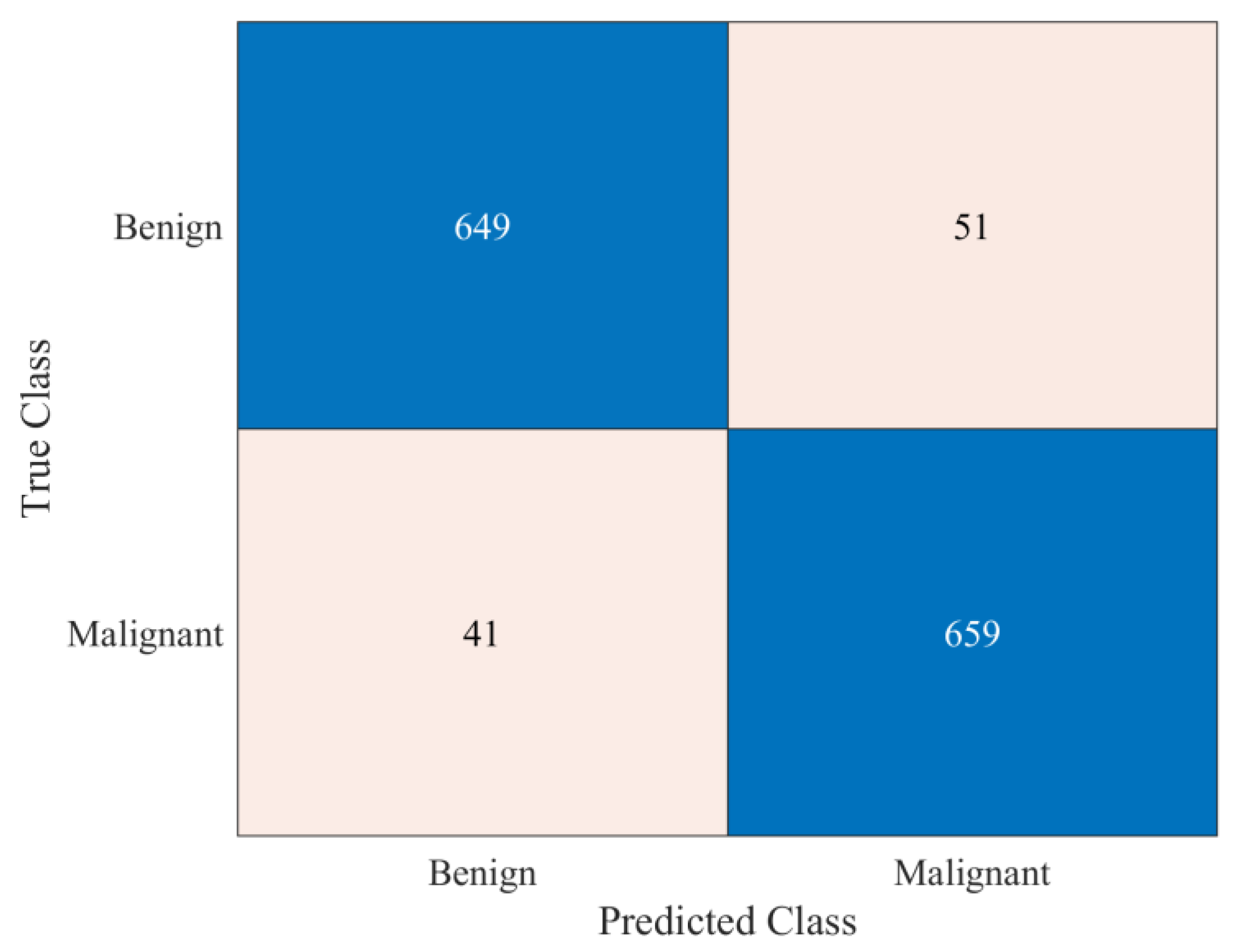
| Sensitivity | Benign | 0.927 |
| Malignant | 0.941 | |
| Specificity | Benign | 0.941 |
| Malignant | 0.927 | |
| Precision | Benign | 0.941 |
| Malignant | 0.928 | |
| F1 Score | Benign | 0.934 |
| Malignant | 0.935 | |
| Parameters | CNNs | ||
|---|---|---|---|
| CNN-2 | CNN-3 | ||
| Training Loss | 3.73 × 10−4 | 2.63 × 10−3 | |
| Training Accuracy (%) | 100 | 100 | |
| Validation Loss | 0.2576 | 0.1956 | |
| Validation Accuracy (%) | 96.25 | 96.20 | |
| Training Time | 389 min 46 s | 513 min 43 s | |
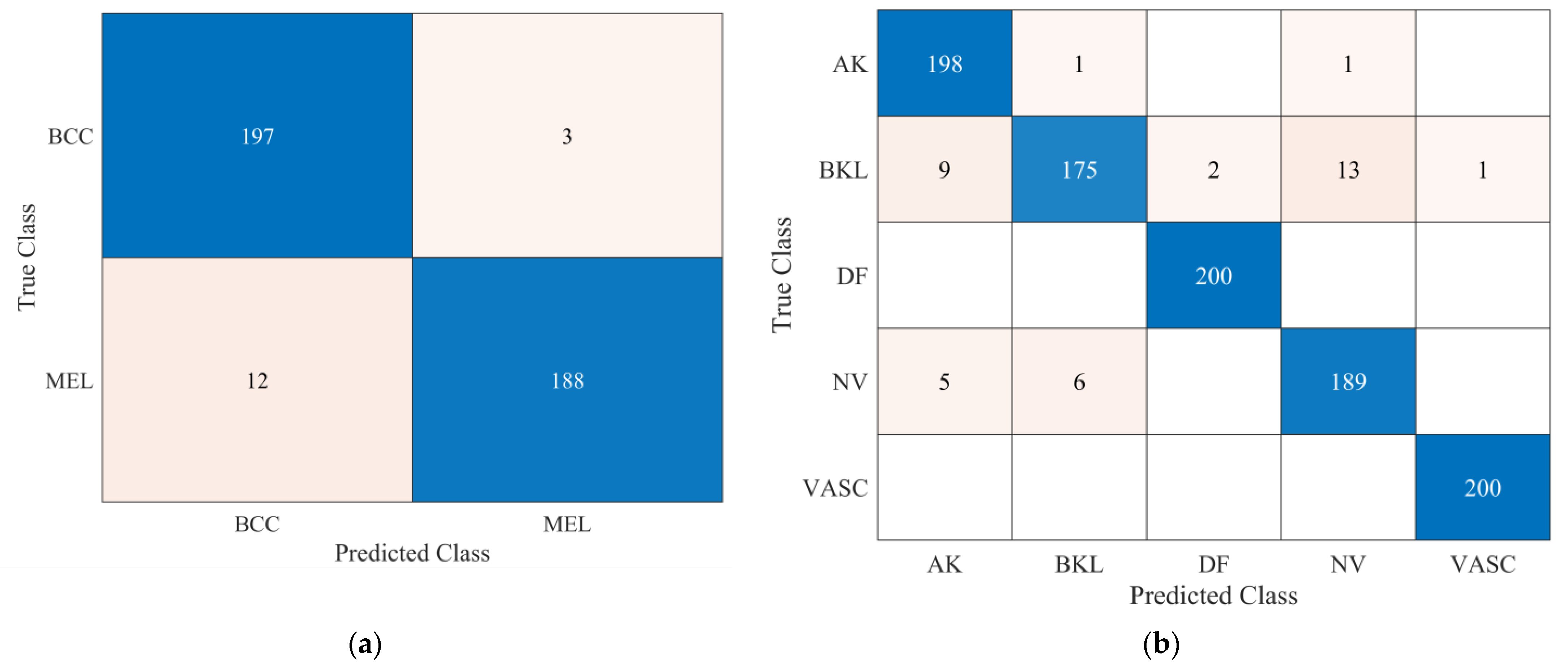
| Sensitivity | AK | - | 0.990 |
| BCC | 0.985 | - | |
| BKL | - | 0.875 | |
| DF | - | 1 | |
| MEL | 0.940 | - | |
| NV | - | 0.945 | |
| VASC | - | 1 | |
| Specificity | AK | - | 0.983 |
| BCC | 0.940 | - | |
| BKL | - | 0.991 | |
| DF | - | 0.998 | |
| MEL | 0.985 | - | |
| NV | - | 0.983 | |
| VASC | - | 0.999 | |
| Precision | AK | - | 0.934 |
| BCC | 0.943 | - | |
| BKL | 0.962 | ||
| DF | - | 0.990 | |
| MEL | 0.984 | - | |
| NV | - | 0.931 | |
| VASC | - | 0.995 | |
| F1 Score | AK | - | 0.961 |
| BCC | 0.963 | - | |
| BKL | - | 0.916 | |
| DF | - | 0.995 | |
| MEL | 0.962 | - | |
| NV | - | 0.938 | |
| VASC | - | 0.998 | |
| Parameters | Developed CNNs | |||
|---|---|---|---|---|
| 22-Layer | 26-Layer | 30-Layer | 34-Layer | |
| Training Loss | 0.0412 | 0.0074 | 0.0051 | 0.0264 |
| Training Accuracy (%) | 98.43 | 100 | 100 | 100 |
| Validation Loss | 0.5645 | 0.4563 | 0.498 | 0.5549 |
| Validation Accuracy (%) | 89.7 | 93.4 | 92.2 | 90.5 |
| Training Time | 11 min 10 s | 11 min 41 s | 13 min 35 s | 13 min 43 s |
| Study | Accuracy (%) |
|---|---|
| Budhiman et al. [39] | 87 (for normal and melanoma class) |
| Bibi et al. [32] | 85.4 |
| Mahbod et al. [40] | 86.2 |
| Ali et al. [41] | 87.9 |
| Carcagnì et al. [42] | 88 |
| Sevli [43] | 91.51 |
| Mehwish et al. [44] | 92.01 |
| Bansal et al. [45] | 94.9 (for normal and melanoma class) |
| This study | 93.4 (for benign and malignant) 94.2 (for benign and malignant using 10-fold cross-validation) 96.2 (for subclassification of benign and malignant) 97.5 (for subclassification of malignant using 10-fold cross-validation) 95.3 (for subclassification of benign using 10-fold cross-validation) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.U.; Khalid, M.; Alshanbari, H.; Zafar, A.; Lee, S.W. Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework. Bioengineering 2023, 10, 1430. https://doi.org/10.3390/bioengineering10121430
Ali MU, Khalid M, Alshanbari H, Zafar A, Lee SW. Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework. Bioengineering. 2023; 10(12):1430. https://doi.org/10.3390/bioengineering10121430
Chicago/Turabian StyleAli, Muhammad Umair, Majdi Khalid, Hanan Alshanbari, Amad Zafar, and Seung Won Lee. 2023. "Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework" Bioengineering 10, no. 12: 1430. https://doi.org/10.3390/bioengineering10121430
APA StyleAli, M. U., Khalid, M., Alshanbari, H., Zafar, A., & Lee, S. W. (2023). Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework. Bioengineering, 10(12), 1430. https://doi.org/10.3390/bioengineering10121430








