Authentication of Ginkgo biloba Herbal Products by a Novel Quantitative Real-Time PCR Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Species and Commercial Samples
2.2. DNA Extraction
2.3. DNA Quality and Purity
2.4. Target Gene Selection, Oligonucleotide Primers and Probes
2.5. Qualitative PCR
2.6. Real-Time PCR
3. Results and Discussion
3.1. DNA Quality and Selection of Target Region
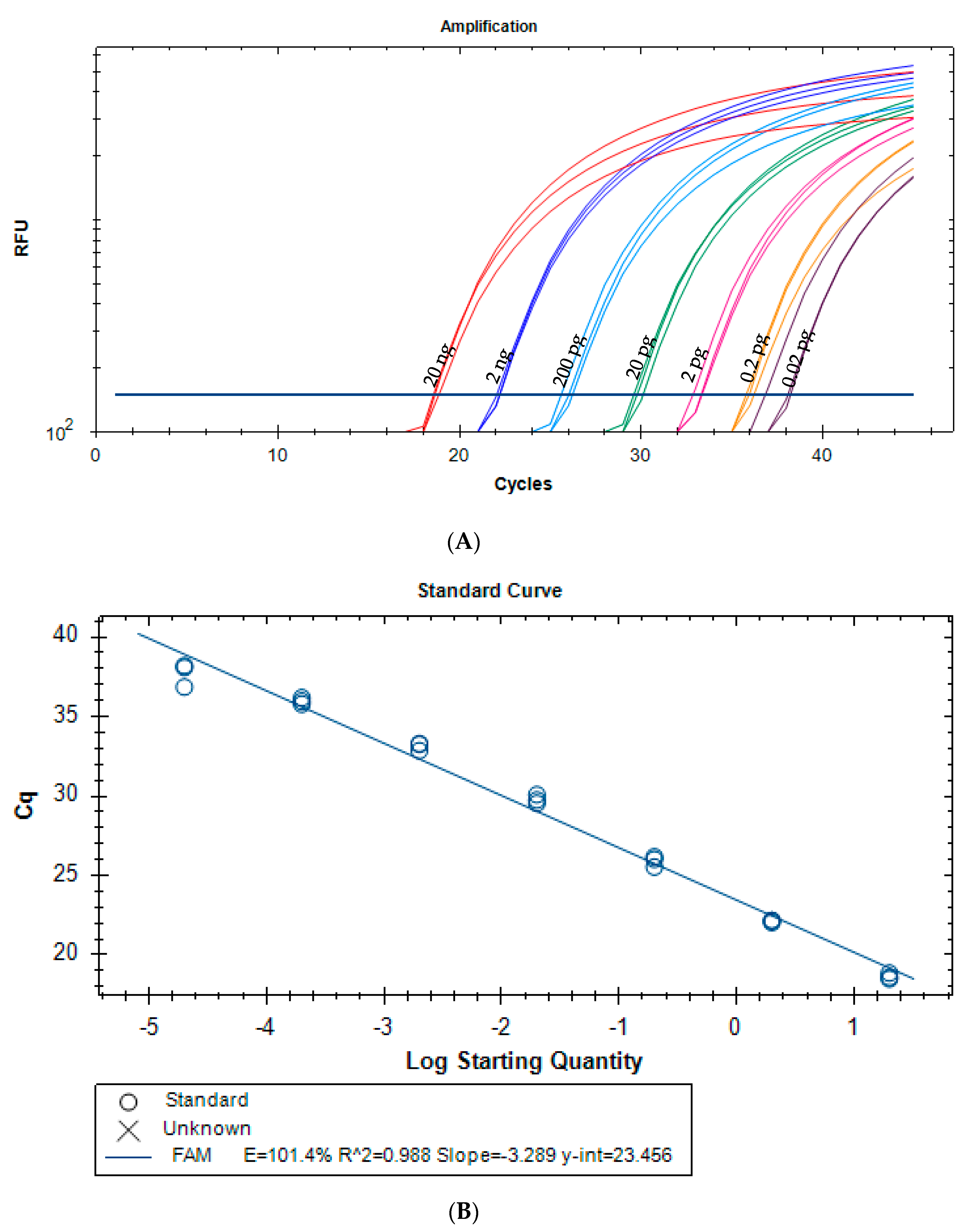
3.2. Quantitative Real-Time PCR
3.2.1. Method Development
3.2.2. Method Validation
3.2.3. Analysis of Commercial Herbal Infusions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Booker, A.; Frommenwiler, D.; Reich, E.; Horsfield, S.; Heinrich, M. Adulteration and poor quality of Ginkgo biloba supplements. J. Herbal Med. 2016, 6, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Kaur, P.; Gopichand; Singh, R.D.; Ahuja, P.S. Biology and chemistry of Ginkgo biloba. Fitoterapia 2008, 79, 401–418. [Google Scholar] [CrossRef] [PubMed]
- Wohlmuth, H.; Savage, K.; Dowell, A.; Mouatt, P. Adulteration of Ginkgo biloba products and a simple method to improve its detection. Phytomedicine 2014, 21, 912–918. [Google Scholar] [CrossRef]
- WHO. New WHO Guidelines to Promote Proper Use of Alternative Medicines. 2004. Available online: https://www.who.int/mediacentre/news/releases/2004/pr44/en/ (accessed on 2 December 2019).
- EMA. Ginkgo Folium—Committee on Herbal Medicinal Products (HMPC/321097/2012). 2015. Available online: https://www.ema.europa.eu/en/medicines/herbal/ginkgo-folium (accessed on 2 December 2019).
- Gafner, S. Botanical Adulterants Prevention Program—Adulteration of Ginkgo Biloba Leaf Extract. 2018. Available online: http://cms.herbalgram.org/BAP/BAB/GinkgoBulletin.html (accessed on 3 December 2019).
- Garcia-Alvarez, A.; Egan, B.; de Klein, S.; Dima, L.; Maggi, F.M.; Isoniemi, M.; Ribas-Barba, L.; Raats, M.M.; Meissner, E.M.; Badea, M.; et al. Usage of plant food supplements across six European countries: Findings from the PlantLIBRA consumer survey. PLoS ONE 2014, 9, e92265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CRN. Consumer Survey on Dietary Supplements. Available online: https://www.crnusa.org/sites/default/files/images/CRN-2017-ConsumerSurvey-4-page-highlights.pdf (accessed on 18 December 2019).
- Market.us. Ginkgo biloba Extract Market by Form (Tablets, Capsules, Liquid Extracts, Others), by Application (Food and Beverages, Cosmetics, Pharmaceuticals), and by Region—Global Forecast to 2029. 2019. Available online: https://www.marketwatch.com/press-release/global-ginkgo-biloba-extract-market-expected-to-observe-major-growth-with-cagr-of-41-2019-08-29?mod=mw_quote_news (accessed on 18 December 2019).
- Pharmacopeia, E. European Directorate for the Quality of Medicines and HealthCare, 8th ed.; Oxford Public International Law: Strasbourg, France, 2013. [Google Scholar]
- Ko, Y.-C.; Lee, R.-J.; Feng, H.-T.; Lee, M.-R. Development of a liquid chromatography tandem mass spectrometry method for identification of flavonoids in Ginkgo biloba. J. Chin. Chem. Soc. 2013, 60, 1333–1338. [Google Scholar] [CrossRef]
- Avula, B.; Sagi, S.; Gafner, S.; Upton, R.; Wang, Y.-H.; Wang, M.; Khan, I.A. Identification of Ginkgo biloba supplements adulteration using high performance thin layer chromatography and ultra high performance liquid chromatography-diode array detector-quadrupole time of flight-mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 7733–7746. [Google Scholar] [CrossRef]
- Frommenwiler, D.A.; Booker, A.; Vila, R.; Heinrich, M.; Reich, E.; Cañigueral, S. Comprehensive HPTLC fingerprinting as a tool for a simplified analysis of purity of ginkgo products. J. Ethnopharmacol. 2019, 243, 112084. [Google Scholar] [CrossRef]
- Walkowiak, A.; Ledziński, Ł.; Zapadka, M.; Kupcewicz, B. Detection of adulterants in dietary supplements with Ginkgo biloba extract by attenuated total reflectance Fourier transform infrared spectroscopy and multivariate methods PLS-DA and PCA. Spectrochim. Acta Part A Mol. Biomol. Spect. 2019, 208, 222–228. [Google Scholar] [CrossRef]
- Franz, C.; Chizzola, R.; Novak, J.; Sponza, S. Botanical species being used for manufacturing plant food supplements (PFS) and related products in the EU member states and selected third countries. Food Funct. 2011, 2, 720–730. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Sakaridis, I.; Argiriou, A.; Tsaftaris, A. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res. Int. 2014, 60, 163–172. [Google Scholar] [CrossRef]
- Grazina, L.; Amaral, J.S.; Mafra, I. Botanical origin authentication of dietary supplements by DNA-based approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1080–1109. [Google Scholar] [CrossRef] [Green Version]
- Ganie, S.H.; Upadhyay, P.; Das, S.; Prasad Sharma, M. Authentication of medicinal plants by DNA markers. Plant Gene 2015, 4, 83–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, D.P. Authentication of Ginkgo biloba herbal dietary supplements using DNA barcoding. Genome 2014, 57, 513–516. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, X.-Y.; Wei, X.-M.; Gao, Z.-T.; Han, J.-P. Rapid authentication of Ginkgo biloba herbal products using the recombinase polymerase amplification assay. Sci. Rep. 2018, 8, 8002. [Google Scholar] [CrossRef] [PubMed]
- Dhivya, S.; Ragupathy, S.; Kesanakurti, P.; Jeevitha, S.; Noce, I.D.; Newmaster, S.G. Validated identity test method for Ginkgo biloba NHPs using dna-based species-specific hydrolysis pcr probe. J. AOAC Int. 2019, 102, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Campos, B.; Amaral, J.S.; Nunes, M.E.; Oliveira, M.B.P.P.; Mafra, I. HRM analysis targeting ITS1 and matK loci as potential DNA mini-barcodes for the authentication of Hypericum perforatum and Hypericum androsaemum in herbal infusions. Food Control 2016, 61, 105–114. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Oliveira, M.B.P.P.; Mafra, I. Novel quantitative real-time PCR approach to determine safflower (Carthamus tinctorius) adulteration in saffron (Crocus sativus). Food Chem. 2017, 229, 680–687. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Gondar, C.; Oliveira, M.B.P.P.; Mafra, I. Effect of food matrix and thermal processing on the performance of a normalised quantitative real-time PCR approach for lupine (Lupinus albus) detection as a potential allergenic food. Food Chem. 2018, 262, 251–259. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- ENGL. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing European Network of GMO Laboratories. European Network of GMO Laboratories, Join Research Centre, EURL, 2015. Available online: https://gmo-crl.jrc.ec.europa.eu/doc/MPR%20Report%20Application%2020_10_2015.pdf (accessed on 3 December 2019).
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Maurizio, R.; Yitao, W.; Shilin, C. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Sun, W.; Wang, B.; Zhao, H.; Li, Y.; Cai, S.; Xiang, L.; Zhu, Y.; Yao, H.; Song, J.; et al. An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci. Rep. 2015, 5, 11318. [Google Scholar] [CrossRef] [PubMed]
- Doganay-Knapp, K.; Orland, A.; König, G.M.; Knöss, W. The potential of three different PCR-related approaches for the authentication of mixtures of herbal substances and finished herbal medicinal products. Phytomedicine 2018, 43, 60–67. [Google Scholar] [CrossRef] [PubMed]
- RBG (Royal Botanic Garden). Kew, Plant DNA C-Values Database, Surrey, Canada. Available online: http://data.kew.org/cvalues/ (accessed on 12 February 2020).
- López-Andreo, M.; Lugo, L.; Garrido-Pertierra, A.; Prieto, M.I.; Puyet, A. Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction. Anal. Biochem. 2005, 339, 73–82. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Quantitative detection of soybean in meat products by a TaqMan real-time PCR assay. Meat Sci. 2014, 98, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.; Amaral, J.S.; Grazina, L.; Oliveira, M.B.P.P.; Mafra, I. Matrix-normalised real-time PCR approach to quantify soybean as a potential food allergen as affected by thermal processing. Food Chem. 2017, 221, 1843–1850. [Google Scholar] [CrossRef]
- Kang, T.S. Basic principles for developing real-time PCR methods used in food analysis: A review. Trends Food Sci. Technol. 2019, 91, 574–585. [Google Scholar] [CrossRef]

| Samples | Label | Qualitative PCR a | Quantitative Real-Time PCR | ||||
|---|---|---|---|---|---|---|---|
| EG-F/EG-R | Gkb2-F/Gkb2-R | Cq ± SD b EG-F/EG-R | Cq± SD b Gkb2-F/Gkb2-R | Estimated Ginkgo (%, w/w, Mean ± SD) c | |||
| 1 | Ginkgo infusion (leaves) | ginkgo | + | + | NA d | NA | NA |
| 2 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| 3 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| 4 | Herbal infusion (sachets) | 15% ginkgo | + | + | 17.03 ± 0.05 | 24.53 ± 0.32 | 2.98 ± 0.60 |
| 5 | Ginkgo infusion (sachets) | 100% ginkgo | + | + | NA | NA | NA |
| 6 | Ginkgo infusion (sachets) | 100% ginkgo | + | + | NA | NA | NA |
| 7 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| 8 | Ginkgo bio-infusion (leaves) (Agr. non EU) | 100% ginkgo | + | + | NA | NA | NA |
| 9 | Ginkgo infusion (leaves) (Agr. non EU) | 100% ginkgo | + | + | NA | NA | NA |
| 10 | Tisana MC (Plant mixture) | 15% ginkgo | + | +/− | 17.85 ± 0.49 | 34.00 ± 1.11 | 0.01 ± 0.01 |
| 11 | Tisana TB (Plant mixture) | 15% ginkgo | + | + | 15.50 ± 0.18 | 23.50 ± 0.05 | 1.98 ± 0.06 |
| 12 | Ginkgo infusion (sachets) | 30% ginkgo | + | + | 18.46 ± 0.13 | 24.12 ± 0.12 | 9.95 ± 0.79 |
| 13 | Ginkgo infusion (sachets) | 100% ginkgo | + | + | NA | NA | NA |
| 14 | Herbal infusion (sachets) | ginkgo (% NL e) | + | + | 17.19 ± 0.07 | 24.80 ± 0.13 | 2.68 ± 0.24 |
| 15 | Herbal infusion (sachets) | ginkgo (% NL) | + | + | 16.71 ± 0.02 | 24.75 ± 0.02 | 2.00 ± 0.26 |
| 16 | Herbal infusion (sachets) (Agr. non EU) | 15% ginkgo | + | + | 16.33 ± 0.11 | 26.64 ± 0.35 | 0.47 ± 0.03 |
| 17 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| 18 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| 19 | Ginkgo infusion (sachets) | 100% ginkgo | + | + | NA | NA | NA |
| 20 | Ginkgo infusion (leaves) | ginkgo | + | + | NA | NA | NA |
| Species | Target | Primer | Sequence (5′→3′) | Length | Reference |
|---|---|---|---|---|---|
| G. biloba | ITS1 | Gkb2-F | GCGGTAAGCCCATCTCTCGA | 175 bp | This work |
| Gkb2-R | CCGAAGCGAACCCGAACAAC | ||||
| Gkb2-P | FAM-ATGCCAAGGTCGCCGGACCGTC-BHQ1 | ||||
| Eukaryote | Nuclear 18S rRNA | EG-F | TCGATGGTAGGATAGTGGCCTACT | 109 bp | [23] |
| EG-R | TGCTGCCTTCCTTGGATGTGGTA | ||||
| EG-P | FAM-ACGGGTGACGGAGAATTAGGGTTCGATTC-BHQ-1 | [24] |
| Samples | Ginkgo (%, w/w) | CV b (%) | Error c (%) | |
|---|---|---|---|---|
| Real Value (%) | Estimated Value a (%) | |||
| CG–A | 20.0 | 23.58 ± 2.22 | 9.41 | 17.9 |
| CG–B | 8.0 | 6.86 ± 0.66 | 9.55 | −14.3 |
| CG–C | 2.0 | 2.11 ± 0.15 | 7.03 | 5.6 |
| CG–D | 0.2 | 0.15 ± 0.02 | 16.88 | −27.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grazina, L.; Amaral, J.S.; Costa, J.; Mafra, I. Authentication of Ginkgo biloba Herbal Products by a Novel Quantitative Real-Time PCR Approach. Foods 2020, 9, 1233. https://doi.org/10.3390/foods9091233
Grazina L, Amaral JS, Costa J, Mafra I. Authentication of Ginkgo biloba Herbal Products by a Novel Quantitative Real-Time PCR Approach. Foods. 2020; 9(9):1233. https://doi.org/10.3390/foods9091233
Chicago/Turabian StyleGrazina, Liliana, Joana S. Amaral, Joana Costa, and Isabel Mafra. 2020. "Authentication of Ginkgo biloba Herbal Products by a Novel Quantitative Real-Time PCR Approach" Foods 9, no. 9: 1233. https://doi.org/10.3390/foods9091233
APA StyleGrazina, L., Amaral, J. S., Costa, J., & Mafra, I. (2020). Authentication of Ginkgo biloba Herbal Products by a Novel Quantitative Real-Time PCR Approach. Foods, 9(9), 1233. https://doi.org/10.3390/foods9091233