Molecular Characterization and Enological Potential of A High Lactic Acid-Producing Lachancea thermotolerans Vineyard Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Fermentations in Pasteurized Grape Must
2.3. Fermentations in Natural Grape Must
2.4. Microbiological Analysis
2.5. Chemical Analysis
2.6. RNA Preparation and First-Strand cDNA Synthesis
2.7. RT-qPCR Analysis
2.8. Sensory Analysis
2.9. Statistical Analysis
3. Results
3.1. Fermentation Kinetics and Yeast Population Dynamics in Pasteurized Grape Must
3.2. Fermentation Kinetics and Yeast Population Dynamics in Natural Grape Must
3.3. The Effect of Different Inoculation Schemes on the Chemical Profiles of Wines Produced from Pasteurized Grape Must
3.4. The Effect of Different Inoculation Schemes on the Chemical Profiles of Wines Produced from Natural Grape Must
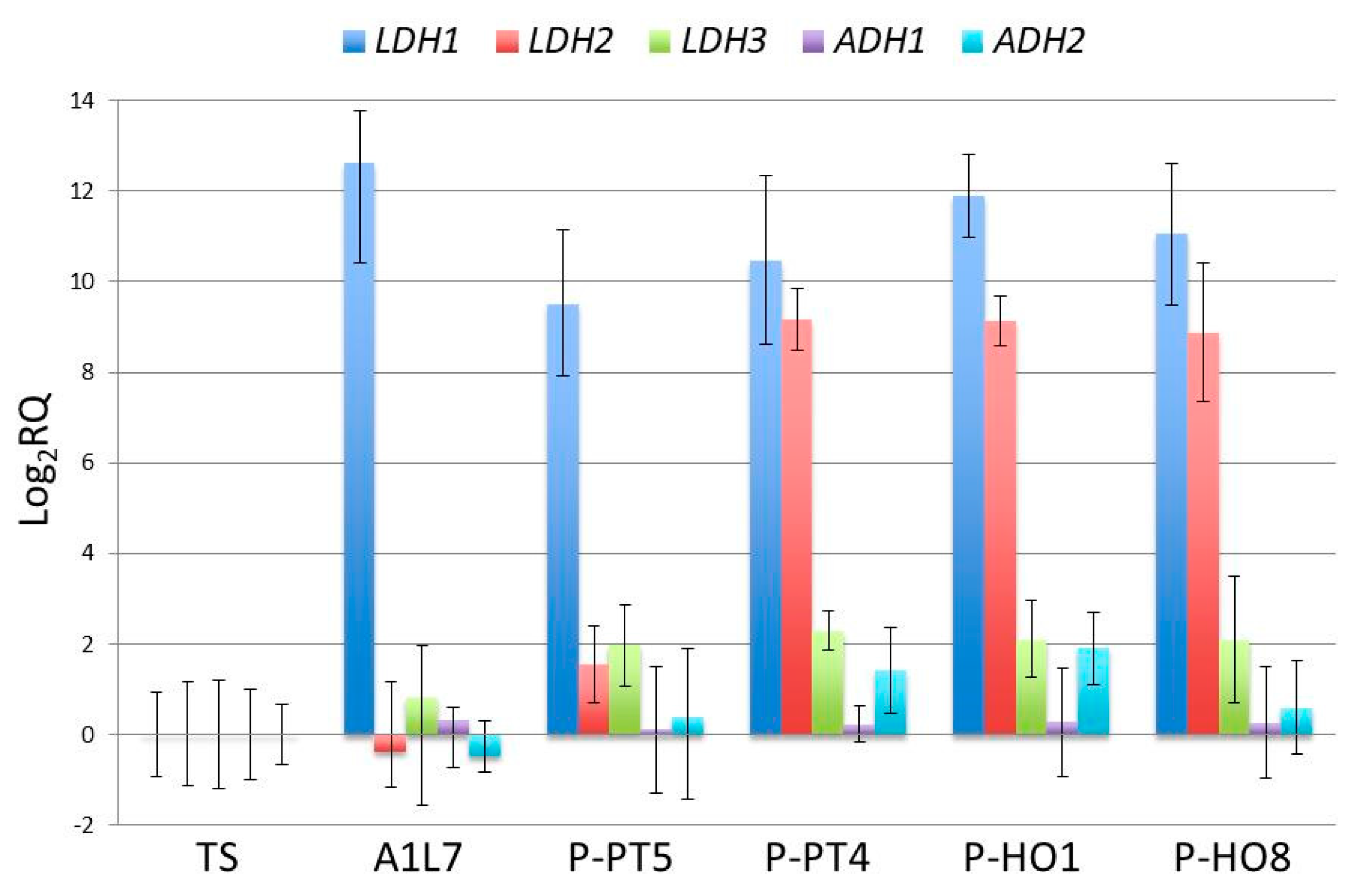
3.5. Transcriptional Analysis of L. thermotolerans LDH and ADH Genes
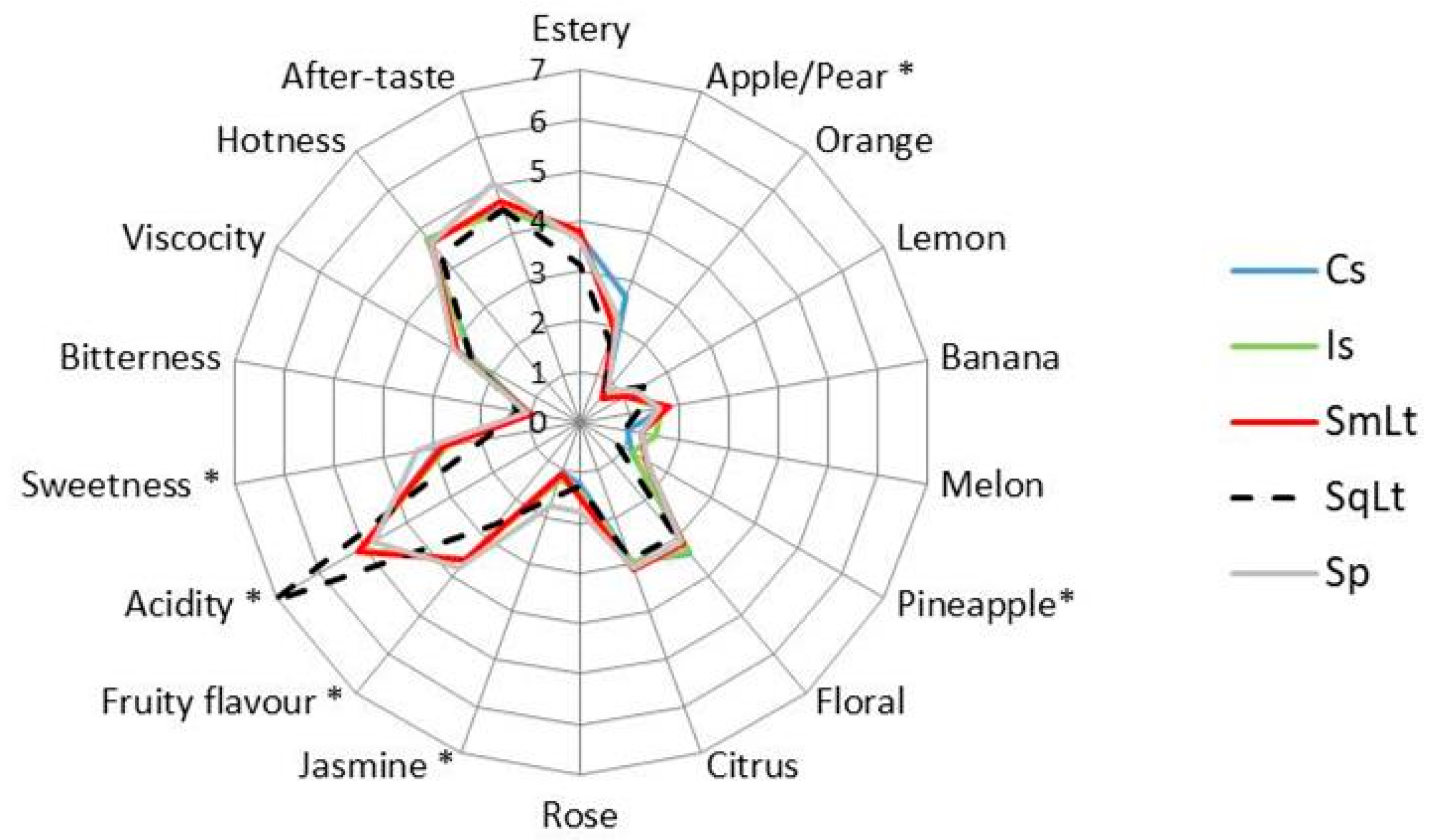
3.6. Sensory Analysis
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nisiotou, A.A.; Spiropoulos, A.E.; Nychas, G.J.E. Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl. Environ. Microbiol. 2007, 73, 6705–6713. [Google Scholar] [CrossRef] [PubMed]
- Sgouros, G.; Chalvantzi, I.; Mallouchos, A.; Paraskevopoulos, Y.; Banilas, G.; Nisiotou, A. Biodiversity and enological potential of non-Saccharomyces yeasts from Nemean vineyards. Fermentation 2018, 4, 32. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Sgouros, G.; Mallouchos, A.; Nisiotis, C.S.; Michaelidis, C.; Tassou, C.; Banilas, G. The use of indigenous Saccharomyces cerevisiae and Starmerella bacillaris strains as a tool to create chemical complexity in local wines. Food Res. Int. 2018, 111, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Nisiotou, A.; Mallouchos, A.; Tassou, C.; Banilas, G. Indigenous Yeast Interactions in Dual-Starter Fermentations May Improve the Varietal Expression of Moschofilero Wine. Front. Microbiol. 2019, 10, 1712. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Tesfaye, W.; Bañuelos, M.A.; González, C.; Suárez Lepe, J.A. Lachancea thermotolerans Applications in Wine Technology. Fermentation 2018, 4, 53. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- Kapsopoulou, K.; Mourtzini, A.; Anthoulas, M.; Nerantzis, E. Biological acidification during grape must fermentation using mixed cultures of Kluyveromyces thermotolerans and Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2007, 23, 735–739. [Google Scholar] [CrossRef]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef]
- Banilas, G.; Sgouros, G.; Nisiotou, A. Development of microsatellite markers for Lachancea thermotolerans typing and population structure of wine-associated isolates. Microbiol. Res. 2016, 193, 1–10. [Google Scholar] [CrossRef]
- Benito, A.; Calderón, F.; Palomero, F.; Benito, S. Quality and composition of Airén wines fermented by sequential inoculation of Lachancea thermotolerans and Saccharomyces cerevisiae. Food Technol. Biotechnol. 2016, 54, 135–144. [Google Scholar] [CrossRef]
- Roudil, L.; Russo, P.; Berbegal, C.; Albertin, W.; Spano, G.; Capozzi, V. Non-Saccharomyces commercial starter cultures: Scientific trends, recent patents and innovation in the wine sector. Recent Pat. Food Nutr. Agric. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Whiting, C. Organic acid metabolism of yeasts during fermentation of alcoholic beverages—A review. J. Inst. Brew. 1976, 86, 2009. [Google Scholar] [CrossRef]
- Hranilovic, A.; Gambetta, J.M.; Schmidtke, L.; Boss, P.K.; Grbin, P.R.; Masneuf-Pomarede, I.; Bely, M.; Albertin, W.; Jiranek, V. Oenological traits of Lachancea thermotolerans show signs of domestication and allopatric differentiation. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Sauer, M.; Porro, D.; Mattanovich, D.; Branduardi, P. 16 years research on lactic acid production with yeast—Ready for the market? Biotechnol. Genet. Eng. Rev. 2010, 27, 229–256. [Google Scholar] [CrossRef]
- Bajpai, P. Biorefinery in the Pulp and Paper Industry; Academic Press: London, UK, 2013. [Google Scholar]
- Budhavaram, N.K.; Fan, Z. Production of lactic acid from paper sludge using acid-tolerant, thermophilic Bacillus coagulan strains. Bioresour. Technol. 2009, 100, 5966–5972. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, J.H. Lactic acid, microbially produced. In Encyclopedia of Microbiology; Schaechter Mosel, O., Ed.; Academic Press: Oxford, UK, 2009; pp. 362–372. [Google Scholar]
- Maas, R.H.; Bakker, R.R.; Eggink, G.; Weusthuis, R.A. Lactic acid production from xylose by the fungus Rhizopus oryzae. Appl. Microbiol. Biotechnol. 2006, 72, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Jina, B.; Kelly, J.M. Production of lactic acid from renewable materials by Rhizopus fungi. Biochem. Eng. J. 2007, 35, 251–263. [Google Scholar] [CrossRef]
- Branduardi, P.; Sauer, M.; De Gioia, L.; Zampella, G.; Valli, M.; Mattanovich, D.; Porro, D. Lactate production yield from engineered yeasts is dependent from the host background, the lactate dehydrogenase source and the lactate export. Microb. Cell Fact. 2006, 5, 4. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterization. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- OIV. Compendium of International Methods of Wine and Must Analysis; International Organisation of Vine and Wine: Paris, France, 2015. [Google Scholar]
- Gump, B.H.; Zoecklein, B.W.; Fugelsang, K.C. Food Microbiology Protocols. In Methods in Biotechnology; Spencer, J.F.T., Ragout de Spencer, A.L., Eds.; Humana Press: Totowa, NJ, USA, 2001; pp. 283–296. [Google Scholar]
- Hjelmeland, A.K.; King, E.S.; Ebeler, S.E.; Heymann, H. Characterizing the chemical and sensory profiles of United States Cabernet Sauvignon wines and blends. Am. J. Enol. Vitic. 2013, 64, 169–179. [Google Scholar] [CrossRef]
- Teste, M.A.; Duquenne, M.; Francois, J.M.; Parrou, J.L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Andorrà, I.; Berradre, M.; Mas, A.; Esteve-Zarzoso, B.; Guillamón, J.M. Effect of mixed culture fermentations on yeast populations and aroma profile. LWT Food Sci. Technol. 2012, 49, 8–13. [Google Scholar] [CrossRef]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: Role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Goold, H.D.; Kroukamp, H.; Williams, T.C.; Paulsen, I.T.; Varela, C.; Pretorius, I.S. Yeast’s balancing act between ethanol and glycerol production in low-alcohol wines. Microb. Biotechnol. 2017, 10, 264–278. [Google Scholar] [CrossRef]
- Balikci, E.K.; Tanguler, H.; Jolly, N.P.; Erten, H. Influence of Lachancea thermotolerans on cv. Emir wine fermentation. Yeast 2016, 33, 313–321. [Google Scholar] [CrossRef]
- Beckner Whitener, M.E.; Stanstrup, J.; Panzeri, V.; Carlin, S.; Divol, B.; Du Toit, M.; Vrhovsek, U. Untangling the wine metabolome by combining untargeted SPME–GCxGC-TOF-MS and sensory analysis to profile Sauvignon blanc co-fermented with seven different yeasts. Metabolomics 2016, 12, 53. [Google Scholar] [CrossRef]
- Whitener, M.E.B.; Stanstrup, J.; Carlin, S.; Divol, B.; Du Toit, M.; Vrhovsek, U. Effect of non-Saccharomyces yeasts on the volatile chemical profile of Shiraz wine. Aust. J. Grape Wine Res. 2017, 23, 179–192. [Google Scholar] [CrossRef]
- Talla, E.; Anthouard, V.; Bouchier, C.; Frangeul, L.; Dujon, B. The complete mitochondrial genome of the yeast Kluyveromyces thermotolerans. FEBS Lett. 2005, 579, 30–40. [Google Scholar] [CrossRef]
- Leskovac, V.; Trivic, S.; Pericin, D. The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 2002, 2, 481–494. [Google Scholar] [CrossRef]
- Turunen, O.; Seelke, R.; Macosko, J. In silico evidence for functional specialization after genome duplication in yeast. FEMS Yeast Res. 2009, 9, 16–31. [Google Scholar] [CrossRef] [PubMed][Green Version]
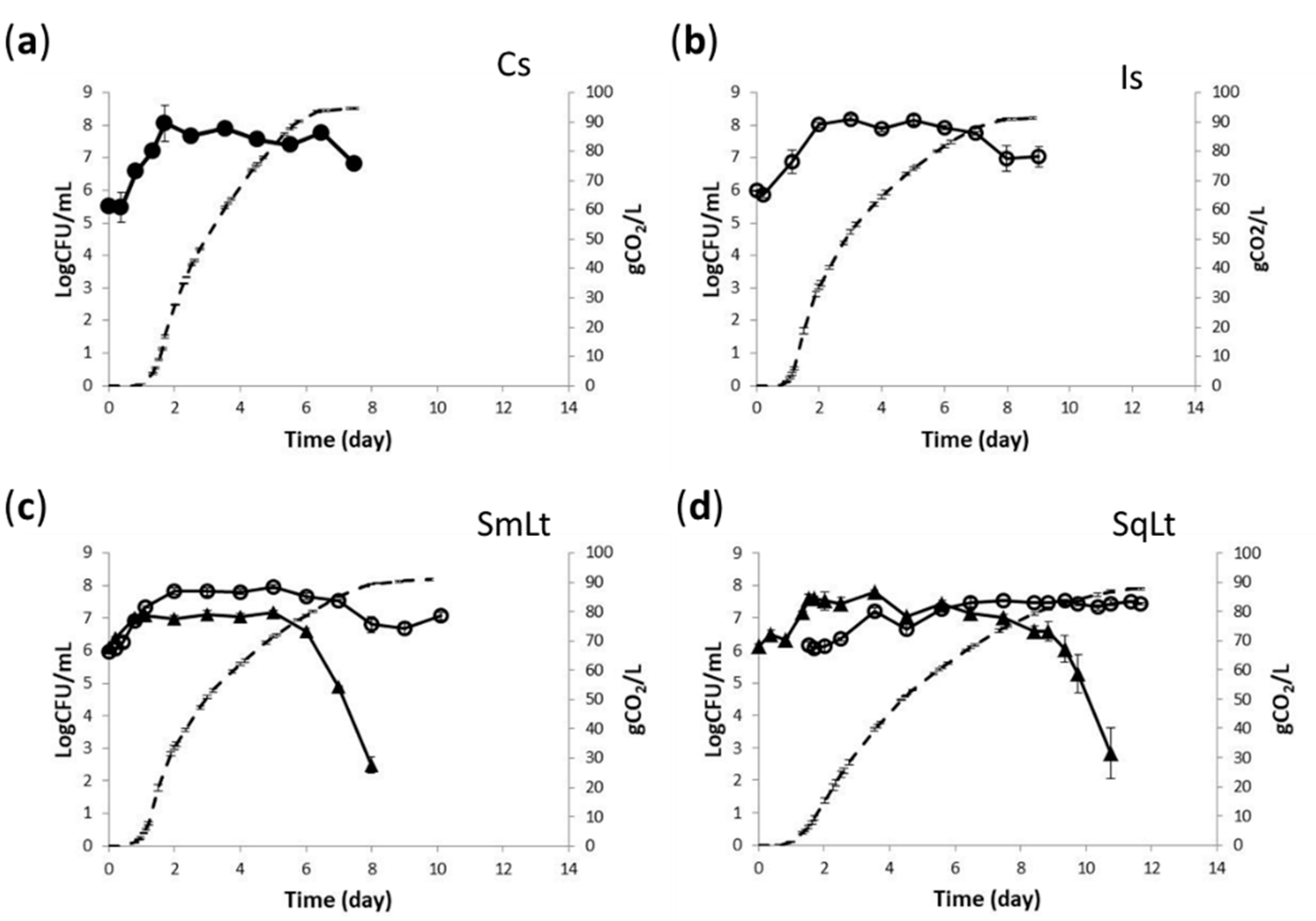
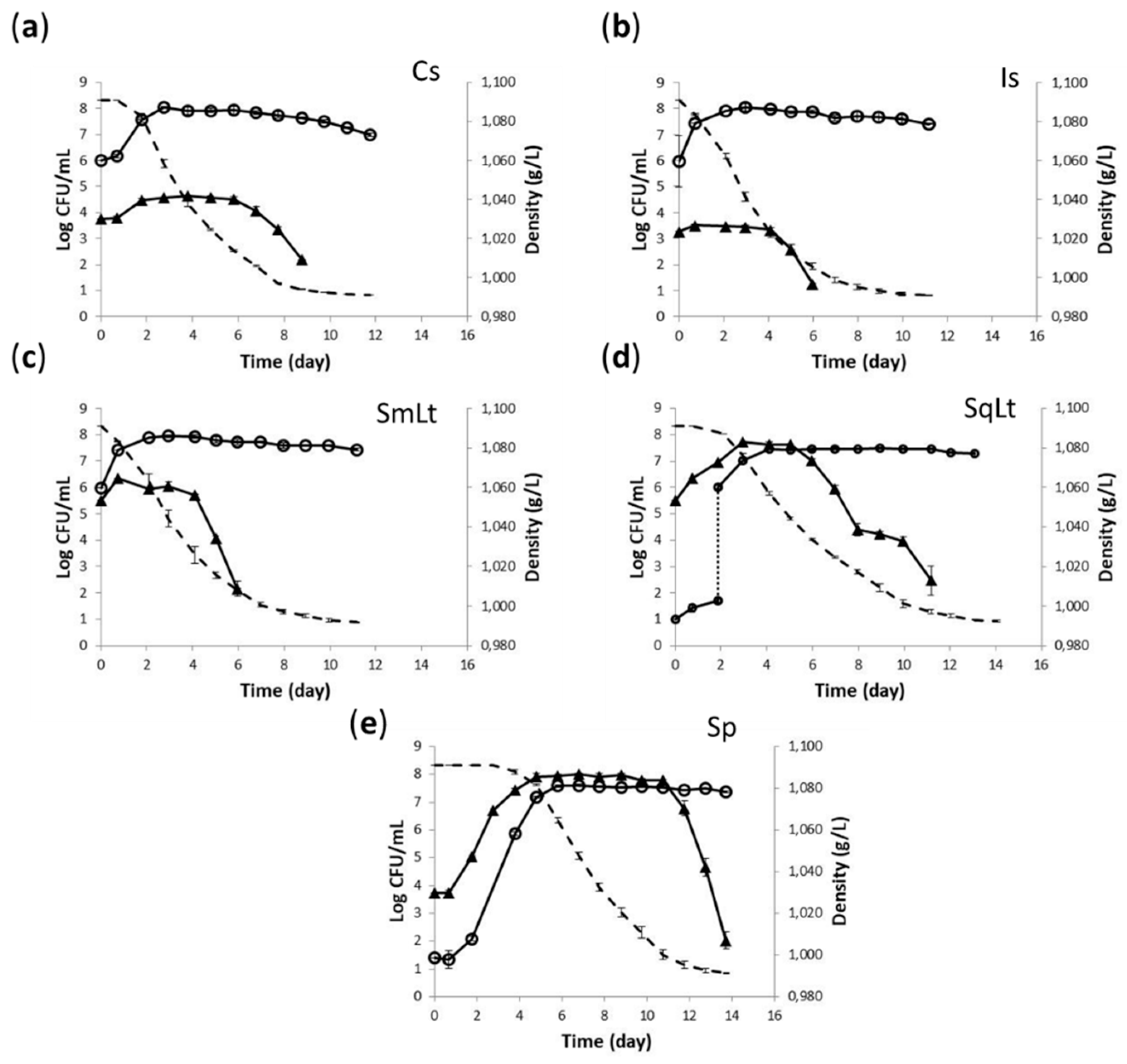
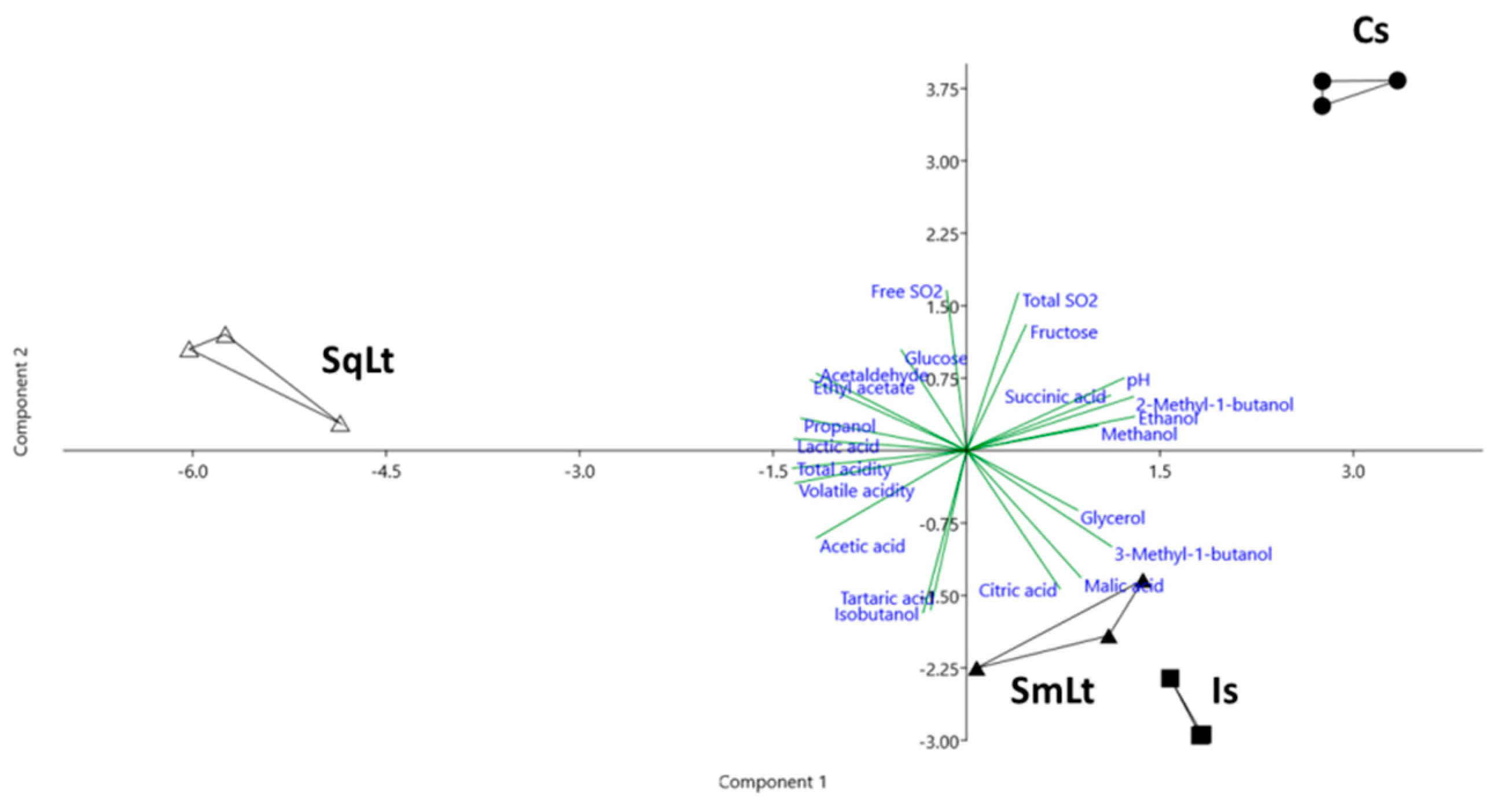
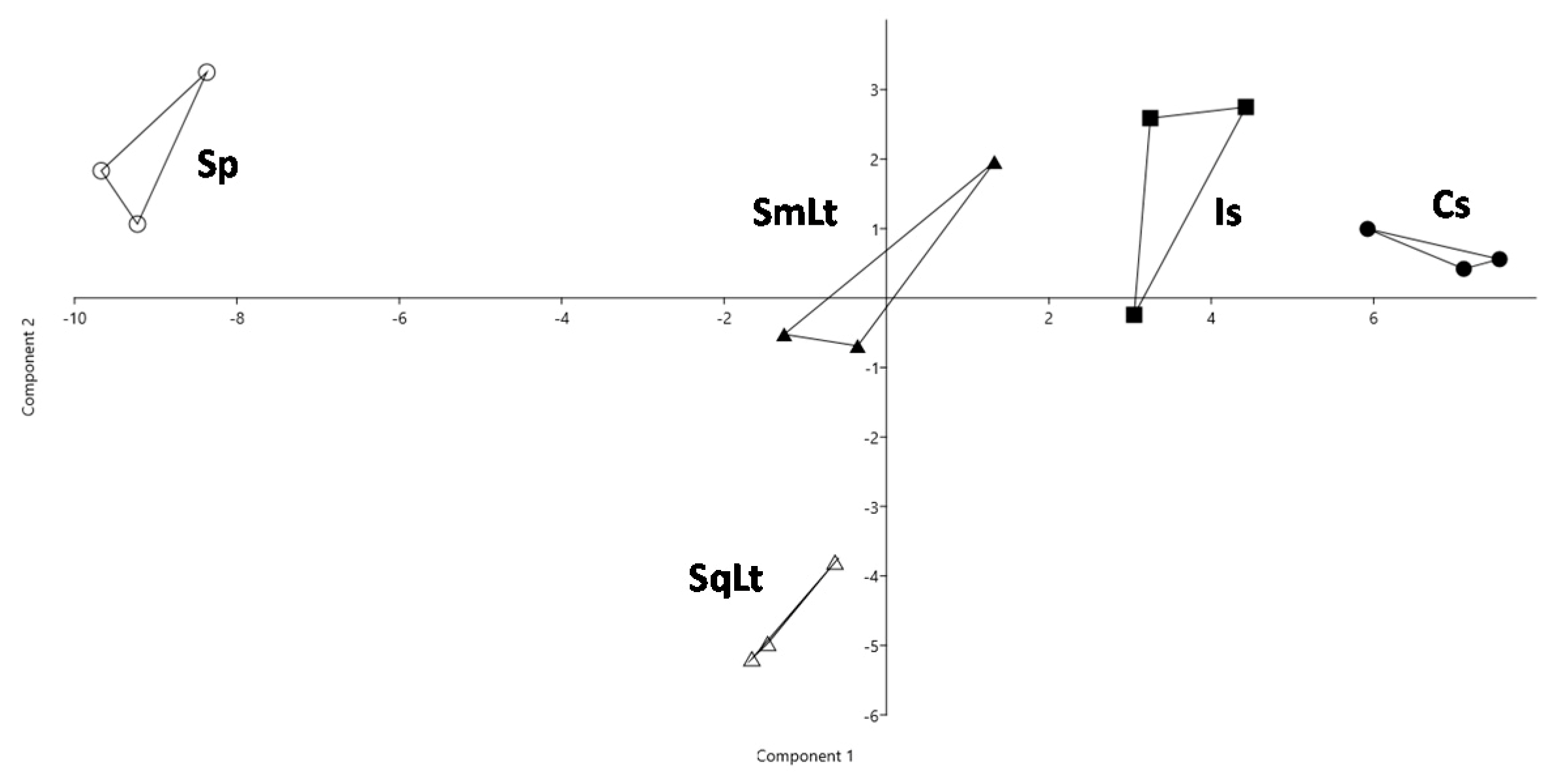

| KEGG Entry/ Gene Name | Protein [KEGG ENZYME] | Forward Primer/ Reverse Primer (5′–3′) |
|---|---|---|
| KLTH0D00440g/LDH1 | L-lactate dehydrogenase [EC:1.1.1.27] | ATTATCACAGGAGGAGCCAATC/ AGAAGCAAAATGGTGTCAGGAG |
| KLTH0G19536g/LDH2 | ATCTACCCACCAGCCTCAAC/ TTTCGCCCTCTGCTTTATGT | |
| KLTH0G19558g/LDH3 | GCAGAGTTATCGGCTCAGGTA/ TGGCTTTTTCGCAGTAATCC | |
| KLTH0B04972g/ADH1 | Alcohol dehydrogenase [EC:1.1.1.2] | GGTGTCTTCGTGCTTGGACT/ TAGGTGCGTTGGGATTTTCT |
| KLTH0E07964g/ADH2 | GCTTCCCTATCCCACCAGAG/ ACGCTTGGAGTAACCACGAG | |
| KLTH0G11352g/TAF10 | Transcription initiation factor TFIID subunit 10 | CACGGTTCACAACTCCAACA/ CGCTGCTTAGGTCGTTCACT |
| Chemical Component | Inoculation Protocol 1 | |||
|---|---|---|---|---|
| Cs | Is | SmLt | SqLt | |
| Total acidity (as tartaric acid g/L) | 6.3 ± 0.1 d | 7.8 ± 0.1 c | 9.2 ± 0.1 b | 15.5 ± 0.1 a |
| pH | 3.53 ± 0.01 a | 3.38 ± 0.01 b | 3.38 ± 0.01 b | 3.24 ± 0.01 c |
| Volatile acidity (as acetic acid g/L) | 0.11 ±0.01 c | 0.16 ±0.00 b | 0.15 ±0.01 b | 0.28 ±0.02 a |
| Free SO2 (mg/L) | 10.0 ± 0.0 a | 6.8 ± 0.8 b | 7.3 ± 0.8 b | 9.0 ± 0.0 a |
| Total SO2 (mg/L) | 22.2 ± 0.7 a | 16.3 ± 0.6 b | 17.5 ± 1.5 b | 18.0 ± 0.1 b |
| Citric acid (mg/L) | 387 ± 3 c | 446 ± 3 a | 418 ± 9 b | 377 ± 10 c |
| Tartaric acid (g/L) | 1.0 ± 0.1 c | 1.3 ± 0.1 a | 1.2 ± 0.1 a | 1.2 ± 0.0 a,b |
| Malic acid (g/L) | 1.9 ± 0.0 c | 2.6 ± 0.0 a | 2.3 ± 0.1 b | 1.6 ± 0.0 d |
| Glucose (g/L) | <0.6 | <0.6 | <0.6 | <0.6 |
| Fructose (g/L) | 1.0 ± 0.0 | <0.6 | <0.6 | <0.6 |
| Succinic acid (g/L) | 1.2 ± 0.0 a | 1.1 ± 0.0 b | 1.2 ± 0.0 a | 1.0 ± 0.0 c |
| Lactic acid (g/L) | < 0.6 | < 0.6 | 2.3 ± 0.1 b | 10.4 ± 0.3 a |
| Glycerol (g/L) | 6.9 ± 0.1 a | 7.0 ± 0.0 a | 7.2 ± 0.2 a | 6.7 ± 0.2 b |
| Acetic acid (mg/L) | < 10 | 68 ± 2 c | 80 ± 5 b | 193 ± 1 a |
| Ethanol (g/L) | 99.4 ± 0.9 a | 96.3 ± 0.4 a | 93.0 ± 0.3 b | 87.0 ± 1.6 c |
| Chemical Component | Inoculation Protocol 1 | |||
|---|---|---|---|---|
| Cs | Is | SmLt | SqLt | |
| Acetaldehyde | 7.5 ± 0.5 b | 6.4 ± 0.7 b | 6.3 ± 0.3 b | 11.0 ± 1.2 a |
| Ethyl acetate | 35.8 ± 2.0 b | 29.8 ± 0.9 c | 32.3 ± 1.1 b | 54.4 ± 1.6 a |
| Methanol | 46.9 ± 2.5 a | 45.7 ± 1.7 a | 43.8 ± 2.9 a | 41.6 ± 2.4 a |
| Propanol | 26.2 ± 1.7 b | 24.4 ± 0.5 c | 28.8 ± 0.2 b | 39.5 ± 1.3 a |
| Isobutanol | 40.2 ± 2.5 c | 69.1 ± 2.0 a | 63.4 ± 1.8 b | 58.9 ± 2.0 b |
| 2-Methyl-1-butanol | 92.8 ± 6.1 a | 71.7 ± 1.3 b | 66.6 ± 2.1 b | 44.4 ± 1.1 c |
| 3-Methyl-1-butanol | 233.2 ± 14.6 c | 306.7 ± 9.3 a | 278.8 ± 6.8 b | 153.4 ± 4.1 d |
| Chemical Component | Inoculation Protocol 1 | ||||
|---|---|---|---|---|---|
| Cs | Is | SmLt | SqLt | Sp | |
| Total acidity (as tartaric acid g/L) | 6.2 ± 0.1 b,c | 6.1± 0.0 c | 6.1 ± 0.1 b,c | 10.2 ± 0.3 a | 6.5 ± 0.1 b |
| pH | 3.34 ± 0.01 a | 3.29 ± 0.01 b | 3.27 ± 0.01 b | 3.15 ± 0.01 c | 3.32 ± 0.01 a |
| Volatile acidity (as acetic acid g/L) | 0.13 ±0.05 c | 0.35 ±0.04 b | 0.37 ±0.02 b | 0.31 ±0.04 b | 0.59 ± 0.04 a |
| Free SO2 (mg/L) | 15.8 ± 1.5 a | 12.8 ± 0.0 a | 14.1 ± 1.3 a | 15.8 ± 0.7 a | 14.5 ± 1.5 a |
| Total SO2 (mg/L) | 24.3 ± 1.3 c | 29.9 ± 1.5 a | 29.9 ± 0.7 a | 29.4 ± 1.3 b | 25.2 ± 2.7 b,c |
| Citric acid (mg/L) | 668 ± 3 a | 657 ± 10 a | 651 ± 4 a | 621 ± 4 b | 664 ± 9 a |
| Tartaric acid (g/L) | 1.3 ± 0.2 b | 1.9 ± 0.2 a | 1.8 ± 0.2 a | 1.6 ± 0.1 a,b | 1.3 ± 0.0 b |
| Malic acid (g/L) | 2.1 ± 0.0 a | 2.1 ± 0.1 a,b | 2.0 ± 0.0 b | 1.8 ± 0.0 d | 1.9 ± 0.0 c |
| Glucose (g/L) | <0.6 | <0.6 | <0.6 | <0.6 | <0.6 |
| Fructose (g/L) | 1.0 ± 0.1 a | 0.9 ± 0.3 a | 0.9 ± 0.3 a | 1.3 ± 0.3 a | 1.0 ± 0.6 a |
| Succinic acid (g/L) | 0.7 ± 0.0 a | <0.6 | <0.6 | 0.6± 0.0 a | <0.6 |
| Lactic acid (g/L) | < 0.6 | < 0.6 | < 0.6 | 5.5 ± 0.4 | < 0.6 |
| Glycerol (g/L) | 6.6 ± 0.0 b | 5.7 ± 0.1 c | 5.7 ± 0.1 c | 6.7 ± 0.1 b | 8.3 ± 0.2 a |
| Acetic acid (mg/L) | 299 ± 10 c | 525 ± 22 b | 522 ± 8 b | 522 ± 18 b | 737 ± 10 a |
| Ethanol (g/L) | 104.7 ± 0.3 b,c | 106.7 ± 0.9 a | 106.0 ± 0.6 a,b | 102.2 ± 0.5 d | 104.2 ± 0.3 c |
| Compound 2 | Inoculation Protocol 3 | ||||
|---|---|---|---|---|---|
| Cs | Is | SmLt | SqLt | Sp | |
| Esters | |||||
| Ethyl acetate (mg/L) | 90.3 ± 5.3 a | 90.9 ± 12.6 a | 86.2 ± 31.4 a | 87.0 ± 14.9 a | 62.6 ± 4.5 a |
| Isobutyl acetate | 130 ± 13 a,b | 167 ± 28 a | 137 ± 19 a,b | 101 ± 16 b | 45 ± 4 c |
| 3-Methylbutyl acetate (mg/L) | 5.8 ± 0.5 a | 5.5 ± 0.3 a | 3.8 ± 0.4 b | 3.3 ± 0.6 b | 0.5 ± 0.4 c |
| Hexyl acetate | 235 ± 15 a | 212 ± 15 a | 157 ± 17 b | 107 ± 6 c | 83 ± 3 c |
| 2-Phenylethyl acetate (mg/L) | 1.9 ± 0.1 a,b | 2.0 ± 0.1 a | 1.5 ± 0.3 b | 0.6 ± 0.1 c | 0.3 ± 0.0 c |
| Diethyl succinate | 41 ± 1 b | 59 ± 12 a,b | 104 ± 48 a | 47 ± 5 a,b | 66 ± 6 a,b |
| Propyl acetate | 88 ± 11 a | 70 ± 5 a,b | 53 ± 8 b | 68 ± 9 a,b | 26 ± 2 c |
| Butyl acetate | 24 ± 3 a | 18 ± 2 b | 14 ± 2 b | 18 ± 3 a,b | 7 ± 1 c |
| (3Z)-3-Hexenyl acetate | 737 ± 53 a | 741 ± 59 a | 495 ± 73 b | 149 ± 31 c | 55 ± 7 c |
| Ethyl isobutyrate | 93 ± 3 a,b | 92 ± 2 b | 90 ± 2 b | 101 ± 5 a | 96 ± 2 a,b |
| Ethyl butanoate | 578 ± 46 a,b | 609 ± 21 a | 487 ± 37 b,c | 375 ± 34 d | 472 ± 34 c |
| Ethyl hexanoate (mg/L) | 1.1 ± 0.1 a | 0.9 ± 0.1 a,b | 0.8 ± 0.1 b | 0.4 ± 0.1 c | 0.9 ± 0.1 a,b |
| Ethyl octanoate (mg/L) | 1.6 ± 0.1 a | 1.5 ± 0.3 a | 1.3 ± 0.2 a | 0.8 ± 0.1 b | 1.4 ± 0.2 a |
| Ethyl dec-9-enoate (mg/L) | 1.7 ± 0.2 a | 4.1 ± 1.7 a | 4.0 ± 1.1 a | 2.4 ± 0.1 a | 3.8 ± 0.8 a |
| Ethyl decanoate | 938 ± 174 a | 929 ± 192 a | 823 ± 265 a | 767 ± 37 a | 938 ± 153 a |
| Ethyl lactate (mg/L) | 2.2 ± 0.2 b | 2.1 ± 0.3 b | 3.9 ± 1.0 b | 112.4 ± 19.0 a | 1.3 ± 0.1 b |
| Isopentyl hexanoate | 35 ± 2 a | 26 ± 8 a,b | 21 ± 5 b | 20 ± 2 b | 19 ± 4 b |
| Methyl hexanoate | 58 ± 8 a | 42 ± 3 b,c | 33 ± 4 b,c | 31 ± 6 c | 45 ± 4 a,b |
| 3-Methylbutyl octanoate | 281 ± 78 a | 279 ± 67 a | 235 ± 67 a | 162 ± 18 a | 163 ± 30 a |
| Ethyl dodecanoate (mg/L) | 5.5 ± 2.3 a | 4.4 ± 1.2 a | 5.1 ± 2.1 a | 3.1 ± 0.3 a | 6.0 ± 0.8 a |
| Alcohols | |||||
| 1-Propanol (mg/L) | 40.9 ± 3.5 a,b | 30.7 ± 4.7 a,b | 29.2 ± 7.2 b | 44.1 ± 7.4 a | 40.1 ± 0.5 a,b |
| 2-Methyl-1-propanol (mg/L) | 33.0 ± 2.9 a | 55.8 ± 23.2 a | 52.5 ± 12.8 a | 52.8 ± 10.0 a | 32.6 ± 1.8 a |
| 3-Methyl-1-butanol (mg/L) | 159.2 ± 15.7 a,b | 182.8 ± 18.9 a | 123.4 ± 29.0 b,c | 146.2 ± 16.0 a,b | 92.8 ± 9.4 c |
| 1-Butanol | 19 ± 1 b | 12 ± 1 c | 12 ± 3 c | 29 ± 4 a | 9 ± 1 c |
| 1-Hexanol | 318 ± 21 b | 317 ± 19 b | 379 ± 30 b | 553 ± 19 a | 545 ± 45 a |
| (Z)-3-Hexen-1-ol | 437 ± 29 a | 462 ± 34 a | 466 ± 40 a | 490 ± 8 a | 497 ± 35 a |
| 2-Ethyl-1-hexanol | 55 ± 2 a | 53 ± 1 a | 53 ± 1 a | 53 ± 1 a | 52 ± 0 a |
| 2,3-Butanediol | 102 ± 18 a | 67 ± 18 a | 82 ± 36 a | 60 ± 12 a | 91 ± 5 a |
| 2-Phenylethanol (mg/L) | 50.0 ± 4.9 a | 43.5 ± 3.8 a,b | 37.7 ± 5.4 b | 38.0 ± 2.3 b | 22.1 ± 1.8 c |
| 1-Nonanol | 24 ± 2 a | 14 ± 1 b | 13 ± 4 b | 8 ± 0 b | 12 ± 3 b |
| Acids | |||||
| Acetic acid (mg/L) | 1.2 ± 0.5 b | 4.4 ± 2.3 ab | 2.6 ± 0.2 b | 2.3 ± 0.4 b | 7.2 ± 1.5 a |
| 2-Methylpropanoic acid | 162 ± 36 a,b | 173 ± 66 ab | 129 ± 22 b | 252 ± 17 a | 177 ± 9 a,b |
| Butanoic acid | 116 ± 37 a,b,c | 162 ± 17 a | 144 ± 11 a,b | 70 ± 17 c | 93 ± 27 b,c |
| 3-Methylbutanoic acid | 114 ± 45 a | 92 ± 17 a | 63 ± 9 a,b | 63 ± 4 a,b | 26 ± 2 b |
| 2-Methylbutanoic acid | 56 ± 16 a | 38 ± 7 a,b,c | 31 ± 7 b,c | 43 ± 2 a,b | 19 ± 3 c |
| Hexanoic acid (mg/L) | 1.0 ± 0.3 c | 3.0 ± 0.2 a | 2.7 ± 0.1 a,b | 1.6 ± 0.3 c | 2.4 ± 0.2 b |
| Octanoic acid (mg/L) | 0.5 ± 0.3 d | 3.6 ± 0.4 a | 3.1 ± 0.4 a,b | 2.0 ± 0.3 c | 2.4 ± 0.2 b,c |
| Decanoic acid | 50 ± 30 c | 854 ± 187 a | 629 ± 156 a,b | 898 ± 70 a | 482 ± 48 b |
| Terpenes | |||||
| α-Phellandrene | 47 ± 16 a | 40 ± 15 a | 36 ± 6 a | 31 ± 7 a | 47 ± 8 a |
| β-Myrcene | 34 ± 11 a | 28 ± 10 a | 25 ± 4 a | 22 ± 5 a | 34 ± 5 a |
| D-Limonene | 42 ± 8 a | 29 ± 11 a,b | 24 ± 3 a,b | 22 ± 4 b | 33 ± 5 a,b |
| α-Pinene | 44 ± 1 a | 39 ± 1 b | 38 ± 0 b | 38 ± 0 b | 39 ± 1 b |
| γ-Terpinene | 36 ± 3 a | 24 ± 2 b | 23 ± 0 b | 22 ± 1 b | 23 ± 0 b |
| p-Cymene | 35 ± 4 a | 17 ± 4 b | 14 ± 0 b | 14 ± 0 b | 15 ± 0 b |
| Linalool | 62 ± 2 a | 59 ± 0 b | 58 ± 1 b | 60 ± 0 a,b | 60 ± 1 a,b |
| α-Terpineol | 23 ± 4 a | 15 ± 5 ab | 11 ± 3 b | 13 ± 2 a,b | 12 ± 3 b |
| β-Citronellol | 14 ± 6 a | 3 ± 1 b | 3 ± 0 b | 3 ± 0 b | 9 ± 1 a,b |
| β-Damascenone | 72 ± 4 a | 77 ± 11 a | 71 ± 15 a | 97 ± 11 a | 76 ± 7 a |
| Miscellaneous | |||||
| Acetaldehyde | 24 ± 2 c | 72 ± 14 a | 68 ± 18 a,b | 40 ± 7 b,c | 18 ± 2 c |
| 1,1-Diethoxy-ethane | 26 ± 3 a | 14 ± 1 b | 14 ± 4 b | 18 ± 4 a,b | 16 ± 1 b |
| 2,3-Butanedione | 12 ± 1 a | 12 ± 2 a | 8 ± 1 a,b | 7 ± 1 b | 10 ± 1 a,b |
| 2-Octanone | 191 ± 18 a | 180 ± 23 a | 127 ± 2 b | 145 ± 17 a,b | 166 ± 22 a,b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sgouros, G.; Mallouchos, A.; Filippousi, M.-E.; Banilas, G.; Nisiotou, A. Molecular Characterization and Enological Potential of A High Lactic Acid-Producing Lachancea thermotolerans Vineyard Strain. Foods 2020, 9, 595. https://doi.org/10.3390/foods9050595
Sgouros G, Mallouchos A, Filippousi M-E, Banilas G, Nisiotou A. Molecular Characterization and Enological Potential of A High Lactic Acid-Producing Lachancea thermotolerans Vineyard Strain. Foods. 2020; 9(5):595. https://doi.org/10.3390/foods9050595
Chicago/Turabian StyleSgouros, Georgios, Athanasios Mallouchos, Maria-Evangelia Filippousi, Georgios Banilas, and Aspasia Nisiotou. 2020. "Molecular Characterization and Enological Potential of A High Lactic Acid-Producing Lachancea thermotolerans Vineyard Strain" Foods 9, no. 5: 595. https://doi.org/10.3390/foods9050595
APA StyleSgouros, G., Mallouchos, A., Filippousi, M.-E., Banilas, G., & Nisiotou, A. (2020). Molecular Characterization and Enological Potential of A High Lactic Acid-Producing Lachancea thermotolerans Vineyard Strain. Foods, 9(5), 595. https://doi.org/10.3390/foods9050595





