Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Strawberry Fruit
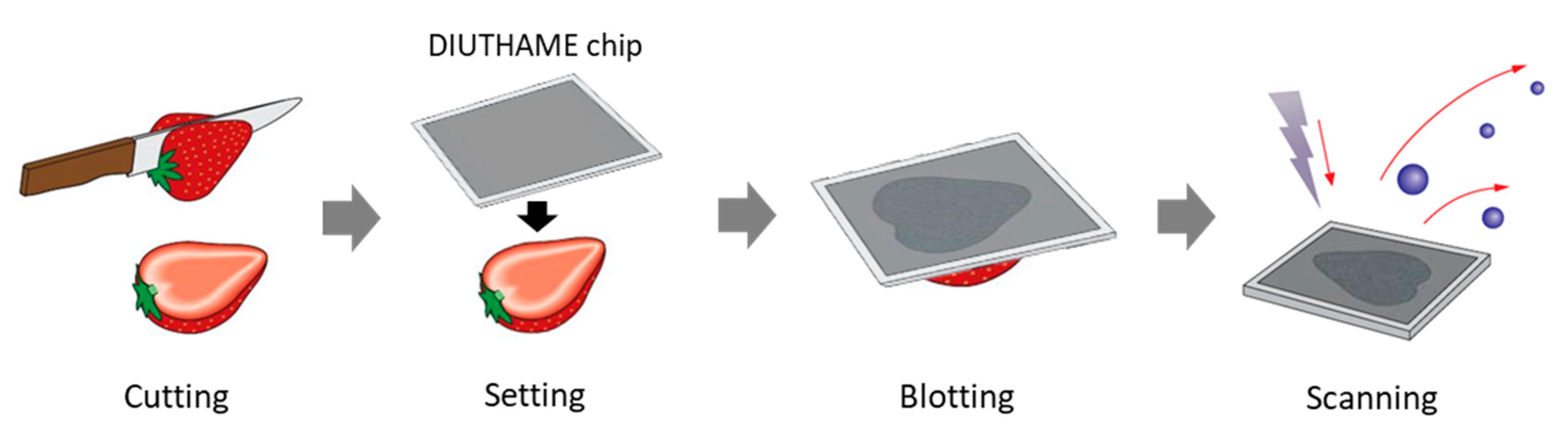
2.3. Preparation of DIUTHAME Chip
2.4. Sample Preparation
2.5. MSI Analysis
3. Results and Discussion
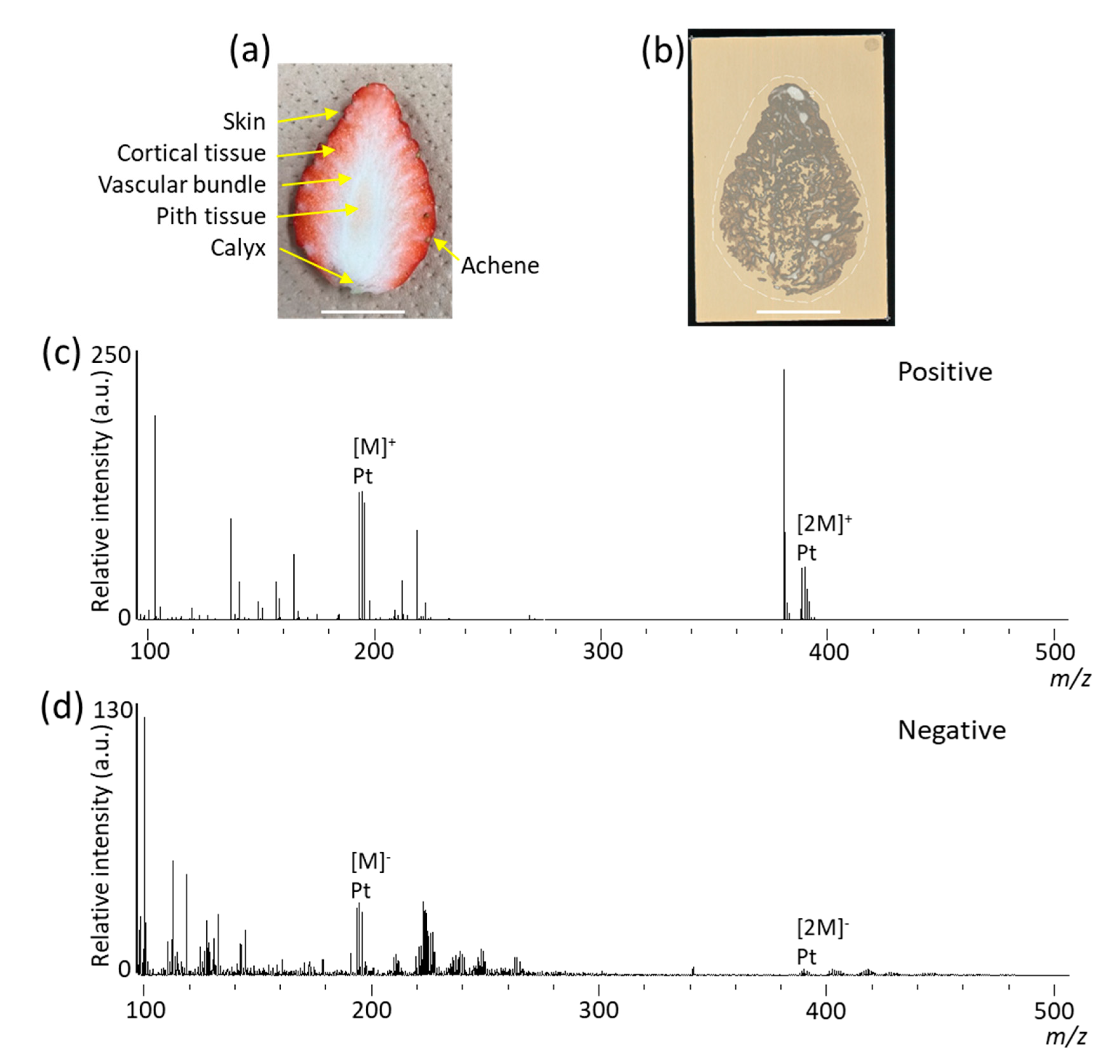
3.1. Blotting on DIUTHAME Chip in MSI Analysis
3.2. Distribution of Metabolites in Strawberry Fruit
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, J.; Liu, X.; Yang, T.; Slovin, J.; Chen, P. Profiling polyphenols of two diploid strawberry (Fragaria vesca) inbred lines using UHPLC-HRMSn. Food Chem. 2014, 146, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Stoeckli, M.; Chaurand, P.; Hallahan, D.E.; Caprioli, R.M. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001, 7, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Cornett, D.S.; Reyzer, M.L.; Chaurand, P.; Caprioli, R.M. MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat. Methods 2007, 4, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Setou, M.; Shrivas, K.; Sroyraya, M.; Yang, H.; Sugiura, Y.; Moribe, J.; Kondo, A.; Tsutsumi, K.; Kimura, Y.; Kurabe, N.; et al. Developments and applications of mass microscopy. Med. Mol. Morphol. 2010, 43, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Sugiura, Y.; Setou, M.; Zaima, N. Visualization of phosphatidylcholine, lysophosphatidylcholine and sphingomyelin in mouse tongue body by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 2011, 400, 1913–1921. [Google Scholar] [CrossRef]
- Enomoto, H.; Sensu, T.; Yumoto, E.; Yokota, T.; Yamane, H. Derivatization for detection of abscisic acid and 12-oxo-phytodienoic acid using matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 2018, 32, 1565–1572. [Google Scholar] [CrossRef]
- Zaima, N.; Sasaki, T.; Tanaka, H.; Cheng, X.W.; Onoue, K.; Hayasaka, T.; Goto-Inoue, N.; Enomoto, H.; Unno, N.; Kuzuya, M.; et al. Imaging mass spectrometry-based histopathologic examination of atherosclerotic lesions. Atherosclerosis 2011, 217, 427–432. [Google Scholar] [CrossRef]
- Zaima, N.; Hayasaka, T.; Goto-Inoue, N.; Setou, M. Matrix-assisted laser desorption/ionization imaging mass spectrometry. Int. J. Mol. Sci. 2010, 11, 5040–5055. [Google Scholar] [CrossRef] [Green Version]
- Morisasa, M.; Sato, T.; Kimura, K.; Mori, T.; Goto-Inoue, N. Application of matrix-assisted laser desorption/ionization mass spectrometry imaging for food analysis. Foods 2019, 8, 633. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, Y.; Goto-Inoue, N.; Moriyama, T.; Zaima, N. Significant advancement of mass spectrometry imaging for food chemistry. Food Chem. 2016, 210, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Enomoto, H.; Moriyama, T.; Kawamura, Y.; Setou, M.; Zaima, N. Visualization of anthocyanin species in rabbiteye blueberry Vaccinium ashei by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Zaima, N.; Goto-Inoue, N.; Hayasaka, T.; Enomoto, H.; Setou, M. Authenticity assessment of beef origin by principal component analysis of matrix-assisted laser desorption/ionization mass spectrometric data. Anal. Bioanal. Chem. 2011, 400, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Sato, K.; Miyamoto, K.; Ohtsuka, A.; Yamane, H. Distribution analysis of anthocyanins, sugars, and organic acids in strawberry fruits using matrix-assisted laser desorption/ionization-imaging mass spectrometry. J. Agric. Food Chem. 2018, 66, 4958–4965. [Google Scholar] [CrossRef]
- Enomoto, H.; Takahashi, S.; Takeda, S.; Hatta, H. Distribution of flavan-3-ol species in ripe strawberry fruit revealed by matrix-assisted laser desorption/ionization-mass spectrometry imaging. Molecules 2020, 25, 103. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, H.; Takeda, S.; Hatta, H.; Zaima, N. Tissue-specific distribution of sphingomyelin species in pork chop revealed by matrix-assisted laser desorption/ionization-imaging mass spectrometry. J. Food Sci. 2019, 84, 1758–1763. [Google Scholar] [CrossRef]
- Enomoto, H.; Furukawa, T.; Takeda, S.; Hatta, H.; Zaima, N. Unique distribution of diacyl-, alkylacyl-, and alkenylacyl-phosphatidylcholine species visualized in pork chop tissues by matrix-assisted laser desorption/ionization–mass spectrometry imaging. Foods 2020, 9, 205. [Google Scholar] [CrossRef] [Green Version]
- Cooks, R.G.; Ouyang, Z.; Takats, Z.; Wiseman, J.M. Ambient mass spectrometry. Science 2006, 311, 1566–1570. [Google Scholar] [CrossRef]
- Enomoto, H.; Sensu, T.; Sato, K.; Sato, F.; Paxton, T.; Yumoto, E.; Miyamoto, K.; Asahina, M.; Yokota, T.; Yamane, H. Visualisation of abscisic acid and 12-oxo-phytodienoic acid in immature Phaseolus vulgaris L. seeds using desorption electrospray ionisation-imaging mass spectrometry. Sci. Rep. 2017, 7, 42977. [Google Scholar] [CrossRef]
- Giampieri, F.; Tulipani, S.; Alvarez-Suarez, J.M.; Quiles, J.L.; Mezzetti, B.; Battino, M. The strawberry: Composition, nutritional quality, and impact on human health. Nutrition 2012, 28, 9–19. [Google Scholar] [CrossRef]
- Giampieri, F.; Alvarez-Suarez, J.M.; Battino, M. Strawberry and human health: Effects beyond antioxidant activity. J. Agric. Food Chem. 2014, 62, 3867–3876. [Google Scholar] [CrossRef]
- Naito, Y.; Kotani, M.; Ohmura, T. A novel laser desorption/ionization method using through hole porous alumina membranes. Rapid Commun. Mass Spectrom. 2018, 32, 1851–1858. [Google Scholar] [CrossRef]
- Kuwata, K.; Itou, K.; Kotani, M.; Ohmura, T.; Naito, Y. DIUTHAME enables matrix-free mass spectrometry imaging of frozen tissue sections. Rapid Commun. Mass Spectrom. 2020, e8729. [Google Scholar] [CrossRef] [PubMed]
- Cabral, E.C.; Mirabelli, M.F.; Perez, C.J.; Ifa, D.R. Blotting assisted by heating and solvent extraction for DESI-MS imaging. J. Am. Soc. Mass Spectrom. 2013, 24, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Nizioł, J.; Misiorek, M.; Ruman, T. Mass spectrometry imaging of low molecular weight metabolites in strawberry fruit (Fragaria x ananassa Duch.) cv. Primoris with 109Ag nanoparticle enhanced target. Phytochemistry 2019, 159, 11–19. [Google Scholar] [CrossRef]
- Ye, H.; Gemperline, E.; Venkateshwaran, M.; Chen, R.; Delaux, P.M.; Howes-Podoll, M.; Ané, J.M.; Li, L. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant J. 2013, 75, 130–145. [Google Scholar] [CrossRef] [PubMed]
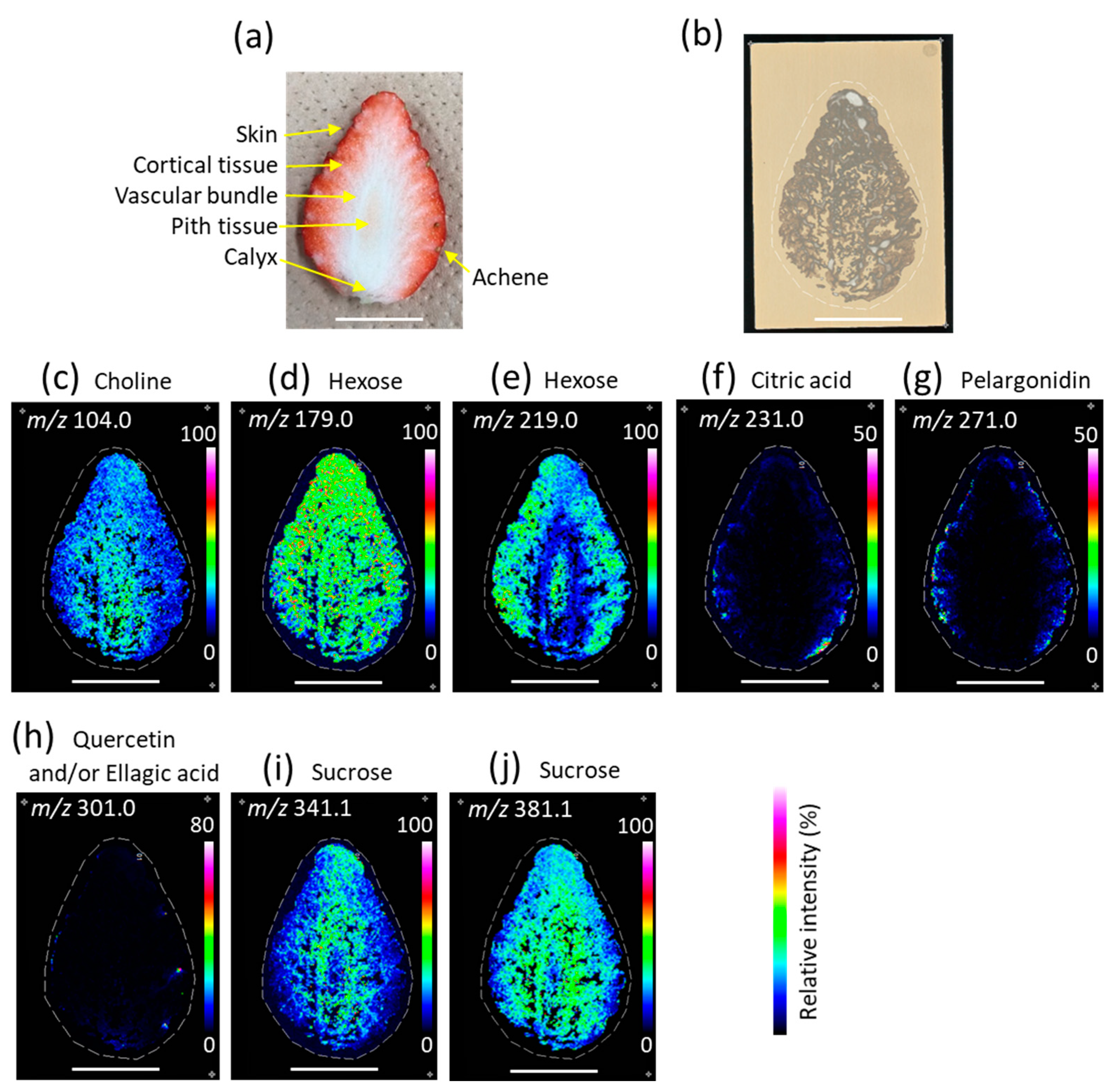
| m/z | Ion type | Metabolites 1 |
|---|---|---|
| 104.0 | [M]+ | Choline |
| 179.0 | [M − H]– | Hexose (glucose and/or fructose) |
| 219.0 | [M + K]+ | Hexose (glucose and/or fructose) |
| 231.0 | [M + K]+ | Citric acid |
| 271.0 | [M]+ | Pelargonidin |
| 301.0 | [M − H]– | Quercetin and/or ellagic acid |
| 341.1 | [M − H]– | Sucrose |
| 381.1 | [M + K]+ | Sucrose |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enomoto, H.; Kotani, M.; Ohmura, T. Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane. Foods 2020, 9, 408. https://doi.org/10.3390/foods9040408
Enomoto H, Kotani M, Ohmura T. Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane. Foods. 2020; 9(4):408. https://doi.org/10.3390/foods9040408
Chicago/Turabian StyleEnomoto, Hirofumi, Masahiro Kotani, and Takayuki Ohmura. 2020. "Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane" Foods 9, no. 4: 408. https://doi.org/10.3390/foods9040408
APA StyleEnomoto, H., Kotani, M., & Ohmura, T. (2020). Novel Blotting Method for Mass Spectrometry Imaging of Metabolites in Strawberry Fruit by Desorption/Ionization Using Through Hole Alumina Membrane. Foods, 9(4), 408. https://doi.org/10.3390/foods9040408






