Application of Mass Spectrometry Imaging for Visualizing Food Components
Abstract
1. Introduction
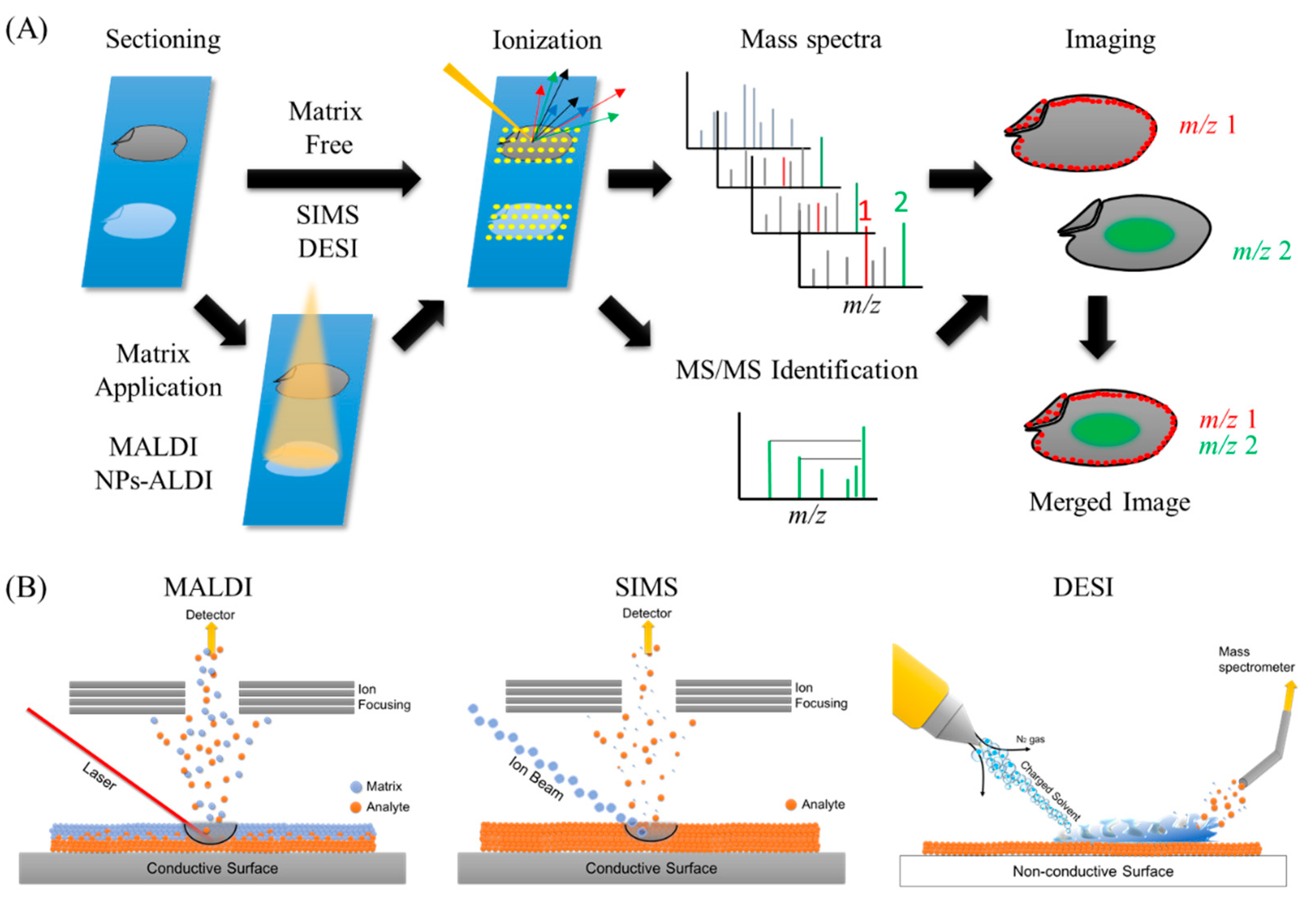
2. History of MSI and Widely Used Ionization Methods
3. Application of MSI in Food Science and Related Fields
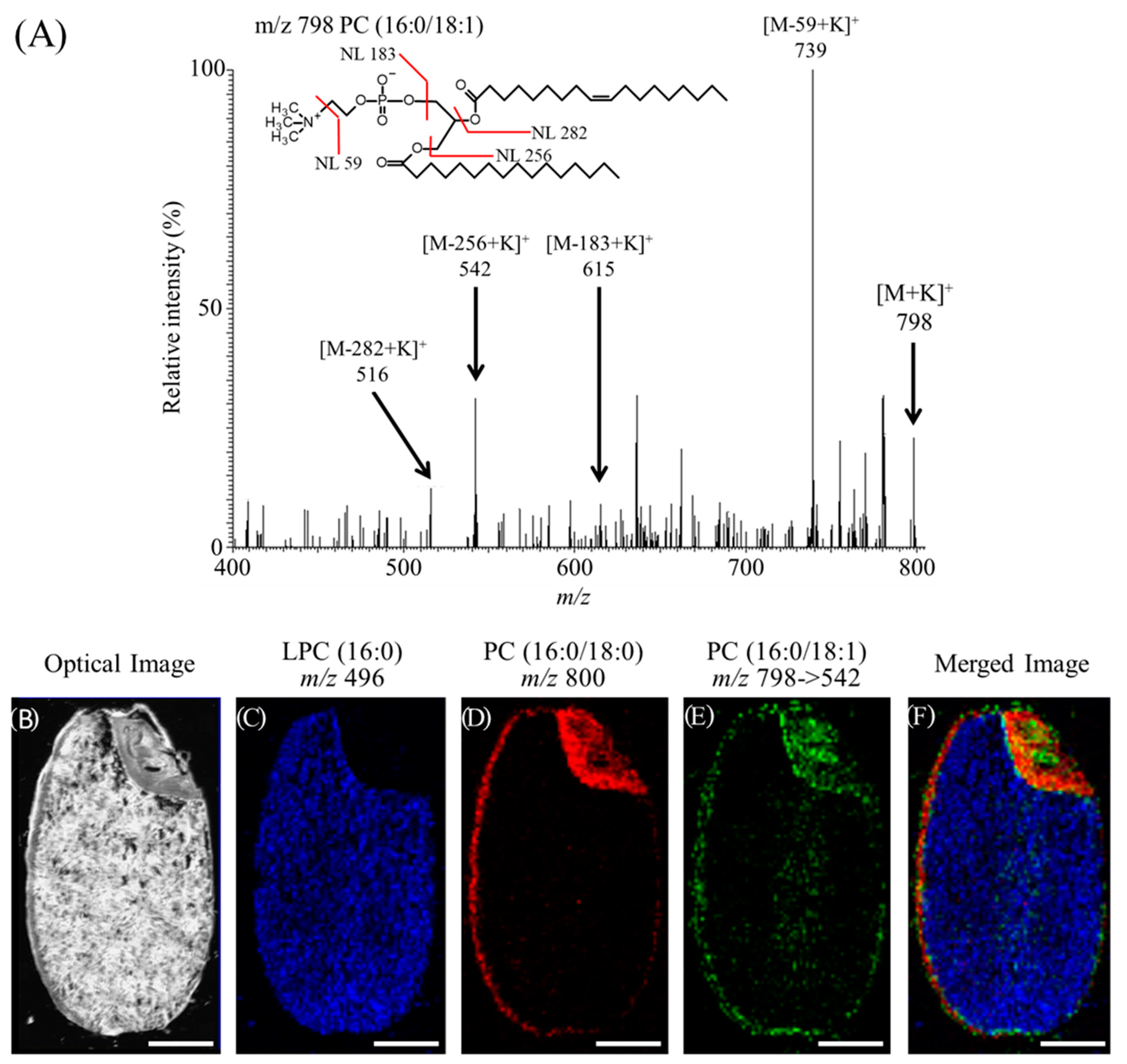
3.1. Lipids
3.2. Carbohydrates
3.3. Proteins and Peptides
3.4. Micronutrients
3.5. Functional Food Factors
3.6. Endogenous Food Toxins and Exogenous Contaminants
3.7. Evaluation of Acidity in Food Product Using MSI
3.8. Imaging of Administered Nutritional Factors
4. Conclusions and Future Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Ahtesh, F.B.; Stojanovska, L.; Apostolopoulos, V. Anti-hypertensive peptides released from milk proteins by probiotics. Maturitas 2018, 115, 103–109. [Google Scholar] [CrossRef]
- Leri, M.; Scuto, M.; Ontario, M.L.; Calabrese, V.; Calabrese, E.J.; Bucciantini, M.; Stefani, M. Healthy Effects of Plant Polyphenols: Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 1250. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Talukdar, S.; Bae, E.J.; Imamura, T.; Morinaga, H.; Fan, W.; Li, P.; Lu, W.J.; Watkins, S.M.; Olefsky, J.M. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanchez, L.; Garcia-Fuentes, E.; Rojo-Martinez, G.; Cardona, F.; Soriguer, F.; Tinahones, F.J. Inverse relation between levels of anti-oxidized-LDL antibodies and eicosapentanoic acid (EPA). Br. J. Nutr. 2008, 100, 585–589. [Google Scholar] [CrossRef]
- Kugo, H.; Zaima, N.; Mouri, Y.; Tanaka, H.; Yanagimoto, K.; Urano, T.; Unno, N.; Moriyama, T. The preventive effect of fish oil on abdominal aortic aneurysm development. Biosci. Biotechnol. Biochem. 2016, 80, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Kugo, H.; Zaima, N.; Onozato, M.; Miyamoto, C.; Hashimoto, K.; Yanagimoto, K.; Moriyama, T. Suppressive effects of dietary EPA-rich fish oil on the degradation of elastin fibers in the aortic wall in nicotine-administered mice. Food Funct. 2017, 8, 2829–2835. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, J.; Lee, Y.J.; Lee, T.G.; Yoon, S. Sample Preparation of Corn Seed Tissue to Prevent Analyte Relocations for Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2017, 28, 1729–1732. [Google Scholar] [CrossRef]
- Seyer, A.; Einhorn, J.; Brunelle, A.; Laprevote, O. Localization of flavonoids in seeds by cluster time-of-flight secondary ion mass spectrometry imaging. Analy. Chem. 2010, 82, 2326–2333. [Google Scholar] [CrossRef]
- Moore, K.L.; Schroder, M.; Lombi, E.; Zhao, F.J.; McGrath, S.P.; Hawkesford, M.J.; Shewry, P.R.; Grovenor, C.R. NanoSIMS analysis of arsenic and selenium in cereal grain. New Phytol. 2010, 185, 434–445. [Google Scholar] [CrossRef]
- Moore, K.L.; Chen, Y.; van de Meene, A.M.; Hughes, L.; Liu, W.; Geraki, T.; Mosselmans, F.; McGrath, S.P.; Grovenor, C.; Zhao, F.J. Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 2014, 201, 104–115. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Goto-Inoue, N.; Moriyama, T.; Zaima, N. Significant advancement of mass spectrometry imaging for food chemistry. Food Chem. 2016, 210, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Morisasa, M.; Sato, T.; Kimura, K.; Mori, T.; Goto-Inoue, N. Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods 2019, 8, 633. [Google Scholar] [CrossRef] [PubMed]
- Gallage, N.J.; Jorgensen, K.; Janfelt, C.; Nielsen, A.J.Z.; Naake, T.; Dunski, E.; Dalsten, L.; Grisoni, M.; Moller, B.L. The Intracellular Localization of the Vanillin Biosynthetic Machinery in Pods of Vanilla planifolia. Plant Cell Physiol. 2018, 59, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Mohana Kumara, P.; Srimany, A.; Arunan, S.; Ravikanth, G.; Uma Shaanker, R.; Pradeep, T. Desorption Electrospray Ionization (DESI) Mass Spectrometric Imaging of the Distribution of Rohitukine in the Seedling of Dysoxylum binectariferum Hook. F. PLoS ONE 2016, 11, e0158099. [Google Scholar] [CrossRef] [PubMed]
- Hinners, P.; O’Neill, K.C.; Lee, Y.J. Revealing Individual Lifestyles through Mass Spectrometry Imaging of Chemical Compounds in Fingerprints. Sci. Rep. 2018, 8, 5149. [Google Scholar] [CrossRef] [PubMed]
- Gouaze, A.; Castaing, J.; Soutoul, J.H.; Chantepie, G. [On the orientation of the scapula and of its glenoid cavity]. Arch. Anat. Pathol. 1962, 10, 175–181. [Google Scholar]
- Agüi-Gonzalez, P.; Jähne, S.; Phan, N.T.N. SIMS imaging in neurobiology and cell biology. J. Anal. Atom. Spectrom. 2019, 34, 1355–1368. [Google Scholar] [CrossRef]
- Caprioli, R.M. Imaging mass spectrometry: Molecular microscopy for enabling a new age of discovery. Proteomics 2014, 14, 807–809. [Google Scholar] [CrossRef]
- Schulz, S.; Becker, M.; Groseclose, M.R.; Schadt, S.; Hopf, C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr. Opin. Biotechnol. 2019, 55, 51–59. [Google Scholar] [CrossRef]
- Mezger, S.T.P.; Mingels, A.M.A.; Bekers, O.; Cillero-Pastor, B.; Heeren, R.M.A. Trends in mass spectrometry imaging for cardiovascular diseases. Anal. Bioanal. Chem. 2019, 411, 3709–3720. [Google Scholar] [CrossRef]
- Yang, F.Y.; Chen, J.H.; Ruan, Q.Q.; Saqib, H.S.A.; He, W.Y.; You, M.S. Mass spectrometry imaging: An emerging technology for the analysis of metabolites in insects. Arch. Insect. Biochem. Physiol. 2019, e21643. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef] [PubMed]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef]
- Hankin, J.A.; Barkley, R.M.; Murphy, R.C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007, 18, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Gemperline, E.; Rawson, S.; Li, L. Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem. 2014, 86, 10030–10035. [Google Scholar] [CrossRef] [PubMed]
- Zaima, N.; Goto-Inoue, N.; Hayasaka, T.; Setou, M. Application of imaging mass spectrometry for the analysis of Oryza sativa rice. Rapid Commun. Mass Spectrom. RCM 2010, 24, 2723–2729. [Google Scholar] [CrossRef]
- Takats, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef]
- Abdelhamid, H.N. Nanoparticle-based surface assisted laser desorption ionization mass spectrometry: A review. Mikrochim. Acta 2019, 186, 682. [Google Scholar] [CrossRef]
- Rudd, D.A.; Benkendorff, K.; Chahal, C.; Guinan, T.; Gustafsson, O.J.R.; Esmaeelian, B.; Krysinska, H.; Pogson, L.; Voelcker, N.H.; Abbott, C.A. Mapping insoluble indole metabolites in the gastrointestinal environment of a murine colorectal cancer model using desorption/ionisation on porous silicon imaging. Sci. Rep. 2019, 9, 12342. [Google Scholar] [CrossRef]
- Niziol, J.; Sekula, J.; Ruman, T. Visualizing spatial distribution of small molecules in the rhubarb stalk (Rheum rhabarbarum) by surface-transfer mass spectrometry imaging. Phytochemistry 2017, 139, 72–80. [Google Scholar] [CrossRef]
- Misiorek, M.; Sekula, J.; Ruman, T. Mass Spectrometry Imaging of low Molecular Weight Compounds in Garlic (Allium sativum L.) with Gold Nanoparticle Enhanced Target. Phytochem. Anal. 2017, 28, 479–486. [Google Scholar] [CrossRef]
- Duenas, M.E.; Larson, E.A.; Lee, Y.J. Toward Mass Spectrometry Imaging in the Metabolomics Scale: Increasing Metabolic Coverage Through Multiple On-Tissue Chemical Modifications. Front. Plant Sci. 2019, 10, 860. [Google Scholar] [CrossRef]
- Hansen, R.L.; Duenas, M.E.; Lee, Y.J. Sputter-Coated Metal Screening for Small Molecule Analysis and High-Spatial Resolution Imaging in Laser Desorption Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 299–308. [Google Scholar] [CrossRef]
- Niziol, J.; Misiorek, M.; Ruman, T. Mass spectrometry imaging of low molecular weight metabolites in strawberry fruit (Fragaria x ananassa Duch.) cv. Primoris with (109)Ag nanoparticle enhanced target. Phytochemistry 2019, 159, 11–19. [Google Scholar] [CrossRef]
- Cha, S.; Zhang, H.; Ilarslan, H.I.; Wurtele, E.S.; Brachova, L.; Nikolau, B.J.; Yeung, E.S. Direct profiling and imaging of plant metabolites in intact tissues by using colloidal graphite-assisted laser desorption ionization mass spectrometry. Plant J. Cell Mol. Biol. 2008, 55, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Knochenmuss, R. MALDI and Related Methods: A Solved Problem or Still a Mystery? Mass Spectrom. 2013, 2, S0006. [Google Scholar] [CrossRef] [PubMed]
- Heard, P.J.; Feeney, K.A.; Allen, G.C.; Shewry, P.R. Determination of the elemental composition of mature wheat grain using a modified secondary ion mass spectrometer (SIMS). Plant J. Cell Mol. Biol. 2002, 30, 237–245. [Google Scholar] [CrossRef]
- Kyriacou, B.; Moore, K.L.; Paterson, D.; de Jonge, M.D.; Howard, D.L.; Stangoulis, J.; Tester, M.; Lombi, E.; Johnson, A.A.T. Localization of iron in rice grain using synchrotron X-ray fluorescence microscopy and high resolution secondary ion mass spectrometry. J. Cereal Sci. 2014, 59, 173–180. [Google Scholar] [CrossRef]
- Marzec, M.E.; Wojtysiak, D.; Poltowicz, K.; Nowak, J.; Pedrys, R. Study of cholesterol and vitamin E levels in broiler meat from different feeding regimens by TOF-SIMS. Biointerphases 2016, 11, 02A326. [Google Scholar] [CrossRef] [PubMed]
- Ifa, D.R.; Srimany, A.; Eberlin, L.S.; Naik, H.R.; Bhat, V.; Cooks, R.G.; Pradeep, T. Tissue imprint imaging by desorption electrospray ionization mass spectrometry. Anal. Methods 2011, 3, 1910. [Google Scholar] [CrossRef]
- De Rijke, E.; Hooijerink, D.; Sterk, S.S.; Nielen, M.W. Confirmation and 3D profiling of anabolic steroid esters in injection sites using imaging desorption electrospray ionisation (DESI) mass spectrometry. Food Addit. Contam. Part A 2013, 30, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Cabral, E.C.; Mirabelli, M.F.; Perez, C.J.; Ifa, D.R. Blotting assisted by heating and solvent extraction for DESI-MS imaging. J. Am. Soc. Mass Spectrom. 2013, 24, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Knudsen, C.; Hansen, N.K.; Jorgensen, K.; Kannangara, R.; Bak, S.; Takos, A.; Rook, F.; Hansen, S.H.; Moller, B.L.; et al. Visualizing metabolite distribution and enzymatic conversion in plant tissues by desorption electrospray ionization mass spectrometry imaging. Plant J. Cell Mol. Biol. 2013, 74, 1059–1071. [Google Scholar] [CrossRef]
- Ekelof, M.; McMurtrie, E.K.; Nazari, M.; Johanningsmeier, S.D.; Muddiman, D.C. Direct Analysis of Triterpenes from High-Salt Fermented Cucumbers Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). J. Am. Soc. Mass Spectrom. 2017, 28, 370–375. [Google Scholar] [CrossRef]
- Enomoto, H.; Sensu, T.; Sato, K.; Sato, F.; Paxton, T.; Yumoto, E.; Miyamoto, K.; Asahina, M.; Yokota, T.; Yamane, H. Visualisation of abscisic acid and 12-oxo-phytodienoic acid in immature Phaseolus vulgaris L. seeds using desorption electrospray ionisation-imaging mass spectrometry. Sci. Rep. 2017, 7, 42977. [Google Scholar] [CrossRef]
- Fowble, K.L.; Okuda, K.; Cody, R.B.; Musah, R.A. Spatial distributions of furan and 5-hydroxymethylfurfural in unroasted and roasted Coffea arabica beans. Food Res. Int. 2019, 119, 725–732. [Google Scholar] [CrossRef]
- Soltwisch, J.; Kettling, H.; Vens-Cappell, S.; Wiegelmann, M.; Muthing, J.; Dreisewerd, K. Mass spectrometry imaging with laser-induced postionization. Science 2015, 348, 211–215. [Google Scholar] [CrossRef]
- Korte, A.R.; Yagnik, G.B.; Feenstra, A.D.; Lee, Y.J. Multiplex MALDI-MS imaging of plant metabolites using a hybrid MS system. Methods Mol. Biol. 2015, 1203, 49–62. [Google Scholar]
- Zhang, Y.; Buchberger, A.; Muthuvel, G.; Li, L. Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics 2015, 15, 3969–3979. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rubel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef]
- Velickovic, D.; Saulnier, L.; Lhomme, M.; Damond, A.; Guillon, F.; Rogniaux, H. Mass Spectrometric Imaging of Wheat (Triticum spp.) and Barley (Hordeum vulgare L.) Cultivars: Distribution of Major Cell Wall Polysaccharides According to Their Main Structural Features. J. Agric. Food Chem. 2016, 64, 6249–6256. [Google Scholar] [CrossRef] [PubMed]
- Pittenauer, E.; Rados, E.; Koulakiotis, N.S.; Tsarbopoulos, A.; Allmaier, G. Processed stigmas of Crocus sativus L. imaged by MALDI-based MS. Proteomics 2016, 16, 1726–1730. [Google Scholar] [CrossRef] [PubMed]
- Cechova, M.; Valkova, M.; Hradilova, I.; Janska, A.; Soukup, A.; Smykal, P.; Bednar, P. Towards Better Understanding of Pea Seed Dormancy Using Laser Desorption/Ionization Mass Spectrometry. Int. J. Mol. Sci. 2017, 18, 2196. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bai, H.; Cai, Z.; Gao, D.; Jiang, Y.; Liu, J.; Liu, H. MALDI imaging for the localization of saponins in root tissues and rapid differentiation of three Panax herbs. Electrophoresis 2016, 37, 1956–1966. [Google Scholar] [CrossRef]
- Peukert, M.; Lim, W.L.; Seiffert, U.; Matros, A. Mass Spectrometry Imaging of Metabolites in Barley Grain Tissues. Curr. Protoc. Plant Biol. 2016, 1, 574–591. [Google Scholar] [CrossRef]
- Gorzolka, K.; Kolling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal Metabolite Profiling of the Barley Germination Process by MALDI MS Imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef]
- Hickert, S.; Cramer, B.; Letzel, M.C.; Humpf, H.U. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry imaging of ochratoxin A and fumonisins in mold-infected food. Rapid Commun. Mass Spectrom. RCM 2016, 30, 2508–2516. [Google Scholar] [CrossRef]
- Hashizaki, R.K.H.; Kazuma, K.; Konno, K.; Kawabata, K.; Kaneko, D.; Katano, H.; Taira, S. Localization Analysis of Natural Toxin of Solanum tuberosum L. via Mass Spectrometric Imaging. Int. J. Biotechnol. Wellness Ind. 2016, 5, 1–5. [Google Scholar]
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and Temporal Mapping of Key Lipid Species in Brassica napus Seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef]
- Bedia, C.; Tauler, R.; Jaumot, J. Analysis of multiple mass spectrometry images from different Phaseolus vulgaris samples by multivariate curve resolution. Talanta 2017, 175, 557–565. [Google Scholar] [CrossRef]
- Crecelius, A.C.; Holscher, D.; Hoffmann, T.; Schneider, B.; Fischer, T.C.; Hanke, M.V.; Flachowsky, H.; Schwab, W.; Schubert, U.S. Spatial and Temporal Localization of Flavonoid Metabolites in Strawberry Fruit (Fragaria x ananassa). J. Agric. Food Chem. 2017, 65, 3559–3568. [Google Scholar] [CrossRef] [PubMed]
- Dopstadt, J.; Vens-Cappell, S.; Neubauer, L.; Tudzynski, P.; Cramer, B.; Dreisewerd, K.; Humpf, H.U. Localization of ergot alkaloids in sclerotia of Claviceps purpurea by matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Bioanal. Chem. 2017, 409, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Morikawa-Ichinose, T.; Fujimura, Y.; Hayakawa, E.; Takahashi, K.; Ishii, T.; Miura, D.; Wariishi, H. Spatially resolved metabolic distribution for unraveling the physiological change and responses in tomato fruit using matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI). Anal. Bioanal. Chem. 2017, 409, 1697–1706. [Google Scholar] [CrossRef]
- Lee, J.W.; Ji, S.H.; Lee, Y.S.; Choi, D.J.; Choi, B.R.; Kim, G.S.; Baek, N.I.; Lee, D.Y. Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages. Int. J. Mol. Sci. 2017, 18, 1114. [Google Scholar] [CrossRef] [PubMed]
- Shiono, K.; Hashizaki, R.; Nakanishi, T.; Sakai, T.; Yamamoto, T.; Ogata, K.; Harada, K.I.; Ohtani, H.; Katano, H.; Taira, S. Multi-imaging of Cytokinin and Abscisic Acid on the Roots of Rice (Oryza sativa) Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Agric. Food Chem. 2017, 65, 7624–7628. [Google Scholar] [CrossRef]
- Feenstra, A.D.; Alexander, L.E.; Song, Z.; Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J. Spatial Mapping and Profiling of Metabolite Distributions during Germination. Plant Physiol. 2017, 174, 2532–2548. [Google Scholar] [CrossRef]
- Usher, S.; Han, L.; Haslam, R.P.; Michaelson, L.V.; Sturtevant, D.; Aziz, M.; Chapman, K.D.; Sayanova, O.; Napier, J.A. Tailoring seed oil composition in the real world: Optimising omega-3 long chain polyunsaturated fatty acid accumulation in transgenic Camelina sativa. Sci. Rep. 2017, 7, 6570. [Google Scholar] [CrossRef]
- Yoshida, S.; Koga, K.; Iwataka, M.; Fuchigami, T.; Haratake, M.; Nakayama, M. Characterization of Selenium Species in the Shijimi Clam. Chem. Pharm. Bull. 2017, 65, 1045–1050. [Google Scholar] [CrossRef][Green Version]
- Enomoto, H.; Sato, K.; Miyamoto, K.; Ohtsuka, A.; Yamane, H. Distribution Analysis of Anthocyanins, Sugars, and Organic Acids in Strawberry Fruits Using Matrix-Assisted Laser Desorption/Ionization-Imaging Mass Spectrometry. J. Agric. Food Chem. 2018, 66, 4958–4965. [Google Scholar] [CrossRef]
- Enomoto, H.; Sensu, T.; Yumoto, E.; Yokota, T.; Yamane, H. Derivatization for detection of abscisic acid and 12-oxo-phytodienoic acid using matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2018, 32, 1565–1572. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Toldra, F. Differences in peptide oxidation between muscles in 12months Spanish dry-cured ham. Food Res. Int. 2018, 109, 343–349. [Google Scholar] [CrossRef]
- Fanuel, M.; Ropartz, D.; Guillon, F.; Saulnier, L.; Rogniaux, H. Distribution of cell wall hemicelluloses in the wheat grain endosperm: A 3D perspective. Planta 2018, 248, 1505–1513. [Google Scholar] [CrossRef]
- Hansen, R.L.; Lee, Y.J. High-Spatial Resolution Mass Spectrometry Imaging: Toward Single Cell Metabolomics in Plant Tissues. Chem. Rec. 2018, 18, 65–77. [Google Scholar] [CrossRef]
- Duenas, M.E.; Feenstra, A.D.; Korte, A.R.; Hinners, P.; Lee, Y.J. Cellular and Subcellular Level Localization of Maize Lipids and Metabolites Using High-Spatial Resolution MALDI Mass Spectrometry Imaging. Methods Mol. Biol. 2018, 1676, 217–231. [Google Scholar] [PubMed]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. Cell Mol. Biol. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Ma, G.; Zhao, X.; Guo, C.; Li, S.; Liu, Y.; He, Q.; Chen, K.; Pan, Y. Glycosylamines-based reactive matrix designed for imaging acidity in Ponkan fruit using matrix assisted laser desorption/ionization mass spectrometry imaging. Anal. Chim. Acta 2018, 1041, 78–86. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.N.; Camargo, A.C.B.; Melo, C.; Catharino, R.R. A fast semi-quantitative screening for cocoa content in chocolates using MALDI-MSI. Food Res. Int. 2018, 103, 8–11. [Google Scholar] [CrossRef]
- Enomoto, H.; Takahashi, S.; Takeda, S.; Hatta, H. Distribution of Flavan-3-ol Species in Ripe Strawberry Fruit Revealed by Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging. Molecules 2019, 25, 103. [Google Scholar] [CrossRef]
- Goto-Inoue, N.; Sato, T.; Morisasa, M.; Igarashi, Y.; Mori, T. Characterization of Metabolite Compositions in Wild and Farmed Red Sea Bream (Pagrus major) Using Mass Spectrometry Imaging. J. Agric. Food Chem. 2019, 67, 7197–7203. [Google Scholar] [CrossRef]
- Resetar Maslov, D.; Svirkova, A.; Allmaier, G.; Marchetti-Deschamann, M.; Kraljevic Pavelic, S. Optimization of MALDI-TOF mass spectrometry imaging for the visualization and comparison of peptide distributions in dry-cured ham muscle fibers. Food Chem. 2019, 283, 275–286. [Google Scholar] [CrossRef]
- Theron, L.; Sayd, T.; Chambon, C.; Venien, A.; Viala, D.; Astruc, T.; Vautier, A.; Sante-Lhoutellier, V. Deciphering PSE-like muscle defect in cooked hams: A signature from the tissue to the molecular scale. Food Chem. 2019, 270, 359–366. [Google Scholar] [CrossRef]
- Horikawa, K.; Hirama, T.; Shimura, H.; Jitsuyama, Y.; Suzuki, T. Visualization of soluble carbohydrate distribution in apple fruit flesh utilizing MALDI-TOF MS imaging. Plant Sci. 2019, 278, 107–112. [Google Scholar] [CrossRef]
- Wisman, A.P.; Tamada, Y.; Hirohata, S.; Gomi, K.; Fukusaki, E.; Shimma, S. Mapping haze-komi on rice koji grains using beta-glucuronidase expressing Aspergillus oryzae and mass spectrometry imaging. J. Biosci. Bioeng. 2019, 129, 296–301. [Google Scholar] [CrossRef]
- Gupta, S.; Rupasinghe, T.; Callahan, D.L.; Natera, S.H.A.; Smith, P.M.C.; Hill, C.B.; Roessner, U.; Boughton, B.A. Spatio-Temporal Metabolite and Elemental Profiling of Salt Stressed Barley Seeds During Initial Stages of Germination by MALDI-MSI and micro-XRF Spectrometry. Front. Plant Sci. 2019, 10, 1139. [Google Scholar] [CrossRef]
- Lu, S.; Aziz, M.; Sturtevant, D.; Chapman, K.D.; Guo, L. Heterogeneous Distribution of Erucic Acid in Brassica napus Seeds. Front. Plant Sci. 2019, 10, 1744. [Google Scholar] [CrossRef]
- Enomoto, H.; Takeda, S.; Hatta, H.; Zaima, N. Tissue-Specific Distribution of Sphingomyelin Species in Pork Chop Revealed by Matrix-Assisted Laser Desorption/Ionization-Imaging Mass Spectrometry. J. Food Sci. 2019, 84, 1758–1763. [Google Scholar] [CrossRef]
- Enomoto, H.; Furukawa, T.; Takeda, S.; Hatta, H.; Zaima, N. Unique Distribution of Diacyl-, Alkylacyl-, and Alkenylacyl-Phosphatidylcholine Species Visualized in Pork Chop Tissues by Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging. Foods 2020, 9, 205. [Google Scholar] [CrossRef]
- Goto-Inoue, N.; Sato, T.; Morisasa, M.; Yamashita, H.; Maruyama, T.; Ikeda, H.; Sakai, R. Mass spectrometry imaging reveals differential localization of natural sunscreens in the mantle of the giant clam Tridacna crocea. Sci. Rep. 2020, 10, 656. [Google Scholar] [CrossRef]
- Sagara, T.; Bhandari, D.R.; Spengler, B.; Vollmann, J. Spermidine and other functional phytochemicals in soybean seeds: Spatial distribution as visualized by mass spectrometry imaging. Food Sci. Nutr. 2020, 8, 675–682. [Google Scholar] [CrossRef]
- Taira, S.; Tokai, M.; Kaneko, D.; Katano, H.; Kawamura-Konishi, Y. Mass Spectrometry Imaging Analysis of Location of Procymidone in Cucumber Samples. J. Agric. Food Chem. 2015, 63, 6109–6112. [Google Scholar] [CrossRef]
- Zaima, N.; Goto-Inoue, N.; Hayasaka, T.; Enomoto, H.; Setou, M. Authenticity assessment of beef origin by principal component analysis of matrix-assisted laser desorption/ionization mass spectrometric data. Anal. Bioanal. Chem. 2011, 400, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Chansela, P.; Goto-Inoue, N.; Zaima, N.; Hayasaka, T.; Sroyraya, M.; Kornthong, N.; Engsusophon, A.; Tamtin, M.; Chaisri, C.; Sobhon, P.; et al. Composition and localization of lipids in Penaeus merguiensis ovaries during the ovarian maturation cycle as revealed by imaging mass spectrometry. PLoS ONE 2012, 7, e33154. [Google Scholar] [CrossRef] [PubMed]
- Goto-Inoue, N.; Morisasa, M.; Machida, K.; Furuichi, Y.; Fujii, N.L.; Miura, S.; Mori, T. Characterization of myofiber-type-specific molecules using mass spectrometry imaging. Rapid Commun. Mass Spectrom. RCM 2019, 33, 185–192. [Google Scholar] [CrossRef]
- Cavatorta, V.; Sforza, S.; Mastrobuoni, G.; Pieraccini, G.; Francese, S.; Moneti, G.; Dossena, A.; Pastorello, E.A.; Marchelli, R. Unambiguous characterization and tissue localization of Pru P 3 peach allergen by electrospray mass spectrometry and MALDI imaging. J. Mass Spectrom. JMS 2009, 44, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Bencivenni, M.; Faccini, A.; Zecchi, R.; Boscaro, F.; Moneti, G.; Dossena, A.; Sforza, S. Electrospray MS and MALDI imaging show that non-specific lipid-transfer proteins (LTPs) in tomato are present as several isoforms and are concentrated in seeds. J. Mass Spectrom. JMS 2014, 49, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Zaima, N.; Moriyama, T.; Kawamura, Y. Different localization patterns of anthocyanin species in the pericarp of black rice revealed by imaging mass spectrometry. PLoS ONE 2012, 7, e31285. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Enomoto, H.; Moriyama, T.; Kawamura, Y.; Setou, M.; Zaima, N. Visualization of anthocyanin species in rabbiteye blueberry Vaccinium ashei by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1885–1895. [Google Scholar] [CrossRef]
- Gong, F.; Yao, S.; Wan, J.; Gan, X. Chocolate Consumption and Risk of Heart Failure: A Meta-Analysis of Prospective Studies. Nutrients 2017, 9, 402. [Google Scholar] [CrossRef]
- Hu, Y.; Pan, Z.J.; Liao, W.; Li, J.; Gruget, P.; Kitts, D.D.; Lu, X. Determination of antioxidant capacity and phenolic content of chocolate by attenuated total reflectance-Fourier transformed-infrared spectroscopy. Food Chem. 2016, 202, 254–261. [Google Scholar] [CrossRef]
- Bansal, S.; Syan, N.; Mathur, P.; Choudhary, S. Pharmacological profile of green tea and its polyphenols: A review. Med. Chem. Res. 2011, 21, 3347–3360. [Google Scholar] [CrossRef]
- Kim, Y.H.; Fujimura, Y.; Hagihara, T.; Sasaki, M.; Yukihira, D.; Nagao, T.; Miura, D.; Yamaguchi, S.; Saito, K.; Tanaka, H.; et al. In situ label-free imaging for visualizing the biotransformation of a bioactive polyphenol. Sci. Rep. 2013, 3, 2805. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Fujimura, Y.; Sasaki, M.; Yang, X.; Yukihira, D.; Miura, D.; Unno, Y.; Ogata, K.; Nakajima, H.; Yamashita, S.; et al. In situ label-free visualization of orally dosed strictinin within mouse kidney by MALDI-MS imaging. J. Agric. Food Chem. 2014, 62, 9279–9285. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Zaima, N.; Yoshimura, Y.; Inaba, S.; Fujimori, T.; Sogon, T.; Moriyama, T. Visualization of the distribution of anthocyanin species in mice eyeball by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. RCM 2018, 32, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N.; van der Hooft, J.J.; Crozier, A. Human studies on the absorption, distribution, metabolism, and excretion of tea polyphenols. Am. J. Clin. Nutr. 2013, 98, 1619S–1630S. [Google Scholar] [CrossRef]
- Matsui, T. Condensed catechins and their potential health-benefits. Eur. J. Pharmacol. 2015, 765, 495–502. [Google Scholar] [CrossRef]
- Nguyen, H.N.; Tanaka, M.; Li, B.; Ueno, T.; Matsuda, H.; Matsui, T. Novel in situ visualisation of rat intestinal absorption of polyphenols via matrix-assisted laser desorption/ionisation mass spectrometry imaging. Sci. Rep. 2019, 9, 3166. [Google Scholar] [CrossRef]
- Hansen, S.E.; Marxen, E.; Janfelt, C.; Jacobsen, J. Buccal delivery of small molecules—Impact of levulinic acid, oleic acid, sodium dodecyl sulfate and hypotonicity on ex vivo permeability and spatial distribution in mucosa. Eur. J. Pharm. Biopharm. 2018, 133, 250–257. [Google Scholar] [CrossRef]

| Instrument | Type of Ionization | Lateral Resolution | Applicable Sample Properties | Favorable Target Molecules |
|---|---|---|---|---|
| SIMS (secondary ion mass soectrometry) | Sputtering of sample surface with the impact of a high-energy primary ion | >1 µm | solid | Elements, fatty acids, lipids, |
| MALDI (matrix assisted laser desorption/ionization) | Laser ablation and desorption/ionization within ablated plume | 5–200 µm | solid | Metabolites, lipids, peptides, carbohydrates |
| DESI (desorption electrospray ionization) | Surface extraction and ionization with highly charged electrospray droplets | 50–200 µm | solid, liquid, frozen, and gaseous | Lipids, peptides, small metabolites |
| Matrices | Analytes |
|---|---|
| N-(1-naphthyl)ethylenediamine dihydrochloride (NEDC) | glucose |
| 3-amino-4-hydroxybenzoic acid (AHBA) | glycan |
| 3-aminoquinoline (3AQ) | glycan |
| 2-,4-,6-trihydroxyacetophenone (THAP) | glycolipids |
| N,N-dimethylaniline (DMA) | hemicellulose |
| 1,5-diaminonapthalene (DAN) | lipids |
| 2-mercaptobenzothiazole (MBT) | lipids |
| dihydroxyacetone phosphate (DHAP) | lipids, glycan |
| 2,5-dihydroxybenzoic acid (DHB) | lipids, glycopeptide, polymer |
| 9-aminoacridine (9-AA) | lipids, metabolites |
| 3-hydroxypicolinic acid (3HPA) | nucleotides |
| anthranilic acid (ANA) | nucleotides |
| nicotinic acid (NA) | nucleotides |
| picolinic acid (PA) | nucleotides |
| sinapinic acid (SA) | peptides, proteins |
| α-cyano-4-hydroxycinnamic acid (CHCA) | peptides, proteins |
| 2-(4-hydroxyphenylazo)-benzoic acid (HABA) | polymer |
| 5-chlorosalycic acid (5CSA) | polymer |
| trans-3-indolacrylic acid (IAA) | polymer, aromatic |
| nifedipine | polyphenols |
| Samples | Imaged Analytes | Instrument/Matrices | Year Published | Reference |
|---|---|---|---|---|
| Wheat grain | Magnesium, calcium, sodium, potassium | SIMS | 2002 | [37] |
| Peas (Pisem sativum) | Flavonoids | SIMS | 2010 | [8] |
| Wheat grain | Arsenic, selenium | SIMS | 2010 | [9] |
| Rice | Minerals | SIMS | 2013 | [10] |
| Rice | Iron, sulfur | SIMS | 2014 | [38] |
| Broiler meat | Phosphatidylcholine, cholesterol | SIMS | 2016 | [39] |
| Corn | Choline, phosphatidylcholine, palmitic acid, phosphatidylinositol | SIMS | 2017 | [7] |
| Myristica malabarica seed | Malabaricone C | DESI | 2011 | [40] |
| Bovine muscle | Anabolic steroid esters | DESI | 2013 | [41] |
| Strawberries (Fragaria x ananassa Duch.) | Hexose, pelargonidin-3-glucoside | DESI | 2013 | [42] |
| Cassava tuber | Hydroxynitrile glucosides | DESI | 2013 | [43] |
| Fermented cucumber | Stigmasterol, β-sitosterol, lupeol | DESI | 2016 | [44] |
| Kidney bean (Phaseolus vulgaris) | Abscisic acid, 12-oxo-phytodienoic acid | DESI | 2017 | [45] |
| Vanilla pod (Vanilla planifolia) | Vanillin, vanillin glucoside, sucrose | DESI | 2018 | [13] |
| Caffea arabica bean | Furan, 5-hydroxymethylfurfural | LDI-DART | 2019 | [46] |
| Braeburn apple | Quercitin pentoside, dihexose | MALD/DHB | 2015 | [47] |
| Corn seed | ADP, malic acid, sulfoquinovosyl diacylglycerol, phosphatidylinositol | MALDI/DHB | 2015 | [48] |
| Crab (Carcinus maenas) | Neuropeptides | MALDI/DHB | 2015 | [49] |
| Flax seed | Flax lignans, cyanogenic glucosides, secoisolariciresinol diglucoside | MALDI/DHB | 2015 | [50] |
| Wheat Barley | Cell wall polysaccharides | MALDI/DHB, DMA | 2016 | [51] |
| Saffron (Crocus sativus L.) | Crocins (tetrahexosyl-crocetin, trihexosyl-crocetin) | MALDI/THAP | 2016 | [52] |
| Pea seed (Pisum sp.) | Hydroxylated long chain fatty acids | MALDI/THAP | 2016 | [53] |
| Panax herbs | Ginsenoside | MALDI/CHCA, DHB | 2016 | [54] |
| Barley Grain | Hexose, sucrose, trisaccharide | MALDI/DHB | 2016 | [55] |
| Barley Grain | Hordatine B, coumaroylagmatine | MALDI/DHB | 2016 | [56] |
| Grape infected with mold | Ochratoxin A, fumonisins | MALDI/CHCA | 2016 | [57] |
| Potato | α-solanine, α-chaconine | MALDI/CHCA | 2016 | [58] |
| Oilseed rape (Brassica napas) | Phosphatidylcholines, triacylglycerols | MALDI/DHB | 2017 | [59] |
| Kidney bean (Phaseolus vulgaris) | Lipids | MALDI/MBT, DHB | 2017 | [60] |
| Strawberry Fruit | Flavonoids | MALDI/THAP | 2017 | [61] |
| Rye plants infected Claviceps purpurea | Alkaloids | MALDI/DHB | 2017 | [62] |
| Tomato fruit | Tomatine, esculeoside A, malate, aspartate, glutamate, AMP, caffeic acid | MALDI/DHB, 9-AA | 2017 | [63] |
| Pandas ginseng root | Ginsenoside | MALDI/DHB | 2017 | [64] |
| Rice root | Cytokinin, abscisic acid | MALDI/CHCA | 2017 | [65] |
| Maize seed (Zea mays) | Polysaccharides, triacylglycerols, amino acids, fatty acids, phospholipids, ceramides, hexose phosphate, glycerol phosphate, citrate | MALDI/DAN, DHB, 9-AA | 2017 | [66] |
| Camelina sativa seed | Triacylglycerols | MALDI/DHB | 2017 | [67] |
| Shijimi clam | selenium species (m/z 534) | MALDI/DHB | 2017 | [68] |
| Strawberry | Anthocyanins, sugars, organic acids | MALDI/DHB | 2018 | [69] |
| Kidney bean (Phaseolus vulgaris) | Abscisic acid, 12-oxo-phytodienoic acid (detivatized with Girard’s T) | MALDI/DAN, DHB | 2018 | [70] |
| Spanish dry-cured ham | Peptides | MALDI/CHCA | 2018 | [71] |
| Wheat grain endosperm | Arabinoxylan beta-glucan (hemicellulose) | MALDI/DHB | 2018 | [72] |
| Maize root, leaf | Molecules related to glycolysis and TCA cycle | MALDI/DAN, 9-AA | 2018 | [73] |
| Maize seed | Rubin, maysin, luteolin/kaempferol, phosphatidylglycerol | MALDI/DHB, DAN, 9-AA | 2018 | [74] |
| Oilseed (Brassica napus) | Phosphatidylcholines, triacylglycerols | MALDI/DHB | 2018 | [75] |
| Ponkan fruit | 3-AQ as an acidity indicator | MALDI/glycosyl-3-aminoquinoline (Gly-3AQ) | 2018 | [76] |
| Chocolate | Catechin | MALDI/CHCA | 2018 | [77] |
| Ripe strawberry fruit | Flavian-3-ol species | MALDI/DAN | 2019 | [78] |
| Res sea bream (Pagrus major) | Lipids, peptides | MALDI/DHB | 2019 | [79] |
| Ham (Biceps femuris muscle) | Peptides | MALDI/CHCA | 2019 | [80] |
| Ham (Biceps femuris muscle) | Peptides | MALDI/SA | 2019 | [81] |
| Apple | Hexose, sorbitol, sucrose | MALDI/DHB | 2019 | [82] |
| Sake rice koji | Glucose | MALDI/NEDC | 2019 | [83] |
| Barley seed stressed with salt | Flavonoids, lipids | MALDI/DHB | 2019 | [84] |
| Oil seed (Brassica napus) | Lipids | MALDI/DHB | 2019 | [85] |
| Pork chop | Sphingomyelin | MALDI/DHB | 2019 | [86] |
| Pork chop | Diacyl, alkylacyl, and alkenylacyl phosphatidylcholine | MALDI/DHB | 2020 | [87] |
| Giant clams (Tridacna crocea) | Mycosporines | MALDI/DHB | 2020 | [88] |
| Soybean | Isoflavones, gamma-tocopherol, spermidine, spermine, arginine | MALDI/DHB | 2020 | [89] |
| Cucumber | Procymidone | FT-ICR/ion oxide nanoparticle | 2015 | [90] |
| Rhubarb stalk | Anthraquinone derivatives, stilbenes, anthocyanins, flavonoids, polyphenols, organic acids, chromenes, chromanones, chromone glycosides, vitamins | AuNPET LDI/gold nanoparticle | 2017 | [30] |
| Garlic (Allium sativum) | Low molecular weight compounds (serine, allyl mercaptan, allyl sulfide etc.) | AuNPET LDI/gold nanoparticle | 2017 | [31] |
| Strawberry fruit | Low molecular weight metabolites | AgNPET/Silver nanoparticle | 2019 | [34] |
| Maize | Amino acids, endogenous metabolites | MALDI/DAN, DHB, gold nanoparticle | 2019 | [32] |
| Maize root seed | Small molecules, neutral lipids | MALDI/DAN, DHB, various metal nanoparticles | 2019 | [33] |
| Samples | Imaged Analytes | Instrument/Matrices | Year Published | Reference |
|---|---|---|---|---|
| Porcine buccal mucosa | Absorbed caffeine and mannitol | MALDI/DAN, DHB | 2018 | [107] |
| Eyeball of mice fed with anthocyanins | Anthocyanins | MALDI/DHB | 2018 | [103] |
| Rat intestine | Polyphenols absorbed in the intestine | MALDI/DAN, nifedipine | 2019 | [106] |
| Murine gastrointestinal tract | Indole derivatives | DIOS | 2019 | [29] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshimura, Y.; Zaima, N. Application of Mass Spectrometry Imaging for Visualizing Food Components. Foods 2020, 9, 575. https://doi.org/10.3390/foods9050575
Yoshimura Y, Zaima N. Application of Mass Spectrometry Imaging for Visualizing Food Components. Foods. 2020; 9(5):575. https://doi.org/10.3390/foods9050575
Chicago/Turabian StyleYoshimura, Yukihiro, and Nobuhiro Zaima. 2020. "Application of Mass Spectrometry Imaging for Visualizing Food Components" Foods 9, no. 5: 575. https://doi.org/10.3390/foods9050575
APA StyleYoshimura, Y., & Zaima, N. (2020). Application of Mass Spectrometry Imaging for Visualizing Food Components. Foods, 9(5), 575. https://doi.org/10.3390/foods9050575





