Flesh ID: Nanopore Sequencing Combined with Offline BLAST Search for the Identification of Meat Source
Abstract
1. Introduction
2. Materials and Methods
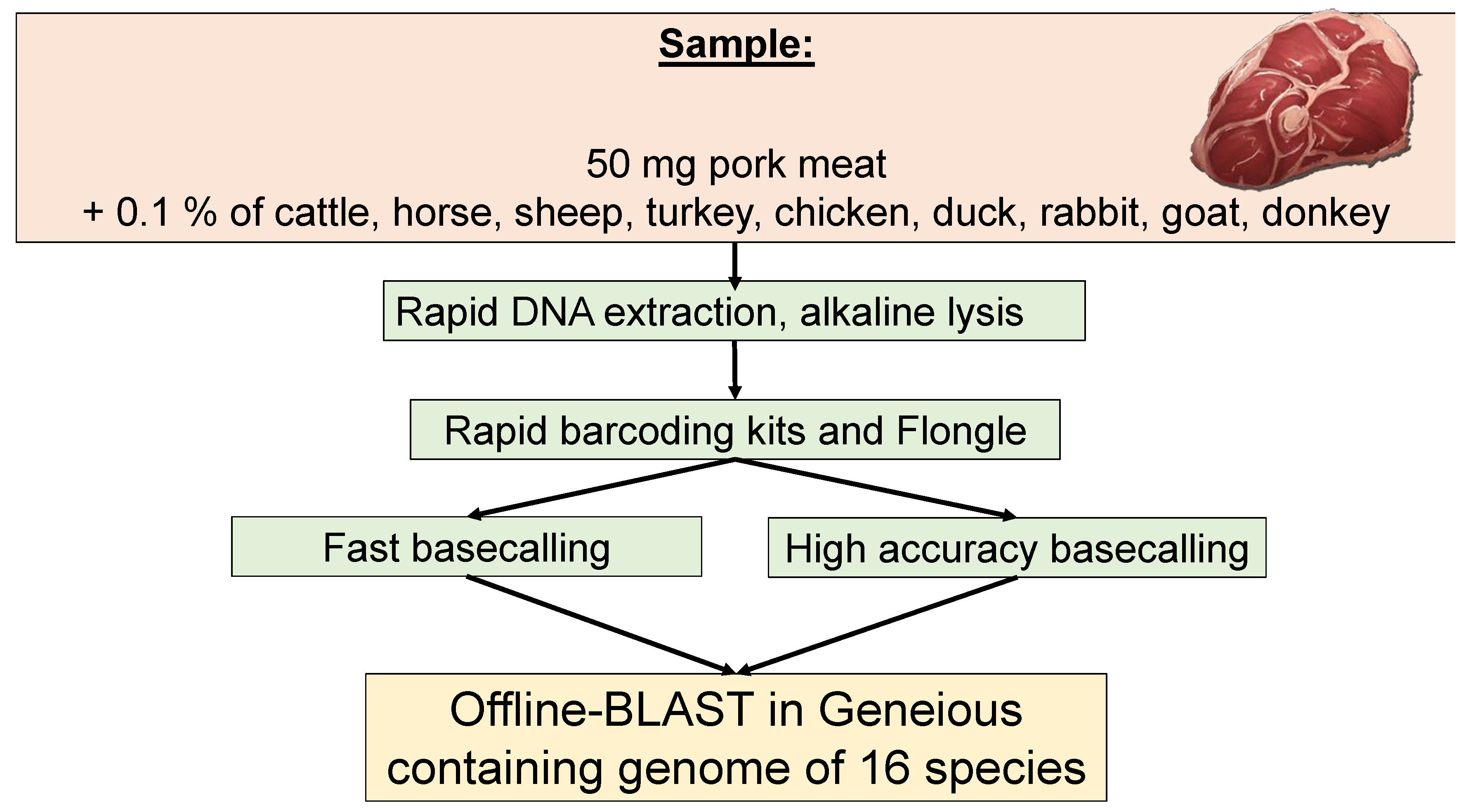
2.1. Meat Samples
2.2. Sequencing Library Preparation
2.3. Sequencing
2.4. Data Analysis
3. Results
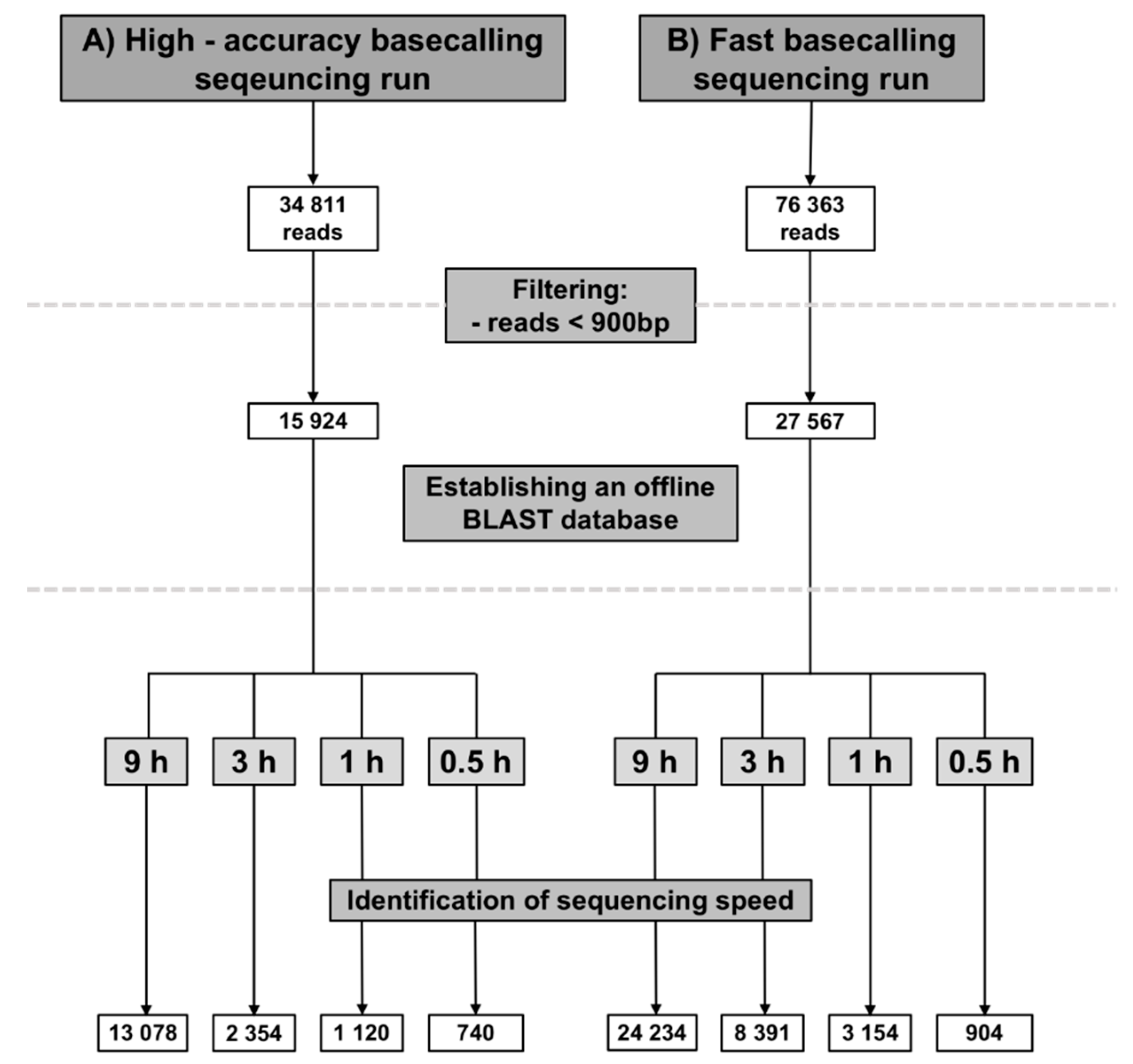
3.1. Data Acquisition
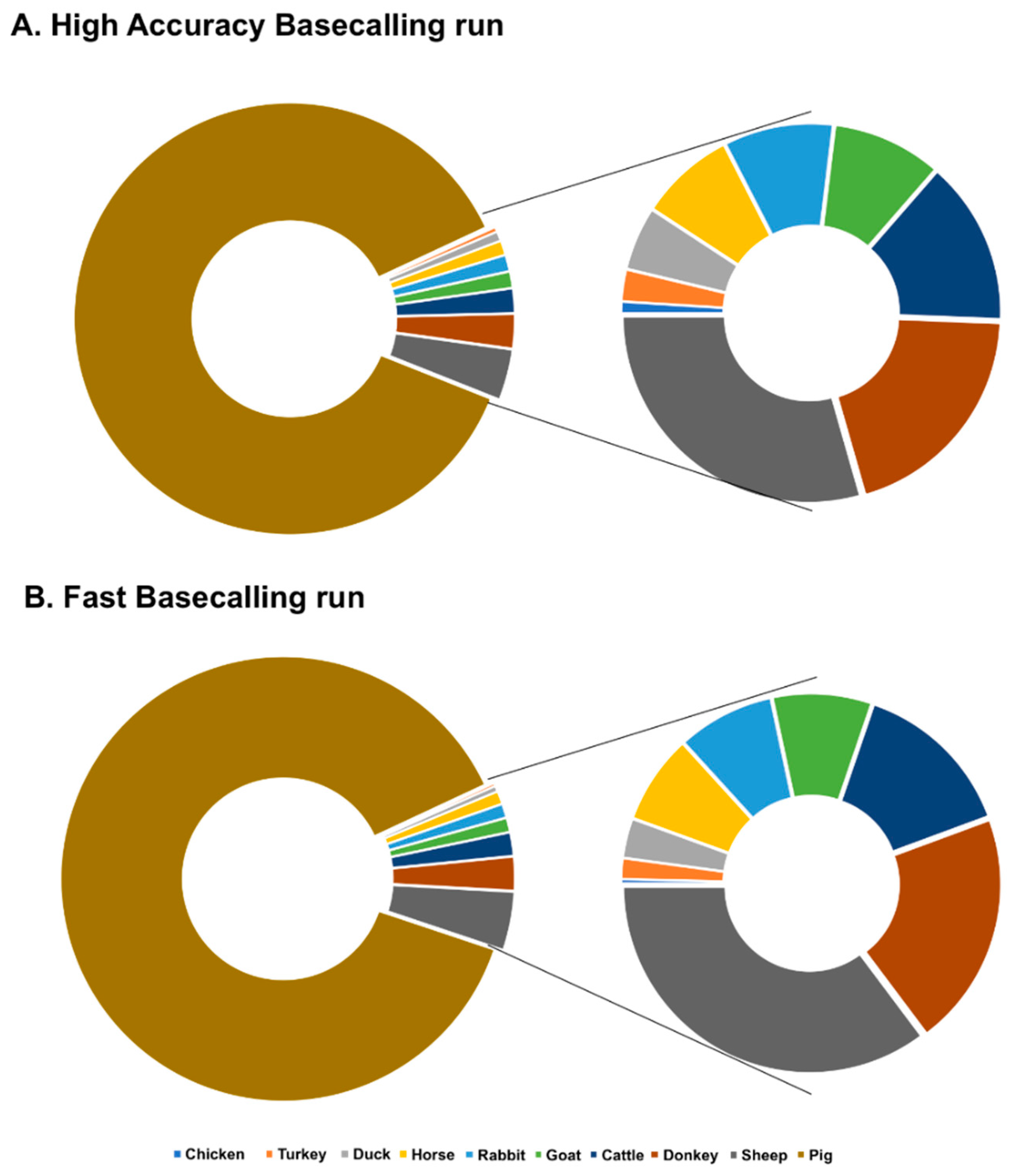
3.2. Offline BLAST-Search
3.3. Identification of Sequencing Speed
3.4. Database Specificity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakyinsige, K.; Man, Y.B.; Sazili, A.Q. Halal authenticity issues in meat and meat products. Meat Sci. 2012, 91, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, D.M.; Steinman, H.A.; Hoffman, L.C. A high incidence of species substitution and mislabelling detected in meat products sold in South Africa. Food Control 2013, 32, 440–449. [Google Scholar] [CrossRef]
- Hargin, K.D. Authenticity issues in meat and meat products. Meat Sci. 1996, 43 (Suppl. 1), 277–289. [Google Scholar] [CrossRef]
- Premanandh, J. Horse meat scandal—A wake-up call for regulatory authorities. Food Control 2013, 34, 568–569. [Google Scholar] [CrossRef]
- Manning, L.; Soon, J.M. Developing systems to control food adulteration. Food Policy 2014, 49, 23–32. [Google Scholar] [CrossRef]
- Asensio, L.; Gonzalez, I.; Garcia, T.; Martin, R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control 2008, 19, 1–8. [Google Scholar] [CrossRef]
- Bai, W.B.; Xu, W.T.; Huang, K.L.; Yuan, Y.F.; Cao, S.S.; Luo, Y.B. A novel common primer multiplex PCR (CP-M-PCR) method for the simultaneous detection of meat species. Food Control 2009, 20, 366–370. [Google Scholar] [CrossRef]
- Zhou, W.; Xia, L.; Huang, C.; Yang, J.; Shen, C.; Jiang, H.; Chu, Y. Rapid analysis and identification of meat species by laser-ablation electrospray mass spectrometry (LAESI-MS). Rapid Commun. Mass Spectrom. 2016, 30 (Suppl. 1), 116–121. [Google Scholar] [CrossRef]
- Sentandreu, M.A.; Sentandreu, E. Authenticity of meat products: Tools against fraud. Food Res. Int. 2014, 60, 19–29. [Google Scholar] [CrossRef]
- Laube, I.; Zagon, J.; Broll, H. Quantitative determination of commercially relevant species in foods by real-time PCR. Int. J. Food Sci. Technol. 2007, 42, 336–341. [Google Scholar] [CrossRef]
- Cho, A.R.; Dong, H.J.; Cho, S. Meat Species Identification using Loop-mediated Isothermal Amplification Assay Targeting Species-Specific Mitochondrial DNA. Korean J. Food Sci. Anim. Resour. 2014, 34, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Kissenkotter, J.; Bohlken-Fascher, S.; Forrest, M.S.; Piepenburg, O.; Czerny, C.P.; Abd El Wahed, A. Recombinase polymerase amplification assays for the identification of pork and horsemeat. Food Chem. 2020, 322, 126759. [Google Scholar] [CrossRef] [PubMed]
- Haynes, E.; Jimenez, E.; Pardo, M.A.; Helyar, S.J. The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control 2019, 101, 134–143. [Google Scholar] [CrossRef]
- Pan, Y.; Qiu, D.; Chen, J.; Yue, Q. Combining a COI Mini-Barcode with Next-Generation Sequencing for Animal Origin Ingredients Identification in Processed Meat Product. J. Food Qual. 2020, 2020, 2907670. [Google Scholar] [CrossRef]
- Rao, M.S.; Chakraborty, G.; Murthy, K.S. Market Drivers and Discovering Technologies in Meat Species Identification. Food Anal. Methods 2019, 12, 2416–2429. [Google Scholar] [CrossRef]
- Hansen, S.; Faye, O.; Sanabani, S.S.; Faye, M.; Bohlken-Fascher, S.; Faye, O.; Sall, A.A.; Bekaert, M.; Weidmann, M.; Czerny, C.P.; et al. Combination random isothermal amplification and nanopore sequencing for rapid identification of the causative agent of an outbreak. J. Clin. Virol. 2018, 106, 23–27. [Google Scholar] [CrossRef]
- Leggett, R.M.; Clark, M.D. A world of opportunities with nanopore sequencing. J. Exp. Bot. 2017, 68, 5419–5429. [Google Scholar] [CrossRef]
- Voorhuijzen-Harink, M.M.; Hagelaar, R.; Van Dijk, J.P.; Prins, T.W.; Kok, E.J.; Staats, M. Toward on-site food authentication using nanopore sequencing. Food Chem. X 2019, 2, 100035. [Google Scholar] [CrossRef]
- Girish, P.S.; Haunshi, S.; Vaithiyanathan, S.; Rajitha, R.; Ramakrishna, C. A rapid method for authentication of Buffalo (Bubalus bubalis) meat by Alkaline Lysis method of DNA extraction and species specific polymerase chain reaction. J. Food Sci. Technol. 2013, 50, 141–146. [Google Scholar] [CrossRef]
- Rapid Sequencing (SQK-RAD004)—Protocol. Available online: https://community.nanoporetech.com/protocols/rapid-sequencing-sqk-rad004/v/RSE_9046_v1_revM_14Aug2019?devices=flongle (accessed on 30 September 2020).
- New Research Algorithms Yield Accuracy Gains for Nanopore Sequencing. Available online: https://nanoporetech.com/about-us/news/new-research-algorithms-yield-accuracy-gains-nanopore-sequencing (accessed on 20 September 2020).
- Deshpande, S.V.; Reed, M.T.; Sullivan, R.F.; Kerjhof, L.J.; Beigel, K.M.; Wade, M.M. Offline Next Generation Metagenomics Sequence Analysis Using MinION Detection Software (MINDS). Genes 2019, 10, 578. [Google Scholar] [CrossRef]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef] [PubMed]
- Forin-Wiart, M.A.; Poulle, M.L.; Piry, S.; Cosson, J.F.; Larose, C.; Galan, M. Evaluating metabarcoding to analyse diet composition of species foraging in anthropogenic landscapes using Ion Torrent and Illumina sequencing. Sci. Rep. 2018, 8, 17091. [Google Scholar] [CrossRef] [PubMed]
- Tillmar, A.O.; Dell’Amico, B.; Welander, J.; Holmlund, G. A universal method for species identification of mammals utilizing next generation sequencing for the analysis of DNA mixtures. PLoS ONE 2013, 8, e83761. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Ghionda, M.C.; D’Alessandro, E.; Geraci, C.; Chiofalo, V.; Fontanesi, L. A next generation semiconductor based sequencing approach for the identification of meat species in DNA mixtures. PLoS ONE 2015, 10, e0121701. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Dill, V.; Shalaby, M.A.; Eschbaumer, M.; Bohlken-Fascher, S.; Hoffmann, B.; Czerny, C.P.; Abd El Wahed, A. Serotyping of foot-and-mouth disease virus using oxford nanopore sequencing. J. Virol. Methods 2019, 263, 50–53. [Google Scholar] [CrossRef]
- Arslan, A.; Ilhak, O.I.; Calicioglu, M. Effect of method of cooking on identification of heat processed beef using polymerase chain reaction (PCR) technique. Meat Sci. 2006, 72, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Sakalar, E.; Abasiyanik, M.F.; Bektik, E.; Tayyrov, A. Effect of heat processing on DNA quantification of meat species. J. Food Sci. 2012, 77, N40–N44. [Google Scholar] [CrossRef]
| Chromo Some | Species | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | Turkey | Goat | Duck | Rabbit | Horse | Cattle | Sheep | |||||||||
| AC | Hits | AC | Hits | AC | Hits | AC | Hits | AC | Hits | AC | Hits | AC | Hits | AC | Hits | |
| 1 | NC_006088.5 | 11 | NC_015011.2 | 12 | NC_030808.1 | 174 | NC_040046.1 | 24 | NC_013669.1 | 47 | NC_009144.3 | 64 | NC_037328.1 | 172 | NC_040252.1 | 219 |
| 2 | NC_006089.5 | 15 | NC_015012.2 | 10 | NC_030809.1 | 170 | NC_040047.1 | 15 | NC_013670.1 | 48 | NC_009145.3 | 26 | NC_037329.1 | 175 | NC_040253.1 | 216 |
| 3 | NC_006090.5 | 10 | NC_015013.2 | 16 | NC_030810.1 | 160 | NC_040048.1 | 17 | NC_013671.1 | 44 | NC_009146.3 | 34 | NC_037330.1 | 147 | NC_040254.1 | 195 |
| 4 | NC_006091.5 | 3 | NC_015014.2 | 4 | NC_030811.1 | 153 | NC_040049.1 | 11 | NC_013672.1 | 28 | NC_009147.3 | 26 | NC_037331.1 | 148 | NC_040255.1 | 151 |
| 5 | NC_006092.5 | 7 | NC_015015.2 | 6 | NC_030812.1 | 158 | NC_040050.1 | 11 | NC_013673.1 | 25 | NC_009148.3 | 24 | NC_037332.1 | 152 | NC_040256.1 | 154 |
| 6 | NC_006093.5 | 4 | NC_015016.2 | 6 | NC_030813.1 | 141 | NC_040051.1 | 4 | NC_013674.1 | 23 | NC_009149.3 | 25 | NC_037333.1 | 143 | NC_040257.1 | 149 |
| 7 | NC_006094.5 | 4 | NC_015017.2 | 5 | NC_030814.1 | 164 | NC_040052.1 | 6 | NC_013675.1 | 51 | NC_009150.3 | 25 | NC_037334.1 | 153 | NC_040258.1 | 144 |
| 8 | NC_006095.5 | 6 | NC_015018.2 | 4 | NC_030815.1 | 154 | NC_040053.1 | 11 | NC_013676.1 | 30 | NC_009151.3 | 29 | NC_037335.1 | 166 | NC_040259.1 | 153 |
| 9 | NC_006096.5 | 2 | NC_015019.2 | 1 | NC_030816.1 | 145 | NC_040054.1 | 7 | NC_013677.1 | 36 | NC_009152.3 | 28 | NC_037336.1 | 153 | NC_040260.1 | 152 |
| 10 | NC_006097.5 | 3 | NC_015020.2 | 5 | NC_030817.1 | 148 | NC_040055.1 | 7 | NC_013678.1 | 24 | NC_009153.3 | 27 | NC_037337.1 | 157 | NC_040261.1 | 137 |
| 11 | NC_006098.5 | 3 | NC_015021.2 | 3 | NC_030818.1 | 145 | NC_040056.1 | 6 | NC_013679.1 | 35 | NC_009154.3 | 14 | NC_037338.1 | 158 | NC_040262.1 | 121 |
| 12 | NC_006099.5 | 3 | NC_015022.2 | 3 | NC_030819.1 | 138 | NC_040057.1 | 6 | NC_013680.1 | 30 | NC_009155.3 | 13 | NC_037339.1 | 135 | NC_040263.1 | 131 |
| 13 | NC_006100.5 | 1 | NC_015023.2 | 3 | NC_030820.1 | 134 | NC_040058.1 | 5 | NC_013681.1 | 34 | NC_009156.3 | 12 | NC_037340.1 | 145 | NC_040264.1 | 135 |
| 14 | NC_006101.5 | 1 | NC_015024.2 | 2 | NC_030821.1 | 149 | NC_040059.1 | 8 | NC_013682.1 | 43 | NC_009157.3 | 40 | NC_037341.1 | 140 | NC_040265.1 | 132 |
| 15 | NC_006102.5 | 0 | NC_015025.2 | 1 | NC_030822.1 | 133 | NC_040060.1 | 9 | NC_013683.1 | 29 | NC_009158.3 | 30 | NC_037342.1 | 145 | NC_040266.1 | 146 |
| 16 | NC_006103.5 | 2 | NC_015026.2 | 2 | NC_030822.1 | 134 | NC_040061.1 | 5 | NC_013684.1 | 32 | NC_009159.3 | 24 | NC_037343.1 | 148 | NC_040267.1 | 140 |
| 17 | NC_006104.5 | 3 | NC_015027.2 | 0 | NC_030824.1 | 124 | NC_040062.1 | 4 | NC_013685.1 | 41 | NC_009160.3 | 27 | NC_037344.1 | 134 | NC_040268.1 | 135 |
| 18 | NC_006105.5 | 2 | NC_015028.2 | 0 | NC_030825.1 | 130 | NC_040063.1 | 4 | NC_013686.1 | 29 | NC_009161.3 | 42 | NC_037345.1 | 128 | NC_040269.1 | 142 |
| 19 | NC_006106.5 | 2 | NC_015029.2 | 5 | NC_030826.1 | 123 | NC_040064.1 | 3 | NC_013687.1 | 26 | NC_009162.3 | 20 | NC_037346.1 | 129 | NC_040270.1 | 136 |
| 20 | NC_006107.5 | 1 | NC_015030.2 | 1 | NC_030827.1 | 136 | NC_040065.1 | 3 | NC_013688.1 | 21 | NC_009163.3 | 16 | NC_037347.1 | 140 | NC_040271.1 | 122 |
| 21 | NC_006108.5 | 0 | NC_015031.2 | 2 | NC_030828.1 | 142 | NC_040066.1 | 4 | NC_013689.1 | 18 | NC_009164.3 | 21 | NC_037348.1 | 138 | NC_040272.1 | 122 |
| 22 | NC_006109.5 | 0 | NC_015032.2 | 0 | NC_030829.1 | 130 | NC_040067.1 | 1 | Not available | - | NC_009165.3 | 14 | NC_037349.1 | 129 | NC_040273.1 | 132 |
| 23 | NC_006110.5 | 1 | NC_015033.2 | 0 | NC_030830.1 | 121 | NC_040068.1 | 2 | Not available | - | NC_009166.3 | 27 | NC_037350.1 | 120 | NC_040274.1 | 142 |
| 24 | NC_006111.5 | 0 | NC_015034.2 | 2 | NC_030831.1 | 132 | NC_040069.1 | 1 | Not available | - | NC_009167.3 | 16 | NC_037351.1 | 142 | NC_040275.1 | 107 |
| 25 | NC_006112.4 | 0 | NC_015035.2 | 0 | NC_030832.1 | 109 | NC_040070.1 | 3 | Not available | - | NC_009168.3 | 11 | NC_037352.1 | 110 | NC_040276.1 | 124 |
| 26 | NC_006113.5 | 1 | NC_015036.2 | 2 | NC_030833.1 | 129 | NC_040071.1 | 3 | Not available | - | NC_009169.3 | 15 | NC_037353.1 | 132 | NC_040277.1 | 114 |
| 27 | NC_006114.5 | 1 | NC_015037.2 | 0 | NC_030834.1 | 115 | NC_040072.1 | 0 | Not available | - | NC_009170.3 | 18 | NC_037354.1 | 117 | Not available | - |
| 28 | NC_006115.5 | 0 | NC_015038.2 | 1 | NC_030835.1 | 124 | NC_040073.1 | 0 | Not available | - | NC_009171.3 | 15 | NC_037355.1 | 125 | Not available | - |
| 29 | Not available | - | NC_015039.2 | 1 | NC_030836.1 | 124 | NC_040074.1 | 0 | Not available | - | NC_009172.3 | 12 | NC_037356.1 | 121 | Not available | - |
| 30 | NC_028739.2 | 0 | NC_015040.2 | 1 | Not available | - | Not available | - | Not available | - | NC_009173.3 | 9 | Not available | - | Not available | - |
| 31 | NC_028740.2 | 1 | Not available | - | Not available | - | Not available | - | Not available | - | NC_009174.3 | 13 | Not available | - | Not available | - |
| 32 | NC_006119.4 | 0 | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - |
| 33 | NC_008465.4 | 1 | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - |
| X | Not available | - | Not available | - | Not available | - | Not available | - | NC_013690.1 | 33 | NC_009175.3 | 39 | NC_037357.1 | 169 | NC_040278.1 | 170 |
| W | NC_006126.5 | 3 | NC_015042.2 | 1 | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - | Not available | - |
| Z | NC_006127.5 | 10 | NC_015041.2 | 9 | Not available | - | NC_040075.1 | 11 | Not available | - | Not available | - | Not available | - | Not available | - |
| mt | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| High Accuracy Basecalling | Fast Basecalling | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 h | 1 h | 3 h | 9 h | 18 h | 0.5 h | 1 h | 3 h | 9 h | 18 h | |||||||||||
| Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | Hits | Pairwise Identity (%) | |
| Chicken | 0 | - | 1 | 84 | 1 | 84 | 3 | 81.37 | 4 | 80.9 | 0 | - | 0 | - | 0 | - | 2 | 81.25 | 2 | 81.25 |
| Turkey | 2 | 87.5 | 2 | 87.5 | 4 | 88.28 | 11 | 85.96 | 11 | 85.96 | 0 | - | 1 | 88.6 | 2 | 84.65 | 10 | 84 | 10 | 84 |
| Duck | 2 | 83.65 | 2 | 83.65 | 2 | 83.65 | 17 | 83.05 | 22 | 83.44 | 1 | 80.8 | 3 | 82.87 | 5 | 83.68 | 16 | 84.31 | 19 | 83.96 |
| Horse | 1 | 79.9 | 2 | 79.4 | 2 | 79.4 | 26 | 80.97 | 33 | 80.82 | 3 | 83.87 | 7 | 82.23 | 16 | 82.9 | 37 | 82.5 | 43 | 82.26 |
| Rabbit | 2 | 87.45 | 3 | 88.3 | 5 | 88.06 | 31 | 84.82 | 38 | 85.14 | 2 | 85.75 | 5 | 86.2 | 19 | 85.08 | 43 | 84.91 | 47 | 84.7 |
| Goat | 1 | 78.4 | 2 | 81.4 | 4 | 84.46 | 32 | 82.26 | 38 | 82.3 | 3 | 84.33 | 7 | 83.7 | 19 | 82.95 | 47 | 82.93 | 48 | 82.94 |
| Cattle | 2 | 87.8 | 2 | 87.8 | 10 | 82.12 | 48 | 82.88 | 57 | 83.2 | 4 | 80.85 | 12 | 81.95 | 26 | 82.21 | 74 | 81.76 | 79 | 81.65 |
| Donkey | 3 | 82.73 | 5 | 81.76 | 13 | 81.42 | 62 | 79.95 | 80 | 80.01 | 1 | 77.8 | 16 | 80.35 | 54 | 79.63 | 110 | 80.3 | 114 | 80.33 |
| Sheep | 4 | 81.05 | 7 | 81.96 | 23 | 82.94 | 100 | 82.94 | 118 | 82.93 | 10 | 82.96 | 27 | 84.35 | 69 | 84.1 | 185 | 82.95 | 197 | 82.93 |
| Pig | 124 | 84.93 | 189 | 84.94 | 405 | 85.28 | 2190 | 84.69 | 2691 | 84.8 | 151 | 82.28 | 485 | 85.35 | 1337 | 85.19 | 3651 | 84.59 | 4067 | 84.47 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kissenkötter, J.; Böhlken-Fascher, S.; Abd El Wahed, A. Flesh ID: Nanopore Sequencing Combined with Offline BLAST Search for the Identification of Meat Source. Foods 2020, 9, 1392. https://doi.org/10.3390/foods9101392
Kissenkötter J, Böhlken-Fascher S, Abd El Wahed A. Flesh ID: Nanopore Sequencing Combined with Offline BLAST Search for the Identification of Meat Source. Foods. 2020; 9(10):1392. https://doi.org/10.3390/foods9101392
Chicago/Turabian StyleKissenkötter, Jonas, Susanne Böhlken-Fascher, and Ahmed Abd El Wahed. 2020. "Flesh ID: Nanopore Sequencing Combined with Offline BLAST Search for the Identification of Meat Source" Foods 9, no. 10: 1392. https://doi.org/10.3390/foods9101392
APA StyleKissenkötter, J., Böhlken-Fascher, S., & Abd El Wahed, A. (2020). Flesh ID: Nanopore Sequencing Combined with Offline BLAST Search for the Identification of Meat Source. Foods, 9(10), 1392. https://doi.org/10.3390/foods9101392






