Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Isolation of the Riboflavin-Producing LAB Strains from Pickle Juices
2.3. Extraction of Riboflavin
2.4. Quantification of Riboflavin Content by HPLC
2.5. Identification of LAB Species
2.6. Inducing LAB Mutation by Roseoflavin
2.7. Identification of LAB Riboflavin Biosynthetic Genes by PCR
2.8. DNA Sequencing Analysis of Riboflavin Operon Upstream Regulatory Region
2.9. Preparation of Soymilk and Soymilk Fermentation
2.10. Statistical Analysis
3. Results and Discussion
3.1. Riboflavin Content Produced by the LAB Isolated from Pickle Juices
3.2. Identification of LAB Strains with High Riboflavin-Producing Capability
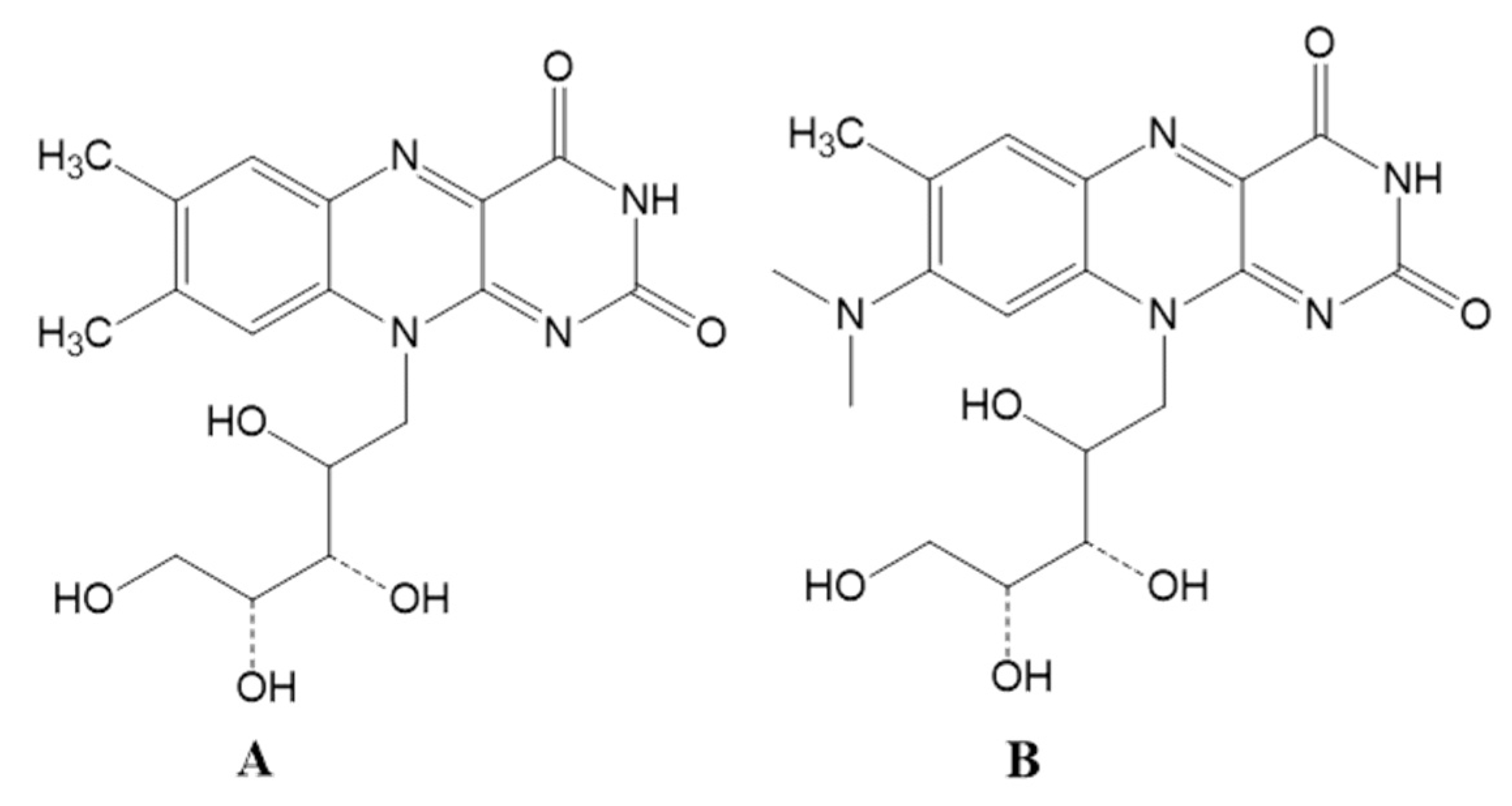
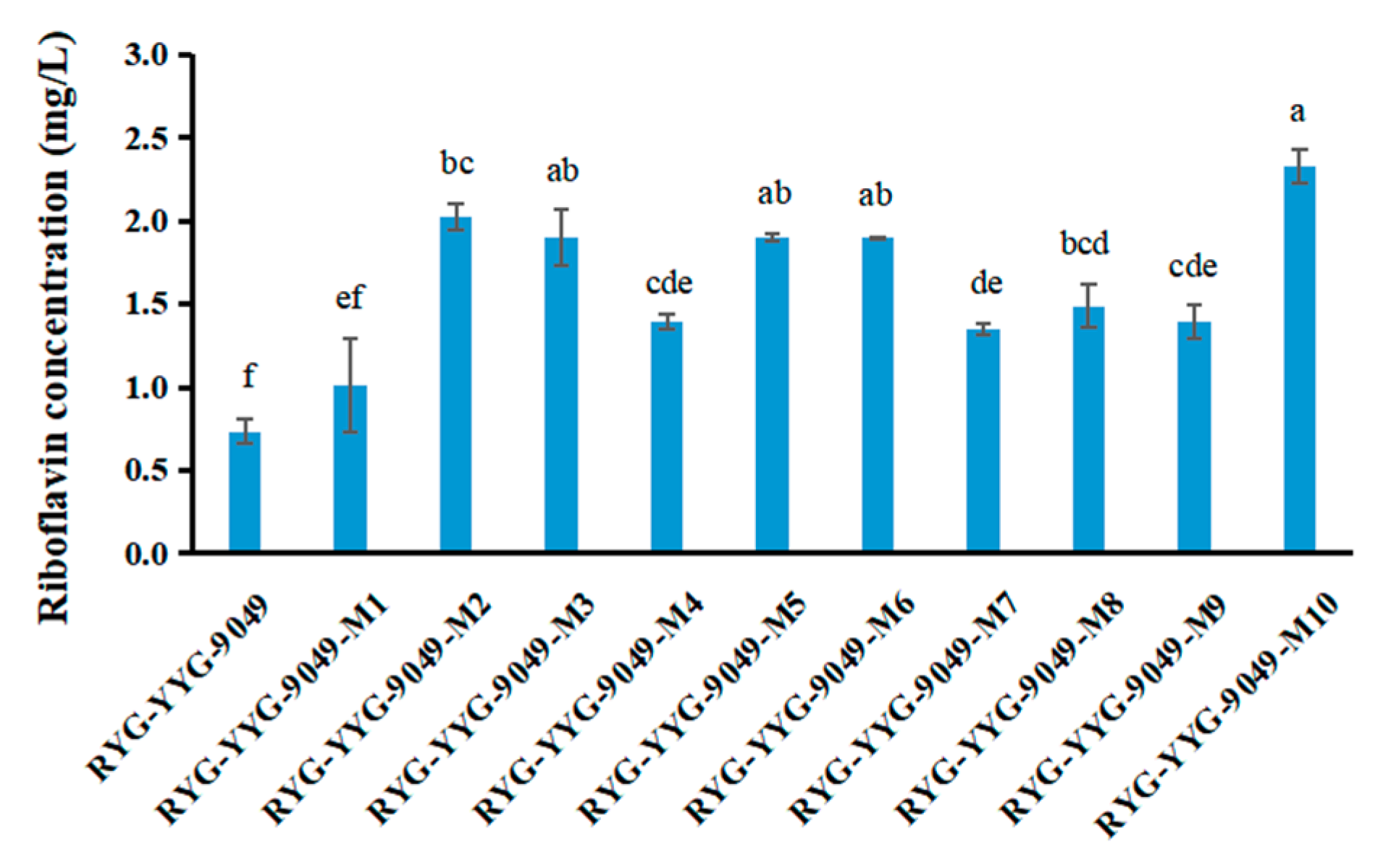
3.3. Riboflavin Production Was Increased in Roseoflavin-Induced Mutant Strains of RYG-YYG-9049
3.4. Roseoflavin-Induced Mutation Did Not Change Riboflavin Biosynthetic Genes in RYG-YYG-9049-M10
3.5. Roseoflavin Induced the Mutation of the Upstream Regulatory Region of Rib Operon in RYG-YYG-9049-M10
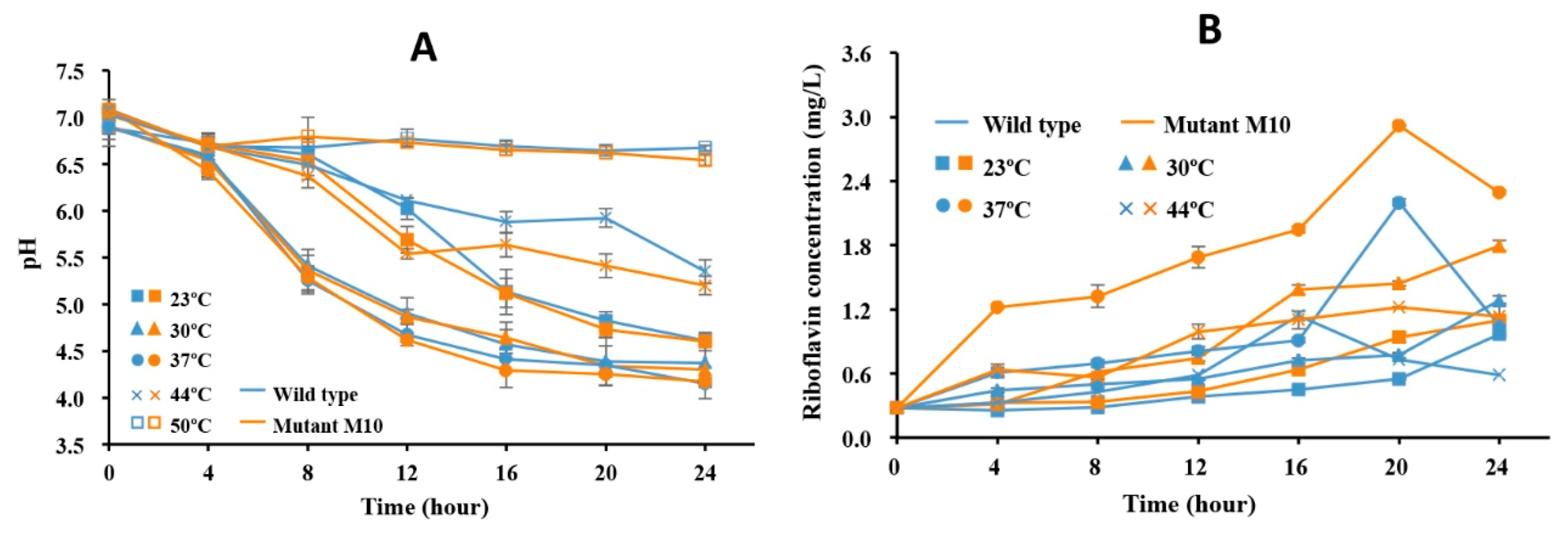
3.6. RYG-YYG-9049 and Its M10 Fermentation Increased Riboflavin Content in Soymilk
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Food and Agriculture Organization; Requirements of Vitamin A, Thiamine, Riboflavine and Niacin; No. 362; World Health Organization: Geneva, Switzerland, 1978. [Google Scholar]
- Northrop-Clewes, C.A.; Thurnham, D.I. The discovery and characterization of riboflavin. Ann. Nutr. Metab. 2012, 61, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, N.; Fein, H.D.; Rosenblum, L.A. Riboflavin deficiency in man. Nutr. Rev. 2010, 34, 275–277. [Google Scholar] [CrossRef]
- Agte, V.V.; Paknikar, K.M.; Chiplonkar, S.A. Effect of riboflavin supplementation on zinc and iron absorption and growth performance in mice. Biol. Trace Elem. Res. 1998, 65, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Mazur-Bialy, A.; Pocheć, E.; Plytycz, B. Immunomodulatory effect of riboflavin deficiency and enrichment-reversible pathological response versus silencing of inflammatory activation. J. Physiol. Pharmacol. 2015, 66, 793–802. [Google Scholar]
- Udhayabanu, T.; Karthi, S.; Mahesh, A.; Varalakshmi, P.; Manole, A.; Houlden, H.; Ashokkumar, B. Adaptive regulation of riboflavin transport in heart: Effect of dietary riboflavin deficiency in cardiovascular pathogenesis. Mol. Cell Biochem. 2018, 440, 147–156. [Google Scholar] [CrossRef]
- Mensink, G.B.M.; Fletcher, R.; Gurinovic, M.; Huybrechts, I.; Lafay, L.; Serra-Majem, L.; Szponar, L.; Tetens, I.; Verkaik-Kloosterman, J.; Baka, A.; et al. Mapping low intake of micronutrients across Europe. Br. J. Nutr. 2013, 110, 755–773. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Ledesma-Amaro, R.; Lozano-Martinez, P.; Díaz-Fernández, D.; Buey, R.M.; Jiménez, A. Bioproduction of riboflavin: A bright yellow history. J. Ind. Microbiol. Biotechnol. 2017, 44, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef] [PubMed]
- Bretzel, W.; Schurter, W.; Ludwig, B.; Kupfer, E.; Doswald, S.; Pfister, M.; van Loon, A.P.G.M. Commercial riboflavin production by recombinant Bacillus subtilis: Down-stream processing and comparison of the composition of riboflavin produced by fermentation or chemical synthesis. J. Ind. Microbiol. Biotechnol. 1999, 22, 19–26. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-group vitamin production by lactic acid bacteria–current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef]
- Russo, P.; Capozzi, V.; Arena, M.P.; Spadaccino, G.; Dueñas, M.T.; López, P.; Fiocco, D.; Spano, G. Riboflavin-overproducing strains of Lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 2014, 98, 3691–3700. [Google Scholar] [CrossRef] [PubMed]
- Pyo, Y.H.; Lee, T.C.; Lee, Y.C. Enrichment of bioactive isoflavones in soymilk fermented with β-glucosidase-producing lactic acid bacteria. Food Res. Int. 2005, 38, 551–559. [Google Scholar] [CrossRef]
- Marazza, J.A.; Nazareno, M.A.; de Giori, G.S.; Garro, M.S. Enhancement of the antioxidant capacity of soymilk by fermentation with Lactobacillus rhamnosus. J. Funct. Foods 2012, 4, 594–601. [Google Scholar] [CrossRef]
- Yang, H.L.; Zhang, L.; Xiao, G.N.; Feng, J.B.; Zhou, H.B.; Huang, F.R. Changes in some nutritional components of soymilk during fermentation by the culinary and medicinal mushroom Grifola frondosa. LWT Food Sci. Technol. 2015, 62, 468–473. [Google Scholar] [CrossRef]
- Achouri, A.; Boye, J.I.; Zamani, Y. Changes in soymilk quality as a function of composition and storage. J. Food Qual. 2007, 30, 731–744. [Google Scholar] [CrossRef]
- Chou, C.C.; Hou, J.W. Growth of bifidobacteria in soymilk and their survival in the fermented soymilk drink during storage. Int. J. Food Microbiol. 2000, 56, 113–121. [Google Scholar] [CrossRef]
- Fernando, S.M.; Murphy, P.A. HPLC determination of thiamin and riboflavin in soybeans and tofu. J. Agric. Food Chem. 1990, 38, 163–167. [Google Scholar] [CrossRef]
- Juarez del Valle, M.; Laiño, J.E.; Savoy de Giori, G.; LeBlanc, J.G. Riboflavin producing lactic acid bacteria as a biotechnological strategy to obtain bio-enriched soymilk. Food Res. Int. 2014, 62, 1015–1019. [Google Scholar] [CrossRef]
- Burgess, C.; O’Connell-Motherway, M.; Sybesma, W.; Hugenholtz, J.; van Sinderen, D. Riboflavin production in Lactococcus lactis: Potential for in situ production of vitamin-enriched foods. Appl. Environ. Microb. 2004, 70, 5769–5777. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Tian, T.; Yue, X. Isolation, identification and preliminary screening of acid-tolerant lactic acid bacteria from naturally fermented pickle juices from daqing. Food Sci. 2014, 35, 150–154. [Google Scholar]
- Zhang, Y.; Lü, J.L.; Yan, Y.M.; Li, W.J. Isolation and Identification of lactic acid bacteria and yeast from traditional fermentative pickle juice. J. Chin. Inst. Food Sci. Technol. 2017, 6, 179–186. [Google Scholar]
- Capozzi, V.; Menga, V.; Digesù, A.M.; De Vita, P.; van Sinderen, D.; Cattivelli, L.; Fares, C.; Spano, G. Biotechnological production of vitamin B2-enriched bread and pasta. J. Agric. Food Chem. 2011, 59, 8013–8020. [Google Scholar] [CrossRef] [PubMed]
- Hüttner, E.K.; Bello, F.D.; Arendt, E.K. Identification of lactic acid bacteria isolated from oat sourdoughs and investigation into their potential for the improvement of oat bread quality. Eur. Food Res. Technol. 2010, 230, 849–857. [Google Scholar] [CrossRef]
- Burgess, C.M.; Smid, E.J.; Rutten, G.; van Sinderen, D. A general method for selection of riboflavin-overproducing food grade micro-organisms. Microb. Cell Fact. 2006, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.Y.; Shah, N.; Wang, M.F.; Lui, W.Y.; Corke, H. Lactobacillus plantarum WCFS1 fermentation differentially affects antioxidant capacity and polyphenol content in mung bean (Vigna radiata) and soya bean (Glycine max) milks. J. Food Process. Preserv. 2017, 41, e12944. [Google Scholar] [CrossRef]
- Lee, K.W.; Shim, J.M.; Kim, D.W.; Yao, Z.; Kim, J.A.; Kin, H.J.; Kim, J.H. Effects of different types of salts on the growth of lactic acid bacteria and yeasts during kimchi fermentation. Food Sci. Biotechnol. 2017, 27, 489–498. [Google Scholar] [CrossRef]
- Cheigh, H.S.; Park, K.Y.; Lee, C.Y. Biochemical, microbiological, and nutritional aspects of kimchi (Korean fermented vegetable products). Crit. Rev. Food Sci. 1994, 34, 175–203. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; Brahma, B.; De, S. Screening of riboflavin-producing lactobacilli by a polymerase-chain-reaction-based approach and microbiological assay. J. Agric. Food Chem. 2016, 64, 1950–1956. [Google Scholar] [CrossRef]
- Jayashree, S.; Jayaraman, K.; Kalaichelvan, G. Isolation, screening and characterization of riboflavin producing lactic acid bacteria from Katpadi, Vellore district. Rec. Res. Sci. Technol. 2010, 2, 83–88. [Google Scholar]
- Kneifel, W.; Kaufmann, M.; Fleischer, A.; Ulberth, F. Screening of commercially available mesophilic dairy starter cultures: Biochemical, sensory, and microbiological properties. J. Dairy Sci. 1992, 75, 3158–3166. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; Savoy de Giori, G.; Sesma, F.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2012, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Wang, H.C.; Hirota, T.; Matsukawa, H.; Kasai, S.; Shinagawa, K.; Otani, S. Riboflavin production by roseoflavin-resistant strains of some bacteria. Agric. Biol. Chem. 1982, 46, 2003–2008. [Google Scholar]
- Thakur, K.; Tomar, S.K.; De, S. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.; van Loon, A.P.; Hohmann, H.P. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 1998, 180, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Yépez, A.; Russo, P.; Spano, G.; Khomenko, I.; Biasioli, F.; Capozzi, V.; Aznar, R. In situ riboflavin fortification of different kefir-like cereal-based beverages using selected Andean LAB strains. Food Microbiol. 2019, 77, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Mohedano, M.L.; Hernández-Recio, S.; Yépez, A.; Requena, T.; Martínez-Cuesta, M.C.; Peláez, C.; Russo, P.; LeBlanc, J.G.; Spano, G.; Aznar, R.; et al. Real-time detection of riboflavin production by Lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using mCherry labeling. Front. Microbiol. 2019, 10, 1748. [Google Scholar] [CrossRef]

| Gene | Primer |
|---|---|
| ribA | f-5′-GAGCGCCGGTATGATATTGC-3′ |
| r-5′-GTGTCCCTCCGTTAGTGTGA-3′ | |
| ribB | f-5′-TGGCAGTTGATGGGGTTAGT-3′ |
| r-5′-CTAATTGCCGGGCCAAGTAC-3′ | |
| ribC | f-5′-CGCTACAGGTGGACCGACTA-3′ |
| r-5′-GCATTCAACCCGACAAGGTA-3′ | |
| ribG | f-5′-ACGTACCAAAATCCCCAGGT-3′ |
| r-5′-ACATCCACCTCAGCATGGTC-3′ | |
| ribH | f-5′-ACCGGGAGCTTTTGAGATTC-3′ |
| r-5′-ATAGTCAAAGTGCGCGGTTG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.-Y.; Zhang, J.-R.; Corke, H.; Gan, R.-Y. Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application. Foods 2020, 9, 88. https://doi.org/10.3390/foods9010088
Ge Y-Y, Zhang J-R, Corke H, Gan R-Y. Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application. Foods. 2020; 9(1):88. https://doi.org/10.3390/foods9010088
Chicago/Turabian StyleGe, Ying-Ying, Jia-Rong Zhang, Harold Corke, and Ren-You Gan. 2020. "Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application" Foods 9, no. 1: 88. https://doi.org/10.3390/foods9010088
APA StyleGe, Y.-Y., Zhang, J.-R., Corke, H., & Gan, R.-Y. (2020). Screening and Spontaneous Mutation of Pickle-Derived Lactobacillus plantarum with Overproduction of Riboflavin, Related Mechanism, and Food Application. Foods, 9(1), 88. https://doi.org/10.3390/foods9010088





