Depicting the Non-Covalent Interaction of Whey Proteins with Galangin or Genistein Using the Multi-Spectroscopic Techniques and Molecular Docking
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Sample Solutions
2.3. Assays of Fluorescence Spectroscopy
2.3.1. Determination of Fluorescent Quenching Mechanism
2.3.2. Assays of Apparent Binding Constants and Site Numbers
2.3.3. Assay of Apparent Thermodynamic Parameters
2.3.4. Assay of Efficiency of Energy Transfer
2.4. Assay of Ultra-Violet Spectroscopy
2.5. Assay of Three-Dimensional Fluorescence Spectra
2.6. Molecular Docking
2.7. Statistical Analysis
3. Results
3.1. The Non-Covalent Interactions between WPI and Galangin or Genistein
3.2. Secondary Conformation Changes of WPI Induced by the Non-Covalent Interactions
3.3. The Binding Sites and Interaction Energy of the WPI-Polyphenol Interactions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bordenave, N.; Hamaker, B.R.; Ferruzzi, M.G. Nature and consequences of non-covalent interactions between flavonoids and macronutrients in foods. Food Funct. 2014, 5, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Ding, H.F.; Hu, X.; Zhang, G.W.; Gong, D.M. Galangin inhibits α-glucosidase activity and formation of non-enzymatic glycation products. Food Chem. 2019, 271, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Patel, S.; Meling, D.D.; Deal, K.; Gao, L.Y.; Helferich, W.G.; Flaws, J.A. The effects of dietary levels of genistein on ovarian follicle number and gene epression. Reprod. Toxicol. 2018, 81, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Russo, G.L.; Daglia, M.; Kasi, P.D.; Ravi, S.; Nabavi, S.F. Understanding genistein in cancer: The “good” and the “bad” effects: A review. Food Chem. 2016, 196, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein-phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Mehranfar, F.; Bordbar, A.K.; Parastar, H. A combined spectroscopic, molecular docking and molecular dynamic simulation study on the interaction of queretin with β-casein nanoparticles. J. Photoch. Photobiol. B 2013, 127, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, V.; Madureira, A.R.; Sarmento, B.; Gomes, A.; Pintado, M.E. Study of the interactions between rosmarinic acid and bovine milk whey protein α-lactalbumin, β-lactoglobulin and lactoferrin. Food Res. Int. 2015, 77, 450–459. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.Y. Binding, stability, and antioxidant activity of quercetin with soy protein isolate particles. Food Chem. 2015, 18, 24–29. [Google Scholar] [CrossRef]
- Al-Hanish, A.; Stanic-Vucinic, D.; Mihailovic, J.; Prodic, I.; Minic, S.; Stojadinovic, M.; Radibratovic, M.; Milcic, M.; Velickovic, T.C. Noncovalent interactions of bovine α-lactalbumin with green tea polyphenol, epigalocatechin-3-gallate. Food Hydrocolloid. 2016, 61, 241–250. [Google Scholar] [CrossRef]
- Poόr, M.; Boda, G.; Kunsági-Máté, S.; Needs, P.W.; Kroon, P.A.; Lemli, B. Fluorescence spectroscopic evaluation of the interactions of quercetin, isorhamnetin, and quercetin-3’-sulfate with different albumins. J. Lumin. 2018, 194, 156–163. [Google Scholar] [CrossRef]
- Rawel, H.M.; Czakka, D.; Rohn, S.; Kroll, J. Interactions of different phenolic acids and flavonoids with soy proteins. Int. J. Biol. Macromol. 2002, 30, 137–150. [Google Scholar] [CrossRef]
- Yuksel, Z.; Avci, E.; Erdem, Y.K. Characteization of binding interactions between green tea flavonoids and milk proteins. Food Chem. 2010, 121, 450–456. [Google Scholar] [CrossRef]
- Jobstl, E.; Howse, J.R.; Fairclough, J.P.A.; Williamson, M.P. Nocovalent cross-linking of casein by epigallocatechingallate characterized by single molecule force microscopy. J. Agric. Food Chem. 2006, 54, 4077–4081. [Google Scholar] [CrossRef] [PubMed]
- Hasni, I.; Bourassa, P.; Hamdani, S.; Samson, G.; Carpentier, R.; Tajmir-Riahi, H.A. Interaction of milk α- and β-caseins with tea polyphenol. Food Chem. 2011, 13, 1837–1845. [Google Scholar] [CrossRef]
- Kanakis, C.D.; Hasni, I.; Bourassa, P.; Tarantilis, P.A.; Polissiou, M.G.; Tajmir-Riahi, H. Milk β-lactoglobulin complexes with tea polyphenols. Food Chem. 2011, 127, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Bose, A. Interaction of tea polyphenols with serum albumins: A fluorescence spectroscopic analysis. J. Lumin. 2016, 169, 220–226. [Google Scholar] [CrossRef]
- Czubinski, J.; Dwiecki, K. A review of methods used for investigation of protein-phenolic compound interactions. Int. J. Food Sci. Tech. 2017, 52, 573–585. [Google Scholar] [CrossRef]
- Arts, M.J.; Haenen, G.R.; Voss, H.P.; Bast, A. Masking of antioxidant capacity by the interaction of flavonoids with protein. Food Chem. Toxicol. 2001, 39, 781–791. [Google Scholar] [CrossRef]
- Haratifar, S.; Meckling, K.A.; Corredig, M. Antiproliferative activity of tea catechins associated with casein micelles, using HT29 colon cancer cells. J. Dairy Sci. 2014, 97, 672–678. [Google Scholar] [CrossRef]
- Staszawski, M.V.; Pilosof, A.M.R.; Jagus, R.J. Antioxidant and antimicrobial performance of different Argentinean green tea varieties as affected by whey proteins. Food Chem. 2011, 125, 186–192. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.H. Degradation kinetics of fisetin and quercetin in solutions affected by medium pH, temperature and coexisted proteins. J. Serb. Chem. Soc. 2016, 81, 243–253. [Google Scholar] [CrossRef]
- Mohanmmadi, F.; Moeeni, M. Analysis of binding interaction of genistein and kaempferol with bovine alpha-lactalbumin. J. Funct. Foods 2015, 12, 458–467. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.; Dai, T.T.; Li, P.Y.; Ye, X.Q.; Chen, J.; Liu, C.M. Comparing the binding interaction between β-lactoglobulin and flavonoids with different structure by multi-spectroscopy analysis and molecular docking. Spectrochim. Acta A 2018, 201, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, D.P.; Mohapatra, S.; Misra, S.; Sahu, P.S. Milk derived bioactive peptides and their impact on human health-A review. Saudi J. Biol. Sci. 2016, 23, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Pike, A.C.; Brew, K.; Acharya, K.R. Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase. Structure 1996, 4, 691–703. [Google Scholar] [CrossRef]
- Vijayalakshmi, L.; Krishna, R.; Sankaranarayanan, R.; Vijayan, M. An asymmetric dimer of beta-lactoglobulin in a low humidity crystal form-structural changes that accompany partial dehydration and protein action. Proteins 2008, 71, 241–249. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstorm, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and Autodock Tools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Lei, S.C.; Xu, D.L.; Saeeduddin, M.; Riaz, A. Characterization of molecular structures of theaflavins and the interactions with bovine serum albumin. J. Food Sci. Technol. 2017, 54, 3421–3432. [Google Scholar] [CrossRef]
- Allahdad, Z.; Varidi, M.; Zadmard, R.; Saboury, A.A. Spectroscopic and docking studies on the interaction between caseins and β-carotene. Food Chem. 2018, 255, 187–196. [Google Scholar] [CrossRef]
- Ren, G.Y.; Sun, H.; Guo, J.Y.; Fan, J.L.; Li, G.; Xu, S.W. Molecular mechanism of interaction between resveratrol and trypsin by spectroscopy and molecular docking. Food Funct. 2019, 10, 3291–3302. [Google Scholar] [CrossRef]
- Bahri, A.; Henriquet, C.; Pugnière, M.; Marchesseau, S.; Chevalier-Lucia, D. Binding analysis between monomeric β-casein and hydrophobic bioactive compounds investigated by surface plasmon resonance and fluorescence spectroscopy. Food Chem. 2019, 286, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.J.; Gao, X.; Hao, M.H.; Tang, L. Comparison of binding interaction between β-lactoglobulin and three common polyphenols using multi-spectroscopy and modeling methods. Food Chem. 2017, 228, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, D.; Wang, G.; Lu, Y. Study of interaction between human serum albumin and three antioxidants: Ascorbic acid, alpha-tocopherol, and proanthocyanidins. Eur. J. Med. Chem. 2013, 70, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Sastry, M.C.S.; Rao, M.S.N. Binding of chlorogenic acid by the isolated polyphenol-free 11 S protein of sunflower (Helianthus annuus) seed. J. Agric. Food Chem. 1990, 38, 2103–2110. [Google Scholar] [CrossRef]
- Prigent, S.V.E.; Koningsveld, G.A.V.; Gruppen, H.; Visser, A.J.W.G.; Voragen, A.G.J. Effects of non-covalent interactions with 5-o-caffeoylquinic acid (CGA) on the heat denaturation and solubility of globular proteins. J. Agric. Food Chem. 2003, 51, 5088–5095. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.B.; Mao, F.F.; Yang, F.; Zhao, Y.L.; Zhang, C.; Yamamoto, K. Interaction of dietary polyphenols with bovine milk proteins: Molecular structure affinity relationship and influencing bioactivity aspects. Mol. Nutr. Food Res. 2011, 55, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.F.; Mcsweeney, P.L.H. Dairy Chemistry and Biochemistry, 1st ed.; Blackie Academic & Professional: Landon, UK, 1998; pp. 157–158. [Google Scholar]
- Sahihi, M.; Ghayeb, Y. An investigation of molecular dynamics simulation and molecular docking: Interaction of citrus flavonoids and bovine β-lactoglobulin in focus. Comput. Biol. Med. 2014, 51, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Ebeler, S.E. Flavonoid effects on DNA oxidation at low concentratios relevant to physiological levels. Food Chem. Toxicol. 2008, 46, 96–104. [Google Scholar] [CrossRef]
- Hatch, F.T.; Lightstone, F.C.; Covin, M.E. Quantitative structure-activity relationship of flavonoids for inhibition of heterocyclic amine mutagenicity. Environ. Mol. Mutagen. 2000, 35, 279–299. [Google Scholar] [CrossRef]
- Strat, K.M.; Rowley, T.J.; Smithson, A.T.; Tessem, J.S.; Hulver, M.W.; Liu, D.; Davya, B.M.; Davy, K.P.; Neilson, A.P. Mechanisms by which cocoa flavanols improve metabolic syndrome and related disorders. J. Nutr. Biochem. 2016, 35, 1–21. [Google Scholar] [CrossRef]
- Shang, S.J.; Liu, Q.L.; Gao, J.D.; Zhu, Y.L.; Liu, J.Y.; Wang, K.Y.; Wei, S.; Zhang, S.D. Insights into in vitro binding of parecoxib to human serum albumin by spectroscopic methods. J. Biochem. Mol. Toxic. 2014, 28, 433–441. [Google Scholar] [CrossRef] [PubMed]
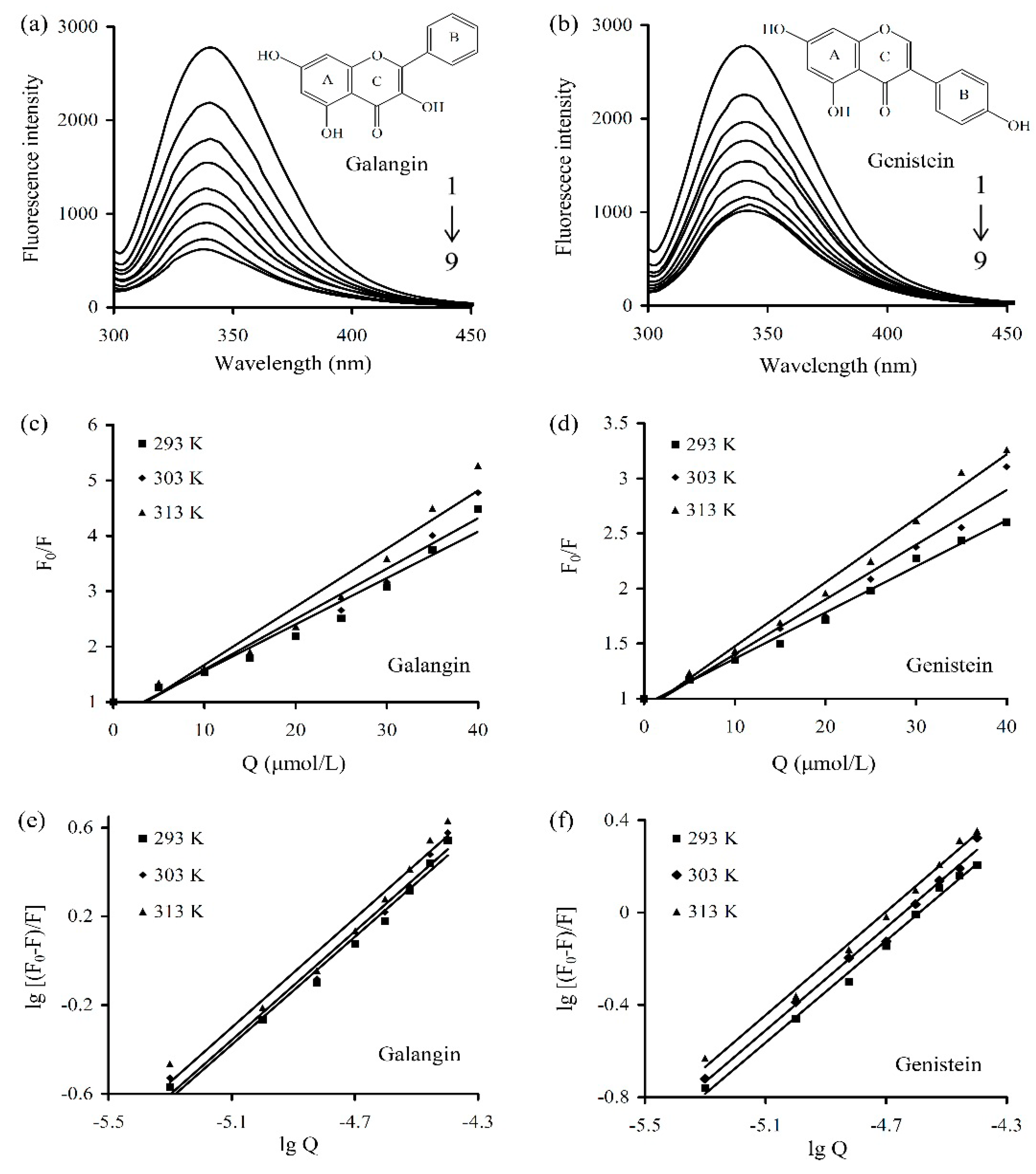
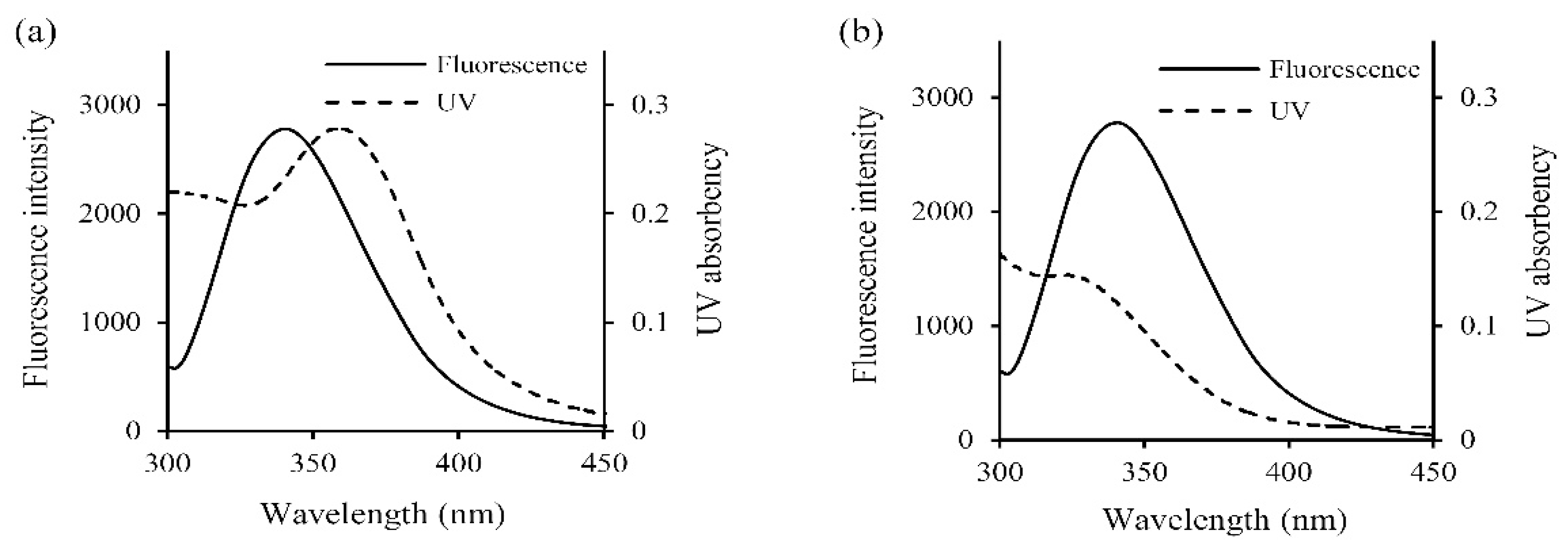


| Polyphenol | T (K) | Equation | Ksv (104 L/mol) | Kq (1012 L/(mol·s)) | R2 |
|---|---|---|---|---|---|
| Galangin | 293 | Y = 0.0838Q + 0.7249 | 8.38 ± 0.20a | 8.38 ± 0.20a | 0.959 |
| 303 | Y = 0.0891Q + 0.6836 | 8.91 ± 0.24b | 8.91 ± 0.24b | 0.953 | |
| 313 | Y = 0.1046Q + 0.6227 | 10.46 ± 0.16c | 10.46 ± 0.16c | 0.955 | |
| Genistein | 293 | Y = 0.0417Q + 0.9484 | 4.17 ± 0.02d | 4.17 ± 0.02d | 0.993 |
| 303 | Y = 0.0496Q + 0.9059 | 4.96 ± 0.06e | 4.96 ± 0.06e | 0.974 | |
| 313 | Y = 0.0581Q + 0.8899 | 5.81 ± 0.08f | 5.81 ± 0.08f | 0.989 |
| Polyphenol | T (K) | n | Ka (105 L/mol) | ΔH (kJ/mol) | ΔG (kJ/mol) | ΔS (J/(mol·K)) |
|---|---|---|---|---|---|---|
| Galangin | 293 | 1.22 ± 0.01 | 6.96 ± 0.65 | 12.5 ± 2.2 | −(32.8 ± 2.5) | 154.6 ± 13.8 |
| 303 | 1.24 ± 0.04 | 9.03 ± 0.54 | −(34.4 ± 2.6) | |||
| 313 | 1.23 ± 0.01 | 9.64 ± 0.47 | −(35.9 ± 2.4) | |||
| Genistein | 293 | 1.10 ± 0.02 | 1.08 ± 0.17 | 41.1 ± 1.8 | −(28.1 ± 1.9) | 236.4 ± 6.0 |
| 303 | 1.12 ± 0.06 | 1.69 ± 0.95 | −(30.5 ± 1.9) | |||
| 313 | 1.15 ± 0.08 | 3.18 ± 0.76 | −(32.9 ± 2.2) |
| Protein-Polyphenol Complex | SDS/Urea Addition | Ka (L/mol) |
|---|---|---|
| WPI-galangin | SDS | (2.24 ± 0.52) × 105 |
| Urea | (8.95 ± 0.42) × 105 | |
| WPI-genistein | SDS | (3.93 ± 0.23) × 104 |
| Urea | (1.59 ± 0.22) × 105 |
| Protein-Polyphenol Complex | J (cm3·L/mol) | R0 (nm) | E | r (nm) |
|---|---|---|---|---|
| WPI-galangin | 5.30×10-15 | 2.41 | 0.542 | 2.34 |
| WPI-genistein | 1.38×10-16 | 1.31 | 0.417 | 1.39 |
| Peak Parameter | WPI | WPI-Galangin | WPI-Genistein | |
|---|---|---|---|---|
| Peak I | Peak position λex/λem (nm/nm) | 240/340 | 240/345 | 240/345 |
| Fluorescence intensity | 311.4 | 130.5 | 115.7 | |
| Peak II | Peak position λex/λem (nm/nm) | 280/340 | 280/345 | 280/345 |
| Fluorescence intensity | 2327 | 639.6 | 1028 | |
| Protein and Polyphenol | Involved Residue | H-Bond Number | ΔG (kJ/mol) |
|---|---|---|---|
| β-Lactoglobulin and galangin | Leu-10, Asp-11, Ile-12, Lys-75, Thr-76, Lys-77, Ile-78, Pro-79, Ala-80, Val-81, Phe-82 | 0 | −30.06 |
| β-Lactoglobulin and genistein | Leu-10, Asp-11, Ile-12, Gln-13, Lys-75, Thr-76, Ile-78, Pro-79, Ala-80, Val-81, Phe-82 | 0 | −29.39 |
| α-Lactalbumin and galangin | Pro-109, Leu-110, Cys-111, Ser-112 *, Asp-113, Lys-114, Leu-115, Gln-117, Trp-118 | 1 | −25.62 |
| α-Lactalbumin and genistein | Gln-2, Leu-3, Thr-4, Lys-5, Phe-31, Tyr-36, Gln-117, Trp-118 | 0 | −27.42 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.-M.; Zhao, X.-H. Depicting the Non-Covalent Interaction of Whey Proteins with Galangin or Genistein Using the Multi-Spectroscopic Techniques and Molecular Docking. Foods 2019, 8, 360. https://doi.org/10.3390/foods8090360
Ma C-M, Zhao X-H. Depicting the Non-Covalent Interaction of Whey Proteins with Galangin or Genistein Using the Multi-Spectroscopic Techniques and Molecular Docking. Foods. 2019; 8(9):360. https://doi.org/10.3390/foods8090360
Chicago/Turabian StyleMa, Chun-Min, and Xin-Huai Zhao. 2019. "Depicting the Non-Covalent Interaction of Whey Proteins with Galangin or Genistein Using the Multi-Spectroscopic Techniques and Molecular Docking" Foods 8, no. 9: 360. https://doi.org/10.3390/foods8090360
APA StyleMa, C.-M., & Zhao, X.-H. (2019). Depicting the Non-Covalent Interaction of Whey Proteins with Galangin or Genistein Using the Multi-Spectroscopic Techniques and Molecular Docking. Foods, 8(9), 360. https://doi.org/10.3390/foods8090360





