HPTLC-PCA Complementary to HRMS-PCA in the Case Study of Arbutus unedo Antioxidant Phenolic Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Sampling Sites and Extraction
2.3. HPTLC Analyses
2.4. LC–ESI–Orbitrap–MS Analysis
2.5. PCA
2.6. Antiradical Activity by Diphenyl-1-Picarylhydrazyl (DPPH) and TEAC Assays
2.7. Determination of Total Phenols
2.8. Statistical Analysis
3. Results and Discussion
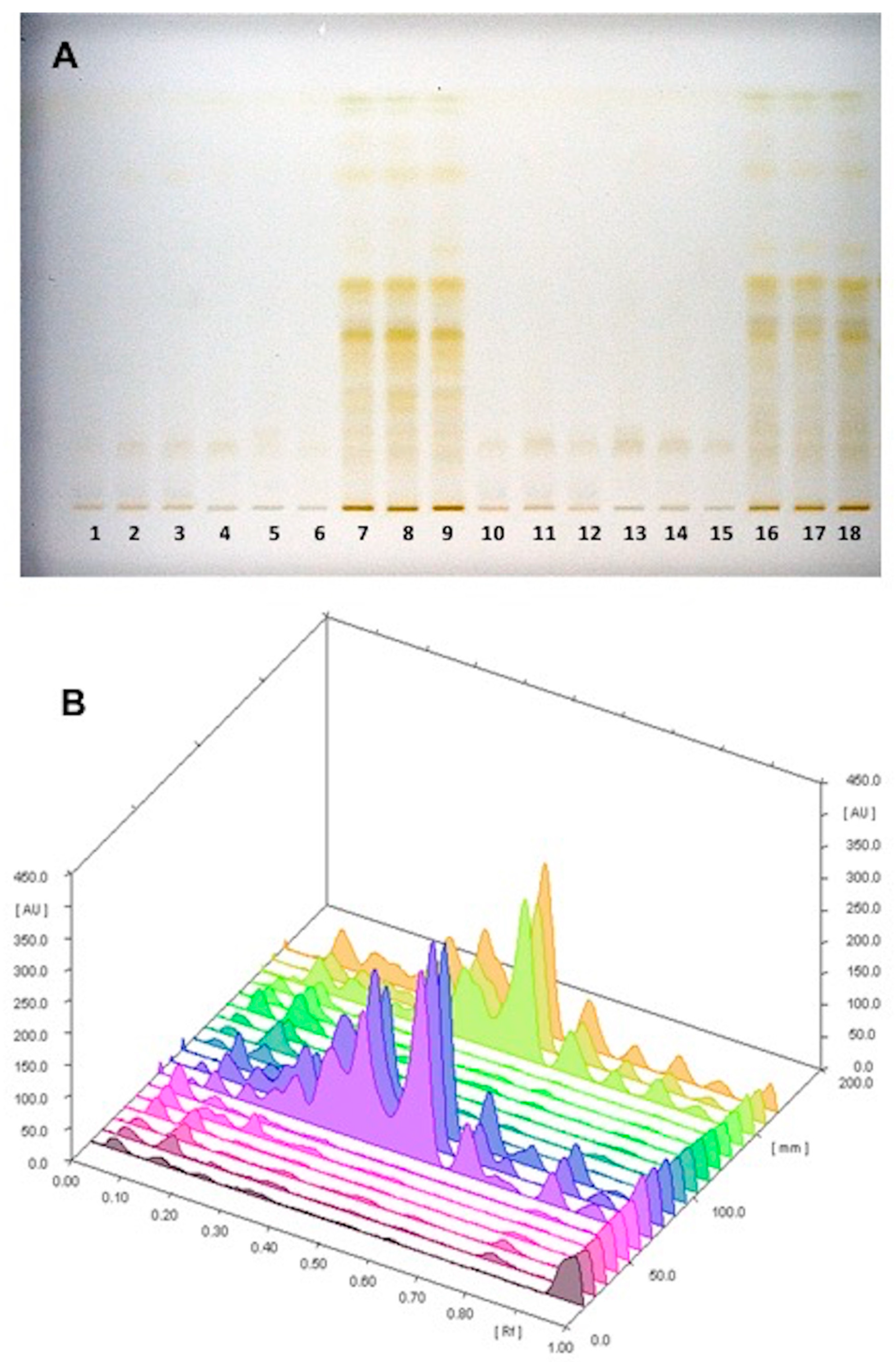
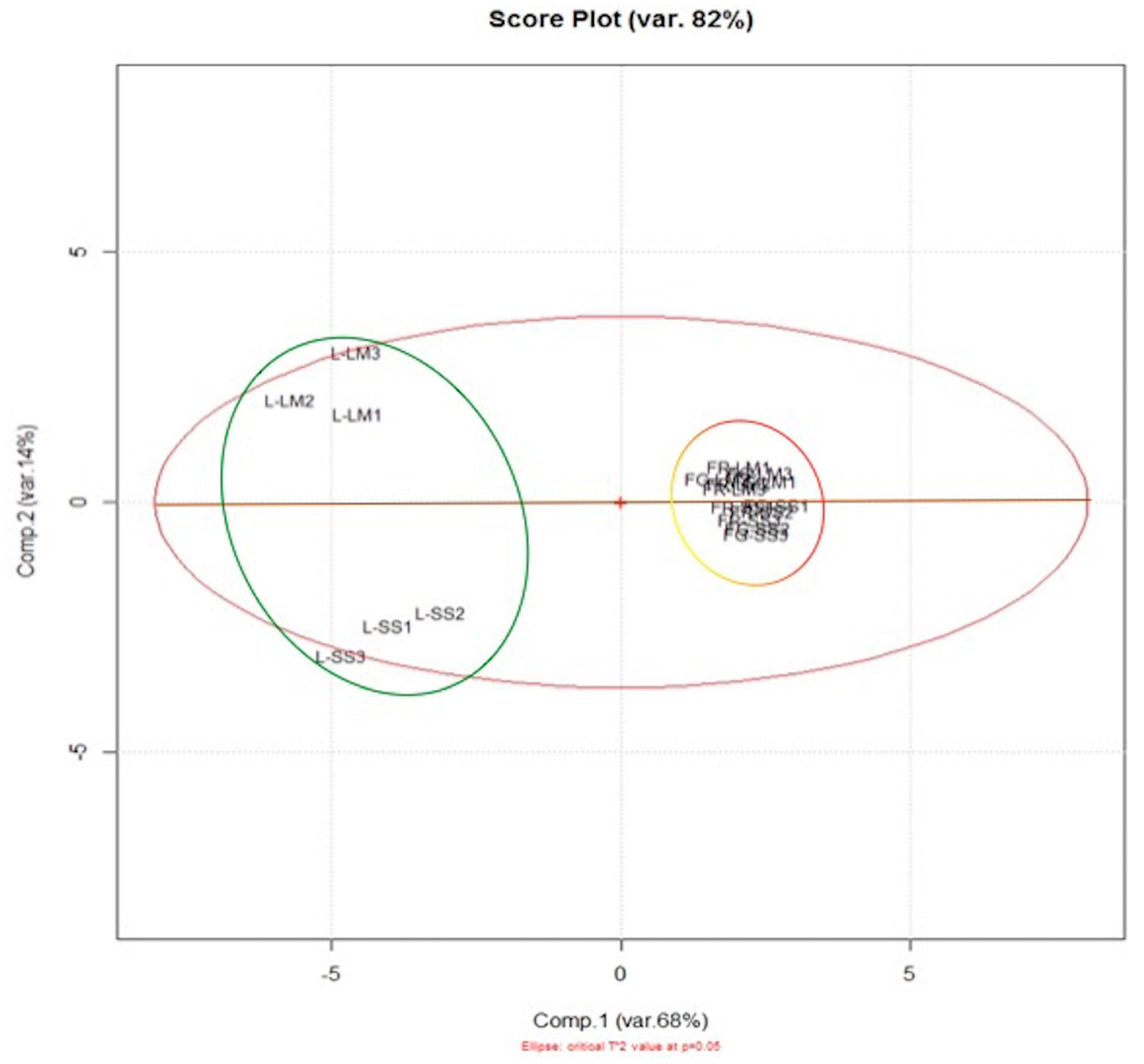
3.1. HPTLC–PCA Analysis
3.2. LC–ESI–Orbitrap–MS Analysis and PCA
3.3. Antioxidant Activity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Astarita, G.; Langridge, J. An Emerging Role for Metabolomics in Nutrition Science. J. Nutr. Nutr. 2013, 6, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.X.; Chen, T.L.; Li, M.; Chen, M.; Zhou, Y.Q.; Cui, G.H.; Zhao, A.H.; Jia, W.; Huang, L.Q.; Qi, X.Q. Use of the Metabolomics Approach to Characterize Chinese Medicinal Material Huangqi. Mol. Plant 2012, 5, 376–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kueger, S.; Steinhauser, D.; Willmitzer, L.; Giavalisco, P. High-resolution plant metabolomics: From mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. Plant J. 2012, 70, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Booker, A.; Frommenwiler, D.; Johnston, D.; Umealajekwu, C.; Reich, E.; Heinrich, M. Chemical variability along the value chains of turmeric (Curcuma longa): A comparison of nuclear magnetic resonance spectroscopy and high performance thin layer chromatography. J. Ethnopharmacol. 2014, 152, 292–301. [Google Scholar] [CrossRef]
- Holtorf, H.; Guitton, M.C.; Reski, R. Plant functional genomics. Naturwissenschaften 2002, 89, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Tolstikov, V.V.; Fiehn, O. Analysis of highly polar compounds of plant origin: Combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal. Biochem. 2002, 301, 298–307. [Google Scholar] [CrossRef]
- Vorst, O.; De Vos, C.H.R.; Lommen, A.; Staps, R.V.; Visser, R.G.F.; Bino, R.J.; Hall, R.D. A non-directed approach to the differential analysis of multiple LC-MS-derived metabolic profiles. Metabolomics 2005, 1, 169–180. [Google Scholar] [CrossRef]
- Avula, B.; Sagi, S.; Gafner, S.; Upton, R.; Wang, Y.H.; Wang, M.; Khan, I.A. Identification of Ginkgo biloba supplements adulteration using high performance thin layer chromatography and ultra high performance liquid chromatography-diode array detector-quadrupole time of flight-mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 7733–7746. [Google Scholar] [CrossRef]
- Piccinonna, S.; Ragone, R.; Stocchero, M.; Del Coco, L.; De Pascali, S.A.; Schena, F.P.; Fanizzi, F.P. Robustness of NMR-based metabolomics to generate comparable data sets for olive oil cultivar classification. An inter-laboratory study on Apulian olive oils. Food Chem. 2016, 199, 675–683. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, Z.D.; Zhao, C.X.; Kong, H.W.; Lu, X.; Xu, G.W. A comparative study of volatile components in green, oolong and black teas by using comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry and multivariate data analysis. J Chromatogr. A 2013, 1313, 245–252. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, P.; Lin, L.Z.; Harnly, J.M.; Yu, L.L.; Li, Z.W. Tentative identification, quantitation, and principal component analysis of green pu-erh, green, and white teas using UPLC/DAD/MS. Food Chem. 2011, 126, 1269–1277. [Google Scholar] [CrossRef]
- Diniz, P.H.G.D.; Barbosa, M.F.; Milanez, K.D.T.D.; Pistonesi, M.F.; De Araujo, M.C.U. Using UV-Vis spectroscopy for simultaneous geographical and varietal classification of tea infusions simulating a home-made tea cup. Food Chem. 2016, 192, 374–379. [Google Scholar] [CrossRef]
- Scoparo, C.T.; De Souza, L.M.; Dartora, N.; Sassaki, G.L.; Gorin, P.A.J.; Iacomini, M. Analysis of Camellia sinensis green and black teas via ultra high performance liquid chromatography assisted by liquid-liquid partition and two-dimensional liquid chromatography (size exclusion x reversed phase). J Chromatogr. A 2012, 1222, 29–37. [Google Scholar] [CrossRef]
- Maldini, M.; Montoro, P.; Addis, R.; Toniolo, C.; Petretto, G.L.; Foddai, M.; Nicoletti, M.; Pintore, G. A new approach to discriminate Rosmarinus officinalis L. plants with antioxidant activity, based on HPTLC fingerprint and targeted phenolic analysis combined with PCA. Ind. Crop. Prod. 2016, 94, 665–672. [Google Scholar] [CrossRef]
- Martelanc, M.; Vovk, I.; Simonovska, B. Separation and identification of some common isomeric plant triterpenoids by thin-layer chromatography and high-performance liquid chromatography. J. Chromatogr. A 2009, 1216, 6662–6670. [Google Scholar] [CrossRef]
- Lakavath, S.; Avula, B.; Wang, Y.H.; Rumalla, C.S.; Gandhe, S.; Venkatrao, A.R.B.; Satishchandra, P.A.; Bobbala, R.K.; Khan, I.A.; Narasimha, A.R.A.V. Differentiating the Gum Resins of Two Closely Related Indian Gardenia Species, G. gummifera and G-lucida, and Establishing the Source of Dikamali Gum Resin Using High-Performance Thin-Layer Chromatography and Ultra-Performance Liquid Chromatography-UV/MS. J. AOAC Int. 2012, 95, 67–73. [Google Scholar] [CrossRef]
- Reich, E.; Schibli, A. High-Performance Thin Layer Chromatography for the Analysis of Medicinal Plants; Thieme: New York, NY, USA, 2007. [Google Scholar]
- Ogegbo, O.L.; Eyob, S.; Parmar, S.; Wang, Z.T.; Bligh, S.W.A. Metabolomics of four TCM herbal products: Application of HPTLC analysis. Anal. Methods UK 2012, 4, 2522–2527. [Google Scholar] [CrossRef]
- Audoin, C.; Holderith, S.; Romari, K.; Thomas, O.P.; Genta-Jouve, G. Development of a Work-Flow for High-Performance Thin-Layer Chromatography Data Processing for Untargeted Metabolomics. JPC J. Planar Chromatogr. 2014, 27, 328–332. [Google Scholar] [CrossRef]
- Wang, J.; Cao, X.S.; Qi, Y.D.; Ferchaud, V.; Chin, K.L.; Tang, F. High-Performance Thin-Layer Chromatographic Method for Screening Antioxidant Compounds and Discrimination of Hibiscus sabdariffa L. by Principal Component Analysis. JPC J. Planar Chromatogr. 2015, 28, 274–279. [Google Scholar] [CrossRef]
- Tuberoso, C.I.G.; Bifulco, E.; Caboni, P.; Cottiglia, F.; Cabras, P.; Floris, I. Floral Markers of Strawberry Tree (Arbutus unedo L.) Honey. J. Agric. Food Chem. 2010, 58, 384–389. [Google Scholar] [CrossRef]
- Kontogianni, V.G.; Tomic, G.; Nikolic, I.; Nerantzaki, A.A.; Sayyad, N.; Stosic-Grujicic, S.; Stojanovic, I.; Gerothanassis, I.P.; Tzakos, A.G. Phytochemical profile of Rosmarinus officinalis and Salvia officinalis extracts and correlation to their antioxidant and anti-proliferative activity. Food Chem. 2013, 136, 120–129. [Google Scholar] [CrossRef]
- Van De Velde, F.; Tarola, A.M.; Guemes, D.; Pirovani, M.E. Bioactive compounds and antioxidant capacity of Camarosa and Selva strawberries (Fragaria x ananassa Duch.). Foods 2013, 2, 120–131. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- D’urso, G.; Pizza, C.; Piacente, S.; Montoro, P. Combination of LC-MS based metabolomics and antioxidant activity for evaluation of bioactive compounds in Fragaria vesca leaves from Italy. J. Pharm. Biomed. 2018, 150, 233–240. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free-Radical Method to Evaluate Antioxidant Activity. Food Sci. Technol. Leb 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Petretto, G.L.; Maldini, M.; Addis, R.; Chessa, M.; Foddai, M.; Rourke, J.P.; Pintore, G. Variability of chemical composition and antioxidant activity of essential oils between Myrtus communis var. Leucocarpa DC and var. Melanocarpa DC. Food Chem. 2016, 197, 124–131. [Google Scholar] [CrossRef]
- Lizcano, L.J.; Bakkali, F.; Ruiz-Larrea, M.B.; Ruiz-Sanz, J.I. Antioxidant activity and polyphenol content of aqueous extracts from Colombian Amazonian plants with medicinal use. Food Chem. 2010, 119, 1566–1570. [Google Scholar] [CrossRef]
- Jardim, C.; Macedo, D.; Figueira, I.; Dobson, G.; Mcdongall, G.J.; Stewart, D.; Ferreira, R.B.; Menezes, R.; Santos, C.N. (Poly)phenol metabolites from Arbutus unedo leaves protect yeast from oxidative injury by activation of antioxidant and protein clearance pathways. J. Funct. Foods 2017, 32, 333–346. [Google Scholar] [CrossRef]
- Pallauf, K.; Rivas-Gonzalo, J.C.; Del Castillo, M.D.; Cano, M.P.; De Pascual-Teresa, S. Characterization of the antioxidant composition of strawberry tree (Arbutus unedo L.) fruits. J. Food Compos. Anal. 2008, 21, 273–281. [Google Scholar] [CrossRef]
- Tuberoso, C.I.G.; Boban, M.; Bifulco, E.; Budimir, D.; Pirisi, F.M. Antioxidant capacity and vasodilatory properties of Mediterranean food: The case of Cannonau wine, myrtle berries liqueur and strawberry-tree honey. Food Chem. 2013, 140, 686–691. [Google Scholar] [CrossRef]
- Nenadis, N.; Llorens, L.; Koufogianni, A.; Diaz, L.; Font, J.; Gonzalez, J.A.; Verdaguer, D. Interactive effects of UV radiation and reduced precipitation on the seasonal leaf phenolic content/composition and the antioxidant activity of naturally growing Arbutus unedo plants. J. Photochem. Photobiol. B 2015, 153, 435–444. [Google Scholar] [CrossRef]
- Fortalezas, S.; Tavares, L.; Pimpao, R.; Tyagi, M.; Pontes, V.; Alves, P.M.; Mcdougall, G.; Stewart, D.; Ferreira, R.B.; Santos, C.N. Antioxidant Properties and Neuroprotective Capacity of Strawberry Tree Fruit (Arbutus unedo). Nutrients 2010, 2, 214–229. [Google Scholar] [CrossRef]
- Longhi, J.G.; Perez, E.; De Lima, J.J.; Candido, L.M.B. In vitro evaluation of Mucuna pruriens (L.) DC. antioxidant activity. Braz. J. Pharm. Sci. 2011, 47, 535–544. [Google Scholar] [CrossRef]
- Erkan, N.; Ayranci, G.; Ayranci, E. Antioxidant activities of rosemary (Rosmarinus Officinalis, L.) extract, blackseed (Nigella sativa L.) essential oil, carnosic acid, rosmarinic acid and sesamol. Food Chem. 2008, 110, 76–82. [Google Scholar] [CrossRef]
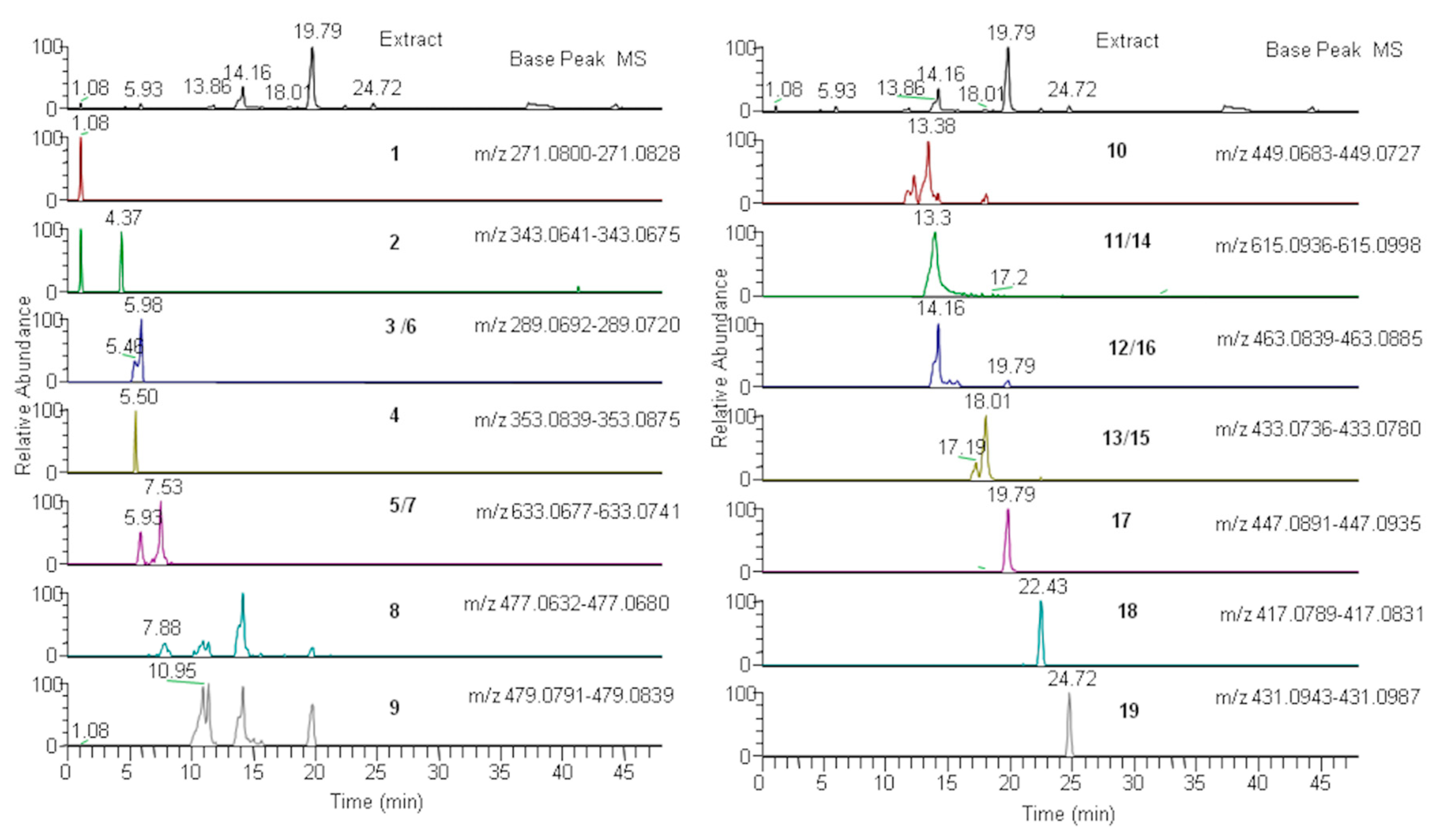

| No. | RT | [M − H]− | Molecular Formula | Δ PPM | MS/MS | Identity | LMFG | LMFR | LML | SSFG | SSFR | SSL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.08 | 271.0814 | C12H15O7 | 0.6 | 139.08 | arbutin | x | x | x | nd | x | x |
| 2 | 4.3 | 343.0658 | C14H15O10 | −0.6 | 283.8/191.1 | galloyl quinic acid | x | x | x | x | x | x |
| 3 | 5.4 | 289.0708 | C27H21O18 | −1.5 | 275.9/245.1/205.1/144.2 | catechin/epicatechin | x | x | x | x | x | x |
| 4 | 5.5 | 353.0857 | C16H17O9 | −2.8 | 262.8/191.01/171.7 | chlorogenic acid | nd | nd | x | nd | nd | x |
| 5 | 5.8 | 633.0712 | C27H21O18 | −1.5 | 463.06/300.9 | strictinin ellagitannin | nd | x | x | x | nd | x |
| 6 | 5.9 | 289.0706 | C27H21O18 | −1.5 | 275.9/245.1/205.1/144.2 | catechin/epicatechin | x | x | x | x | x | x |
| 7 | 7.6 | 633.0709 | C27H21O18 | −1.5 | 463.06/300.9 | strictinin ellagitannin | nd | x | x | x | nd | x |
| 8 | 8.0 | 477.0656 | C21H17O13 | 0.2 | 462.0/366.07/325.05/191.03/164.7 | digalloylquinic shikimic acid | x | x | x | x | nd | x |
| 9 | 10.9 | 479.0815 | C21H19O13 | −1.43 | 451.2/354.7/316.02/297.6/165.8/145.5 | myricetin glucoside | nd | nd | x | nd | nd | x |
| 10 | 13.0 | 449.0705 | C20H17O12 | −1.7 | 317.02/183.3/149.5 | myricetin pentoside | nd | x | x | nd | nd | x |
| 11 | 13.3 | 615.0967 | C28H23O16 | 1.8 | 463.08/301.03 | quercetin galloylhexoside isomer | x | x | x | x | x | x |
| 12 | 14.2 | 463.0862 | C21H19O12 | −1.5 | 316.02/284.8/183.5 | myricetin rhamnoside | nd | x | x | x | nd | x |
| 13 | 17.1 | 433.0758 | C20H17O11 | −1.12 | 355.3/312.7/301.03/283.8/216.2 | quercetin pentoside | x | x | x | x | x | x |
| 14 | 17.2 | 615.0960 | C28H23O16 | 1.02 | 463.08/301.03/265.3/241.08 | quercetin galloylhexoside isomer | x | x | x | x | x | x |
| 15 | 17.7 | 433.0758 | C20H17O11 | −1.1 | 301.03/265.8/247.7/219.2/181.9/134.9 | quercetin pentoside | x | x | x | x | x | x |
| 16 | 19.5 | 463.0860 | C21H19O12 | 0.8 | 404.3/316.02/301.04/282; 2 | isoquercitrin | nd | x | x | x | nd | x |
| 17 | 19.7 | 447,0913 | C21H19O11 | −1.2 | 301.03/287.9/178.9 | quercitrin | x | x | x | x | x | x |
| 18 | 22.4 | 417.0810 | C20H17O10 | −0.9 | 374.9/285.04/175.6 | kaempferol pentoside | nd | x | x | x | x | x |
| 19 | 24.7 | 431.0965 | C21H19O10 | −1.09 | 285.03/235.2/195.9 | Kaempferol-rhamnoside (afzelin) | x | x | x | nd | x | x |
| DPPH IC50 | ABTS IC50 | |||||
|---|---|---|---|---|---|---|
| 0 min | 30 min | p Value | 0 min | 50 min | p Value | |
| Trolox (µg/mL) | 13.50 | 6.15 | 3.34 | 3.28 | ||
| A. unedo Sassari Red fruits (µg/mL) | 229.09 ± 81.75 | 103.59 ± 45.97 | Time 0 min LSS vs. FR SS p < 0.05 LSS vs. FG SS p < 0.05 | 52.71 ± 16.13 | 21.43 ± 7.34 | Time 0 min LSS vs. FR SS p < 0.01 LSS vs. FG SS p < 0.05 |
| A. unedo Sassari Yellow fruits (µg/mL) | 300.92 ± 107.31 | 131.53 ± 44.61 | 101.11 ± 53.44 | 40.33 ± 21.95 | ||
| A. unedo Sassari leaves (µg/mL) | 29.37 ± 5.87 | 15.18 ± 4.11 | Time 30 min LSS vs. FR SS p < 0.05 LSS vs. FG SS p < 0.01 | 7.63 ± 1.38 | 2.35 ± 0.06 | Time 50 min LSS vs. FR SS p < 0.01 LSS vs. FG SS p < 0.05 |
| A. unedo La Maddalena Red fruits (µg/mL) | 235.67 ± 43.72 | 116.84 ± 16.54 | Time 0 min L LM vs. FR LM p < 0.01 L LM vs. FG LM p < 0.01 | 79.36 ± 46.09 | 34.91 ± 23.07 | Time 0 min L LM vs. FG LM p < 0.05 |
| A. unedo La Maddalena Yellow fruits (µg/mL) | 380.14 ± 90.72 | 180.30 ± 75.48 | 96.09 ± 40.98 | 49.89 ± 17.62 | ||
| A. unedo La Maddalena leaves (µg/mL) | 23.35 ± 4.25 | 8.85 ± 7.79 | Time 30 min L LM vs. FR LM p < 0.01 L LM vs. FG LM p < 0.05 | 6.73 ± 2.14 | 2.72 ± 1.68 | Time 50 min L LM vs. FG LM p < 0.01 |
| Extract Concent Ration (µg/mL) | GAE A. Unedo Sassari Red Fruits (µg GAE) | GAE A. Unedo Sassari Yellow Fruits (µg GAE) | GAE A. Unedo Sassari Leaves (µg GAE) | GAE A. Unedo La Maddalena Red Fruits (µg GAE) | GAE A. Unedo La Maddalena Yellow Fruits (µg GAE) | GAE A. Unedo La Maddalena Leaves(µg GAE) | Pearson Product Moment Correlation |
|---|---|---|---|---|---|---|---|
| 100 | 67.10 ± 17.14 *† | 43.62 ± 11.85 *† | 347.56 ± 126.05 | 53.16 ± 35.75 *† | 50.62 ± 20.64 *† | 327.90 ± 126.75 | * vs. DPPH p < 0.01 † vs. ABTS p < 0.05 |
| 50 | 35.33 ± 9.37 *† | 22.72 ± 6.41 *† | 179.52 ± 70.68 | 26.42 ± 11.73 *† | 26.42 ± 8.04 *† | 181.55 ± 84.65 | |
| 25 | 23.67 ± 4.29 *† | 16.13 ± 9.12 *† | 116.46 ± 36.12 | 21.11 ± 11.83 *† | 18.10 ± 5.64 *† | 143.84 ± 67.03 | |
| 10 | 10.15 ± 3.27 *† | 6.78 ± 2.43 *† | 51.50 ± 15.93 | 9.53 ± 3.44 *† | 8.30 ± 2.42 *† | 64.56 ± 33.28 | |
| 5 | 6.63 ± 1.16 *† | 4.24 ± 0.55 *† | 29.01 ± 4.24 | 6.17 ± 3.36 *† | 5.04 ± 3.89 *† | 38.12 ± 17.86 | |
| 1 | 3.17 ± 1.66 *† | 2.40 ± 0.70 *† | 10.04 ± 1.54 | 4.91 ± 1.69 *† | 2.52 ± 1.17 *† | 18.54 ± 14.44 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maldini, M.; D’Urso, G.; Pagliuca, G.; Petretto, G.L.; Foddai, M.; Gallo, F.R.; Multari, G.; Caruso, D.; Montoro, P.; Pintore, G. HPTLC-PCA Complementary to HRMS-PCA in the Case Study of Arbutus unedo Antioxidant Phenolic Profiling. Foods 2019, 8, 294. https://doi.org/10.3390/foods8080294
Maldini M, D’Urso G, Pagliuca G, Petretto GL, Foddai M, Gallo FR, Multari G, Caruso D, Montoro P, Pintore G. HPTLC-PCA Complementary to HRMS-PCA in the Case Study of Arbutus unedo Antioxidant Phenolic Profiling. Foods. 2019; 8(8):294. https://doi.org/10.3390/foods8080294
Chicago/Turabian StyleMaldini, Mariateresa, Gilda D’Urso, Giordana Pagliuca, Giacomo Luigi Petretto, Marzia Foddai, Francesca Romana Gallo, Giuseppina Multari, Donatella Caruso, Paola Montoro, and Giorgio Pintore. 2019. "HPTLC-PCA Complementary to HRMS-PCA in the Case Study of Arbutus unedo Antioxidant Phenolic Profiling" Foods 8, no. 8: 294. https://doi.org/10.3390/foods8080294








