Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Seafood Factories
2.2. Traditional Methods of Diagnostic
- Site 1 (direct food contact): Utensils such as trays, mixer blades, bowls used for grinding, brining tanks and conveyor belts for descaling.
- Site 2 (direct food contact): Utensils such as racks, trays, bowls for mixing, net scoops for fish handling and tables for degutting.
- Site 3 (adjacent food contact): Tables and machines used for forming, salting and rinsing.
- Site 4: Factory floors.
- Site 5: Drains.
- Bacillus cereus: Mannitol Egg Yolk Polymyxin agar (Oxoid, Hampshire, UK) and Mannitol Phenol Deoxycholate agar (Oxoid, Hampshire, UK).
- Listeria monocytogenes: Fraser Broth and Oxford Agar (Oxoid, Hampshire, England.).
- Salmonella spp.: Buffered Peptone Water (Oxoid, Hampshire, UK) and Rappaport-Vassiliadis Soy Peptone (Merck, Darmstadt, Germany).
- Shigella spp.: Shigella broth with novobiocin (HiMedia, Mumbai, India), Xylose Lysine Deoxycholate agar (Oxoid, Hampshire, UK), MacConkey agar (Oxoid, Hampshire, UK), Hektoen Enteric agar (Oxoid, Hampshire, UK) and nutrient agar (Merck, Darmstadt, Germany).
- Vibrio spp. (V. cholerae, V. parahaemolyticus, V. vulnificus): Alkaline Peptone Water (Oxoid, Hampshire, UK), Selenite F Broth (HiMedia, Mumbai, India) and Thiosulfate–Citrate–Bile Salts–Sucrose agar (Merck, Darmstadt, Germany).
2.3. Next-Generation Sequencing Method
2.3.1. Sampling Procedures
2.3.2. NGS Workflow
- Amplicon Primer, Bacterial 16S V3-V4 (5′ to 3′).
- Forward primer (16S V3-V4): CCTACGGGNGGCWGCAG.
- Reverse primer (16S V3-V4): GACTACHVGGGTATCTAATCC.
2.3.3. Bioinformatics and Data Analysis
3. Results and Discussions
3.1. Culturable Pathogens Using Traditional Method
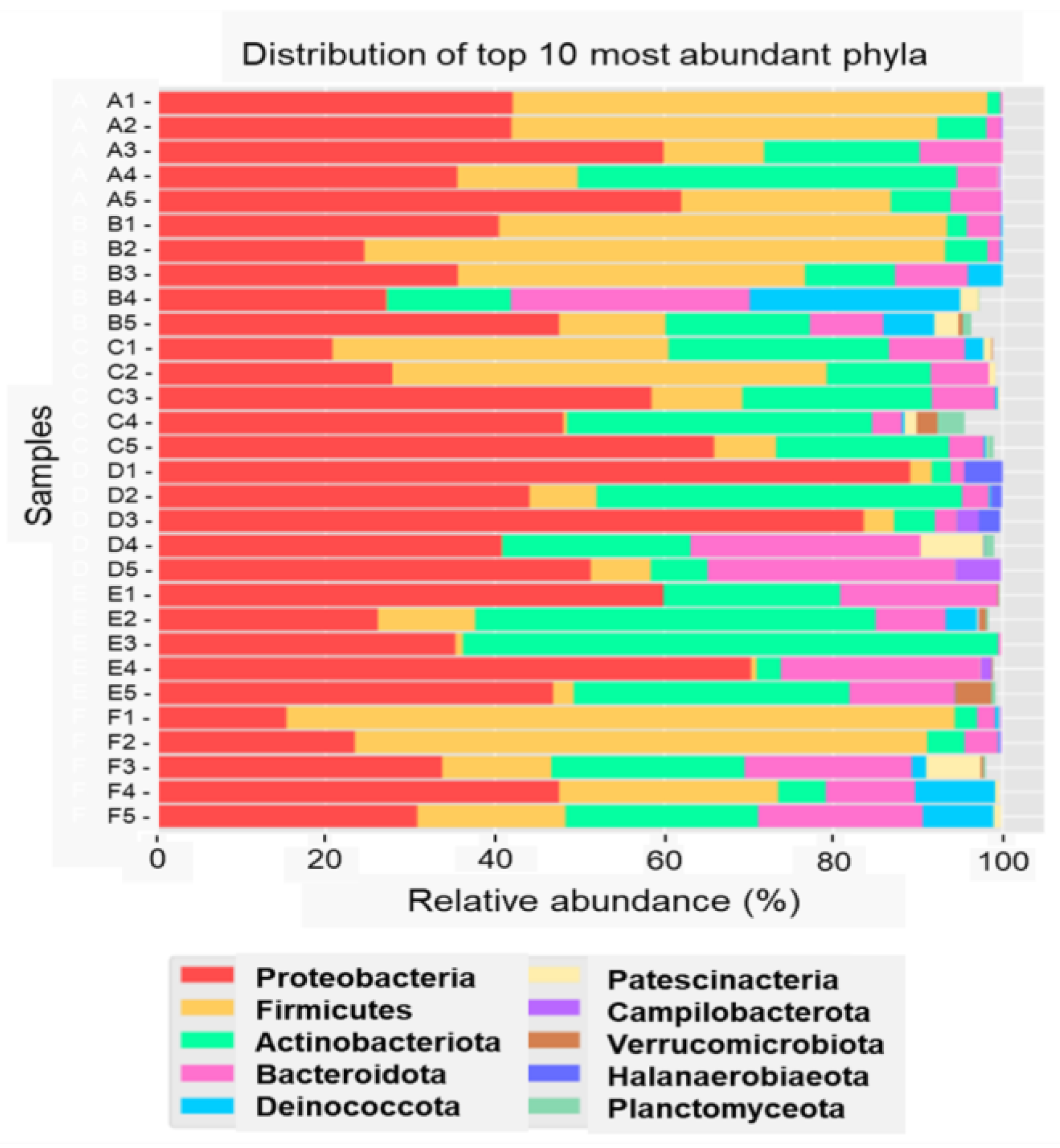
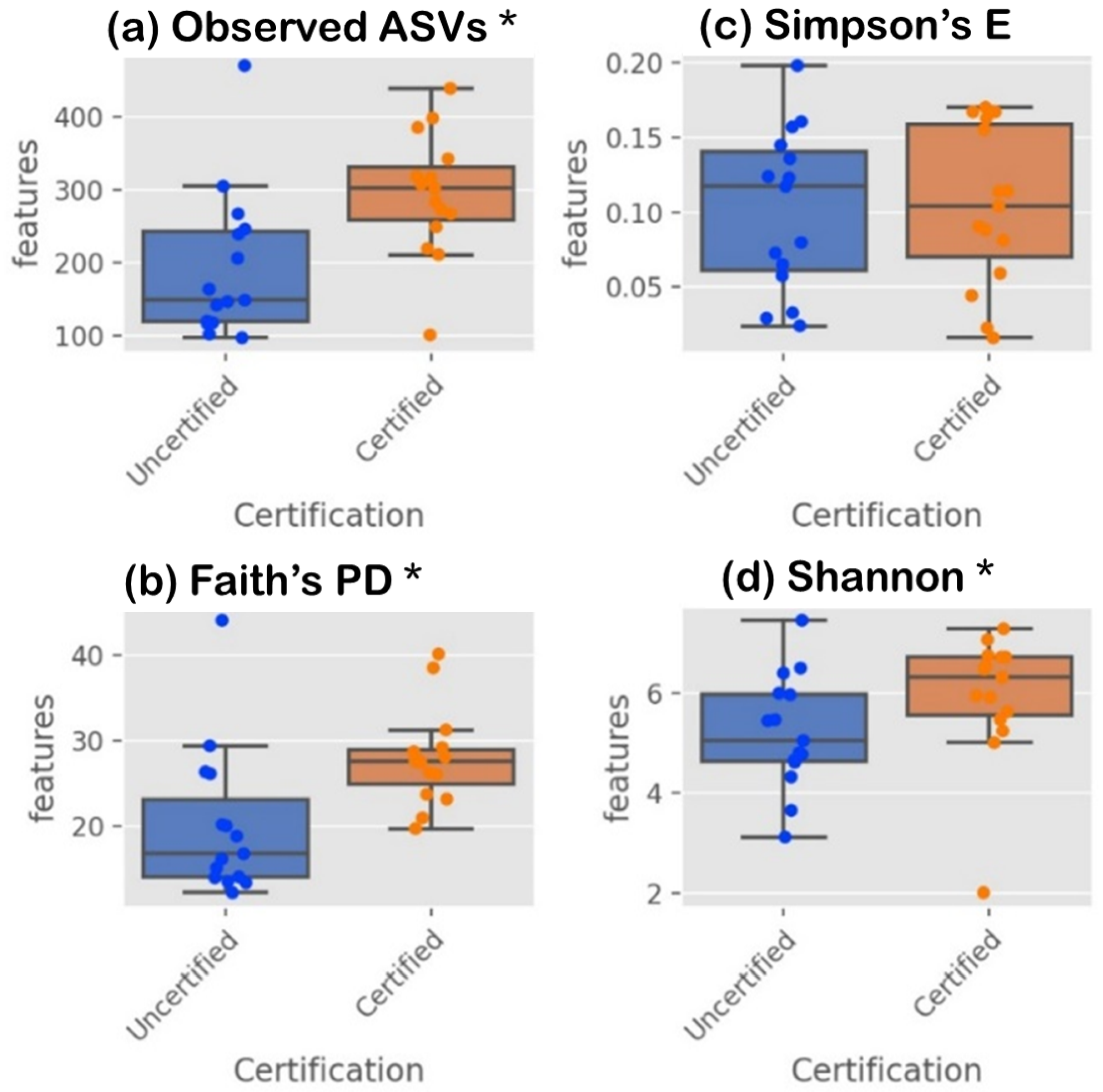
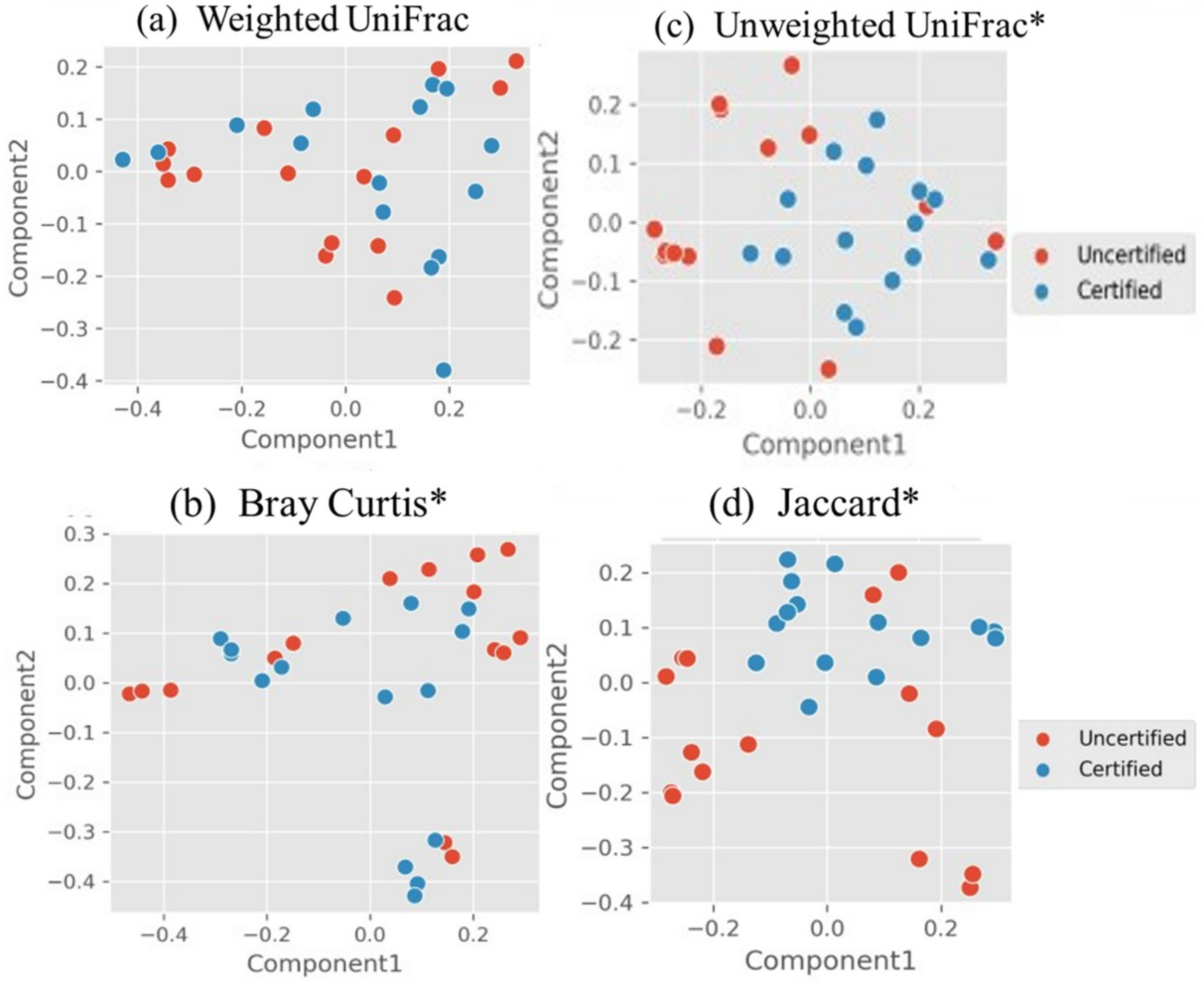
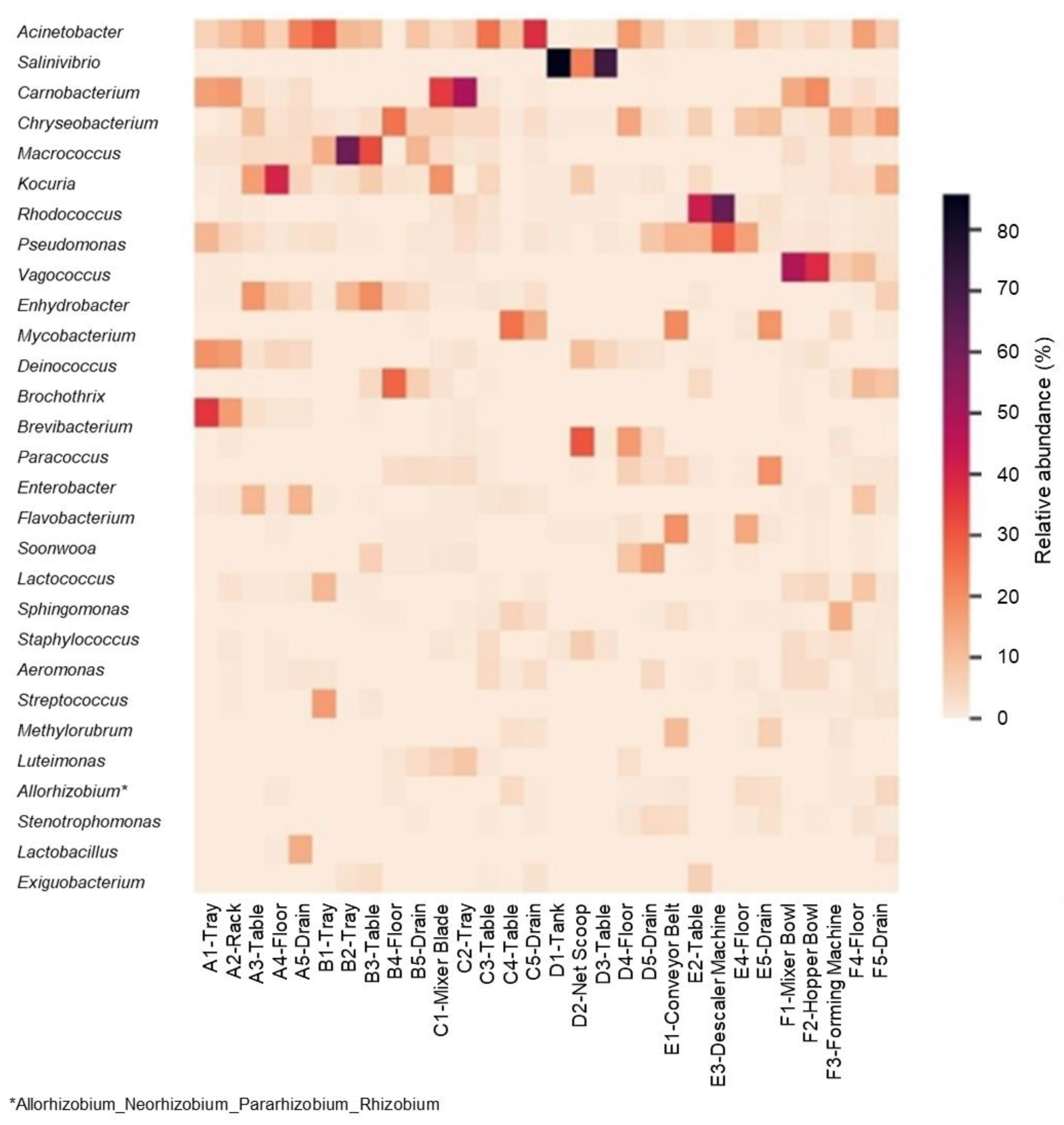
3.2. Microbiome in Seafood Factories Measured Using NGS Method
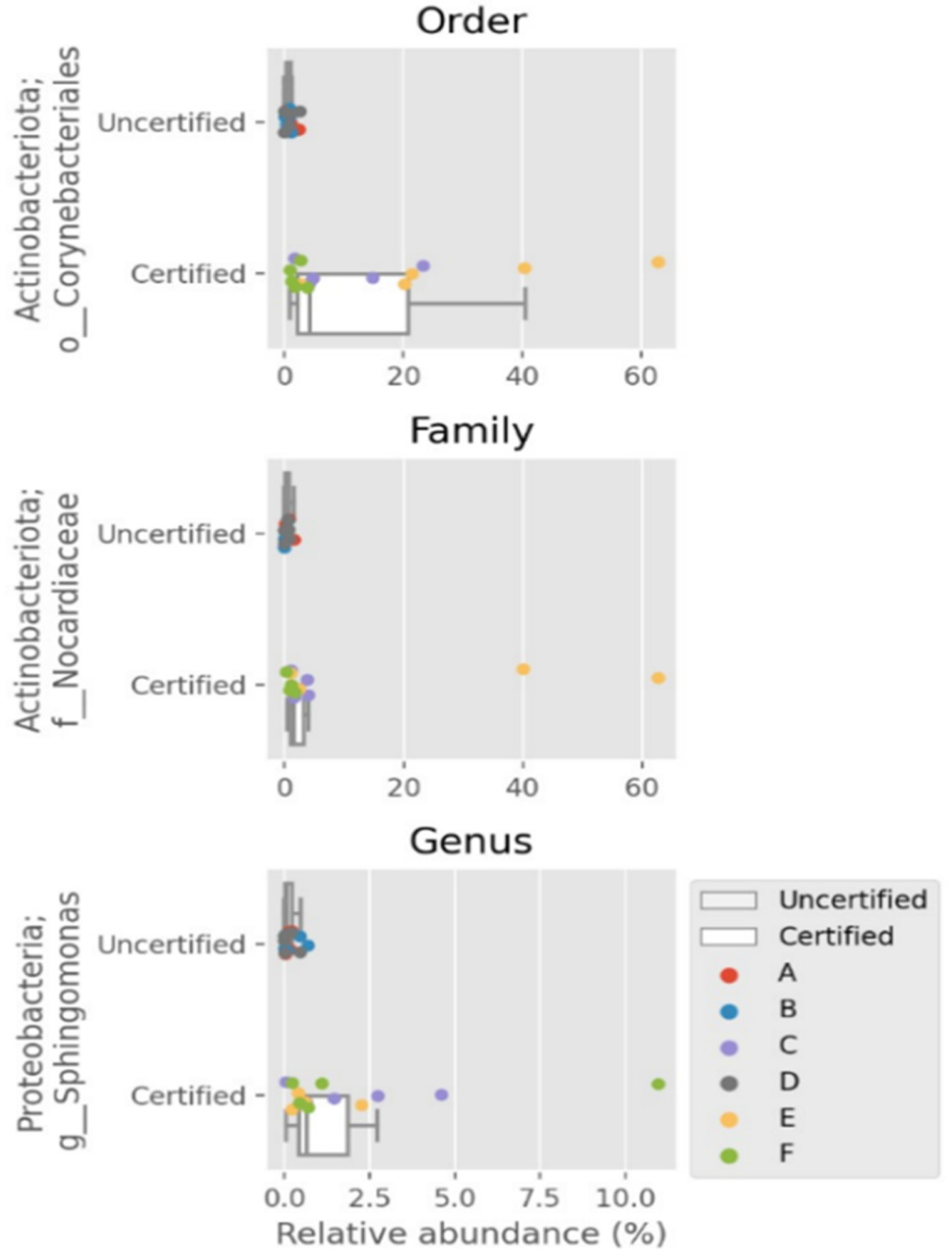
3.3. Distinctive Genera in “Certified” and “Uncertified” Seafood Processing Using NGS Method
3.4. Identification of Lactic Acid Bacteria in Seafood Processing
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAP | Best Aquaculture Practice |
| BRCGS | British Retail Consortium Global Standards |
| Est. | Established |
| FSMS | Food Safety Management System |
| FSSC22000 | Food Safety System Certification (FSSC) 22000 |
| GMP | Good Manufacturing Practices |
| HACCP | Hazard Analysis Critical Control Point |
| HDPE | high-density polyethylene |
| k | thousand (103) |
| m | million (106) |
| MeSTI | Makanan Selamat, Tanggungjawab Industri |
| N/A | not applicable |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| PP | polypropylene |
| pH | potential of hydrogen |
| RM | Ringgit Malaysia |
| SUS | stainless steel |
| VHM | Veterinary Health Mark |
References
- FAO. FAO Fisheries & Aquaculture—Quality and Safety of Fish and Fish Products. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/fishery/quality_safety/en (accessed on 6 January 2021).
- FAO. FAO Fisheries & Aquaculture—Fishery and Aquaculture Country Profiles—Malaysia. FAO. Available online: http://www.fao.org/fishery/facp/MYS/en (accessed on 3 January 2021).
- Lee, J.C.; Daraba, A.; Voidarou, C.; Rozos, G.; El Enshasy, H.A.; Varzakas, T. Implementation of Food Safety Management Systems along with Other Management Tools (HAZOP, FMEA, Ishikawa, Pareto). The Case Study of Listeria monocytogenes and Correlation with Microbiological Criteria. Foods 2021, 10, 2169. [Google Scholar] [CrossRef]
- Fernando, Y.; Ng, H.; Walters, T. Regulatory incentives as a moderator of determinants for the adoption of Malaysian food safety system. Br. Food J. 2015, 117, 1336–1353. [Google Scholar] [CrossRef]
- Ahamat, H.; Manaf, N.H.A.; Manap, N.A.; Rahman, N.A. Regulatory Strategies for Facilitating Exports by Microenterprises in Malaysia. Malays. J. Bus. Econ. (MJBE) 2019, 1, 119–128. [Google Scholar]
- Surya, T.; Jeyasekaran, G.; Shakila, R.J.; Sivaraman, B.; Shalini, R.; Sundhar, S.; Arisekar, U. Prevalence of biofilm forming Salmonella in different seafood contact surfaces of fishing boats, fish landing centres, fish markets and seafood processing plants. Mar. Pollut. Bull. 2022, 185, 114285. [Google Scholar] [CrossRef] [PubMed]
- Sudagidan, M.; Ozalp, V.C.; Öztürk, O.; Yurt, M.N.Z.; Yavuz, O.; Tasbasi, B.B.; Ucak, S.; Mavili, Z.S.; Coban, A.; Aydin, A. Bacterial surface, biofilm and virulence properties of Listeria monocytogenes strains isolated from smoked salmon and fish food contact surfaces. Food Biosci. 2021, 41, 101021. [Google Scholar] [CrossRef]
- Jacxsens, L.; Kussaga, J.; Luning, P.A.; Van der Spiegel, M.; Devlieghere, F.; Uyttendaele, M. A Microbial Assessment Scheme to measure microbial performance of Food Safety Management Systems. Int. J. Food Microbiol. 2009, 134, 113–125. [Google Scholar] [CrossRef]
- Fathurrahman, R.N.; Rukayadi, Y.; Ungku Fatimah, U.Z.A.; Jinap, S.; Abdul-Mutalib, N.A.; Sanny, M. The performance of food safety management system in relation to the microbiological safety of salmon nigiri sushi: A multiple case study in a Japanese chain restaurant. Food Control 2021, 127, 108111. [Google Scholar] [CrossRef]
- Banerjee, G.; Agarwal, S.; Marshall, A.; Jones, D.H.; Sulaiman, I.M.; Sur, S.; Banerjee, P. Application of advanced genomic tools in food safety rapid diagnostics: Challenges and opportunities. Curr. Opin. Food Sci. 2022, 47, 100886. [Google Scholar] [CrossRef]
- Janekrongtham, C.; Dejburum, P.; Sujinpram, S.; Rattanathumsakul, T.; Swaddiwudhipong, W. Outbreak of seafood-related food poisoning from undetectable Vibrio parahaemolyticus-like pathogen, Chiang Mai Province, Thailand, December 2020. Trop. Med. Int. Health 2022, 27, 92–98. [Google Scholar] [CrossRef]
- Harper, S.; Counihan, K.L.; Kanrar, S.; Paoli, G.C.; Tilman, S.; Gehring, A.G. Investigating the Quantification Capabilities of a Nanopore-Based Sequencing Platform for Food Safety Application via External Standards of Lambda DNA and Lambda Spiked Beef. Foods 2024, 13, 3304. [Google Scholar] [CrossRef]
- Imanian, B.; Donaghy, J.; Jackson, T.; Gummalla, S.; Ganesan, B.; Baker, R.C.; Henderson, M.; Butler, E.K.; Hong, Y.; Ring, B.; et al. The power, potential, benefits, and challenges of implementing high-throughput sequencing in food safety systems. Npj Sci. Food 2022, 6, 35. [Google Scholar] [CrossRef]
- De Filippis, F.; Parente, E.; Ercolini, D. Recent Past, Present, and Future of the Food Microbiome. Annu. Rev. Food Sci. Technol. 2018, 9, 589–608. [Google Scholar] [CrossRef] [PubMed]
- Billington, C.; Kingsbury, J.M.; Rivas, L. Metagenomics Approaches for Improving Food Safety: A Review. J. Food Prot. 2022, 85, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Baráti-Deák, B.; Mohácsi-Farkas, C.; Belák, Á. Searching for Antagonistic Activity of Bacterial Isolates Derived from Food Processing Environments on Some Food-Borne Pathogenic Bacteria. Acta Aliment. 2020, 49, 415–423. [Google Scholar] [CrossRef]
- Rizal, N.S.M.; Neoh, H.-M.; Ramli, R.; Periyasamy, P.R.A.K.; Hanafiah, A.; Samat, M.N.A.; Tan, T.L.; Wong, K.K.; Nathan, S.; Chieng, S.; et al. Advantages and Limitations of 16S rRNA Next-Generation Sequencing for Pathogen Identification in the Diagnostic Microbiology Laboratory: Perspectives from a Middle-Income Country. Diagnostics 2020, 10, 816. [Google Scholar] [CrossRef] [PubMed]
- FSQ. Fish & Fish Product to the European Union (EU). Available online: https://hq.moh.gov.my/fsq/ikan-hasilan-ikan-ke-kesatuan-eropah-eu? (accessed on 13 December 2024).
- NSW Food Authority. Environmental Swabbing. Available online: https://www.foodauthority.nsw.gov.au/sites/default/files/2020-01/environmental_swabbing.pdf (accessed on 6 November 2024).
- ISO 7932:2004; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Presumptive Bacillus cereus; Colony-Count Technique at 30 °C. ISO: Geneva, Switzerland, 2004. Available online: https://www.iso.org/standard/38219.html (accessed on 1 September 2022).
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp., Part 1: Detection Method. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/60313.html (accessed on 1 September 2022).
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/56712.html (accessed on 1 September 2022).
- ISO 21872-1:2017; Microbiology of the Food Chain—Horizontal Method for the Determination of Vibrio spp.—Part 1: Detection of Potentially Enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/74112.html (accessed on 1 September 2022).
- ISO 21567:2004; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection of Shigella spp. ISO: Geneva, Switzerland, 2004. Available online: https://www.iso.org/standard/34612.html (accessed on 1 September 2022).
- Grimont, P.A.D.; Weill, F.-X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur: Paris, France, 2007. [Google Scholar]
- Ochman, H.; Selander, R.K. Standard Reference Strains of Escherichia coli from Natural Populations. J. Bacteriol. 1984, 157, 690–693. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Illumina. 16S Metagenomic Sequencing Library Preparation Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2013. Available online: http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 30 January 2021).
- Illumina. Illumina 16S Metagenomics Sequencing Workflow. Illumina 2017, 12, 1–3. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. CSE 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: http://scikit-learn.sourceforge.net (accessed on 14 January 2023).
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.; Guillaume, B.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.H.; et al. Vegan: Community Ecology Package Version: 2.5-6. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 14 January 2023).
- Mandal, S.; van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Münch, S.; Braun, P.; Wernery, U.; Kinne, J.; Pees, M.; Flieger, A.; Tietze, E.; Rabsch, W. Prevalence, serovars, phage types, and antibiotic susceptibilities of Salmonella strains isolated from animals in the United Arab Emirates from 1996 to 2009. Trop. Anim. Health Prod. 2012, 44, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
- Finn, L.; Onyeaka, H.; O’Neill, S. Listeria monocytogenes Biofilms in Food-Associated Environments: A Persistent Enigma. Foods 2023, 12, 3339. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Møretrø, T. Microbial diversity and ecology of biofilms in food industry environments associated with Listeria monocytogenes persistence. Curr. Opin. Food Sci. 2021, 37, 171–178. [Google Scholar] [CrossRef]
- Silva, A.; Silva, V.; Gomes, J.P.; Coelho, A.; Batista, R.; Saraiva, C.; Esteves, A.; Martins, Â.; Contente, D.; Diaz-Formoso, L.; et al. Listeria monocytogenes from Food Products and Food Associated Environments: Antimicrobial Resistance, Genetic Clustering and Biofilm Insights. Antibiotics 2024, 13, 447. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial Biofilms in the Food Industry—A Comprehensive Review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Sánchez, D.; Galvão, J.A.; Oetterer, M. Contamination sources, serogroups, biofilm-forming ability and biocide resistance of Listeria monocytogenes persistent in tilapia-processing facilities. J. Food Sci. Technol. 2017, 54, 3867–3879. [Google Scholar] [CrossRef]
- Surya, T.; Jeyasekaran, G.; Shakila, R.J.; Alsalhi, M.S.; Devanesan, S.; Sivaraman, B.; Arisekar, U.; Pham, T.H. Effect of antibiotics and sanitizers on Salmonella biofilms associated with seafood contact surfaces. Microbiol. Res. 2023, 266, 127213. [Google Scholar] [CrossRef]
- Tee, X.W.; Abdul-Mutalib, N.A. Salmonella Biofilm on Food Contact Surfaces and the Efficacy of Chemical Disinfectants: A Systematic Review. Pertanika J. Sci. Technol. 2023, 31, 2187–2201. [Google Scholar] [CrossRef]
- Holah, J. Minimum hygienic design requirements for food processing factories. In Hygienic Design of Food Factories, 2nd ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead: Cambridge, UK, 2023; pp. 221–240. [Google Scholar] [CrossRef]
- Marriott, N.G.; Schilling, M.W.; Gravani, R.B. Sanitary Design and Construction for Food Processing. In Principles of Food Sanitation; Springer: Cham, Switzerland, 2018; pp. 267–278. [Google Scholar] [CrossRef]
- Faille, C.; Cunault, C.; Dubois, T.; Bénézech, T. Hygienic design of food processing lines to mitigate the risk of bacterial food contamination with respect to environmental concerns. Innov. Food Sci. Emerg. Technol. 2018, 46, 65–73. [Google Scholar] [CrossRef]
- Brauge, T.; Mougin, J.; Ells, T.; Midelet, G. Sources and contamination routes of seafood with human pathogenic Vibrio spp.: A Farm-to-Fork approach. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13283. [Google Scholar] [CrossRef]
- Wan Norhana, M.N.; Poole, S.E.; Deeth, H.C.; Dykes, G.A. Prevalence, persistence and control of Salmonella and Listeria in shrimp and shrimp products: A review. Food Control 2010, 21, 343–361. [Google Scholar] [CrossRef]
- Dima, A.; Radu, E.; Dobrin, C. Exploring Key Barriers of HACCP Certification Adoption in the Meat Industry: A Decision-Making Trial and Evaluation Laboratory Approach. Foods 2024, 13, 1303. [Google Scholar] [CrossRef]
- Lee, J.C.; Neonaki, M.; Alexopoulos, A.; Varzakas, T. Case Studies of Small-Medium Food Enterprises around the World: Major Constraints and Benefits from the Implementation of Food Safety Management Systems. Foods 2023, 12, 3218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, Z.; Feng, L.; Qu, D.; Zhu, J. Identification of microbial communities and multi-species biofilms contamination in seafood processing environments with different hygiene conditions. Food Microbiol. 2024, 122, 104553. [Google Scholar] [CrossRef]
- Anihouvi, D.G.H.; Henriet, O.; Kpoclou, Y.E.; Scippo, M.; Hounhouigan, D.J.; Anihouvi, V.B.; Mahillon, J. Bacterial diversity of smoked and smoked-dried fish from West Africa: A metagenomic approach. J. Food Process. Preserv. 2021, 45, e15919. [Google Scholar] [CrossRef]
- Nikolaev, Y.; Yushina, Y.; Mardanov, A.; Gruzdev, E.; Tikhonova, E.; El-Registan, G.; Beletskiy, A.; Semenova, A.; Zaiko, E.; Bataeva, D.; et al. Microbial Biofilms at Meat-Processing Plant as Possible Places of Bacteria Survival. Microorganisms 2022, 10, 1583. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Cabo, M.L. Tracking bacteriome variation over time in Listeria monocytogenes-positive foci in food industry. Int. J. Food Microbiol. 2020, 315, 108439. [Google Scholar] [CrossRef]
- Ross, S.R.P.J.; Sasaki, T. Limited theoretical and empirical evidence that response diversity determines the resilience of ecosystems to environmental change. Ecol. Res. 2024, 39, 115–130. [Google Scholar] [CrossRef]
- Pimm, S.L. The complexity and stability of ecosystems. Nature 1984, 307, 321–326. [Google Scholar] [CrossRef]
- Sadiq, F.A.; De Reu, K.; Burmølle, M.; Maes, S.; Heyndrickx, M. Synergistic interactions in multispecies biofilm combinations of bacterial isolates recovered from diverse food processing industries. Front. Microbiol. 2023, 14, 1159434. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, V.; Bosilevac, J.M.; Barkhouse, D.A.; Velez, S.E.; Chitlapilly Dass, S. Shotgun-metagenomics reveals a highly diverse and communal microbial network present in the drains of three beef-processing plants. Front. Cell. Infect. Microbiol. 2023, 13, 1240138. [Google Scholar] [CrossRef]
- Klümper, U.; Gionchetta, G.; Catão, E.; Bellanger, X.; Dielacher, I.; Elena, A.X.; Fang, P.; Galazka, S.; Goryluk-Salmonowicz, A.; Kneis, D.; et al. Environmental microbiome diversity and stability is a barrier to antimicrobial resistance gene accumulation. Commun. Biol. 2024, 7, 706. [Google Scholar] [CrossRef] [PubMed]
- Sequino, G.; Cobo-Diaz, J.F.; Valentino, V.; Tassou, C.; Volpe, S.; Torrieri, E.; Nychas, G.-J.; Ordóñez, A.Á.; Ercolini, D.; De Filippis, F. Microbiome mapping in beef processing reveals safety-relevant variations in microbial diversity and genomic features. Food Res. Int. 2024, 186, 114318. [Google Scholar] [CrossRef]
- Eissa, R.A.M.; Salem, M.A.; Elsanat, S.Y. Controlling of Potential Hazard in Potato Chips Processing through Food Safety Management System FSMS (ISO 22000). Menoufia J. Food Dairy Sci. 2019, 4, 171–188. [Google Scholar] [CrossRef]
- Lacorte, G.A.; Cruvinel, L.A.; Ávila, M.d.P.; Dias, M.F.; Pereira, A.d.A.; Nascimento, A.M.A.; Franco, B.D.G.d.M. Investigating the influence of Food Safety Management Systems (FSMS) on microbial diversity of Canastra cheeses and their processing environments. Food Microbiol. 2022, 105, 104023. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Abdelrahman, H.; Kelly, A.; Roy, L.; Wang, L. Profiling and source tracking of the microbial populations and resistome present in fish products. Int. J. Food Microbiol. 2024, 413, 110591. [Google Scholar] [CrossRef]
- Hospodsky, D.; Pickering, A.J.; Julian, T.R.; Miller, D.; Gorthala, S.; Boehm, A.B.; Peccia, J. Hand bacterial communities vary across two different human populations. Microbiology 2014, 160, 1144–1152. [Google Scholar] [CrossRef]
- Edmonds-Wilson, S.L.; Nurinova, N.I.; Zapka, C.A.; Fierer, N.; Wilson, M. Review of human hand microbiome research. J. Dermatol. Sci. 2015, 80, 3–12. [Google Scholar] [CrossRef]
- Goetsch, A.G.; Ufearo, D.; Keiser, G.; Heiss, C.; Azadi, P.; Hershey, D.M. An exopolysaccharide pathway from a freshwater Sphingomonas isolate. J. Bacteriol. 2024, 206. [Google Scholar] [CrossRef] [PubMed]
- Cherifi, T.; Arsenault, J.; Quessy, S.; Fravalo, P. Co-Occurrence of L. monocytogenes with Other Bacterial Genera and Bacterial Diversity on Cleaned Conveyor Surfaces in a Swine Slaughterhouse. Microorganisms 2022, 10, 613. [Google Scholar] [CrossRef] [PubMed]
- Busse, H.J.; Hauser, E.; Kämpfer, P. Description of two novel species, Sphingomonas abaci sp. nov. and Sphingomonas panni sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 2565–2569. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Shen, F.-T.; Lai, W.-A.; Zhu, Z.-L.; Chen, W.-M.; Chou, J.-H.; Lin, Z.-Y.; Young, C.-C. Sphingomonas formosensis sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1581–1586. [Google Scholar] [CrossRef]
- Maillet, A.; Bouju-Albert, A.; Roblin, S.; Vaissié, P.; Leuillet, S.; Dousset, X.; Jaffrès, E.; Combrisson, J.; Prévost, H. Impact of DNA extraction and sampling methods on bacterial communities monitored by 16S rDNA metabarcoding in cold-smoked salmon and processing plant surfaces. Food Microbiol. 2021, 95, 103705. [Google Scholar] [CrossRef]
- Gaillac, A.; Briandet, R.; Delahaye, E.; Deschamps, J.; Vigneau, E.; Courcoux, P.; Jaffrès, E.; Prévost, H. Exploring the Diversity of Biofilm Formation by the Food Spoiler Brochothrix thermosphacta. Microorganisms 2022, 10, 2474. [Google Scholar] [CrossRef]
- de Paiva Anciens Ramos, G.L.; Vigoder, H.C.; dos Santos Nascimento, J. Kocuria spp. in Foods: Biotechnological Uses and Risks for Food Safety. Appl. Food Biotechnol. 2021, 8, 79–88. [Google Scholar] [CrossRef]
- Kloos, W.E.; Ballard, D.N.; George, C.G.; Webster, J.A.; Hubner, R.J.; Ludwig, W.; Schleifer, K.H.; Fiedler, F.; Schubert, K. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., Macrococcus bovicus sp. nov. and Macrococcus carouselicus sp. nov. Int. J. Syst. Bacteriol. 1998, 48, 859–877. [Google Scholar] [CrossRef]
- Götz, F.; Bannerman, T.; Schleifer, K.-H. The Genera Staphylococcus and Macrococcus. In The Prokaryotes; Springer: New York, USA, 2006; pp. 5–75. [Google Scholar] [CrossRef]
- Mazhar, S.; Hill, C.; McAuliffe, O. The Genus Macrococcus: An Insight I#into Its Biology, Evolution, and Relationship with Staphylococcus. Adv. Appl. Microbiol. 2018, 105, 1–50. [Google Scholar] [CrossRef]
- Ramos, G.L.P.A.; Vigoder, H.C.; Nascimento, J.S. Technological Applications of Macrococcus caseolyticus and its Impact on Food Safety. Curr. Microbiol. 2021, 78, 11–16. [Google Scholar] [CrossRef]
- Mašlanová, I.; Wertheimer, Z.; Sedláček, I.; Švec, P.; Indráková, A.; Kovařovic, V.; Schumann, P.; Spröer, C.; Králová, S.; Šedo, O.; et al. Description and comparative genomics of Macrococcus caseolyticus subsp. hominis subsp. nov., Macrococcus goetzii sp. nov., Macrococcus epidermidis sp. nov., and Macrococcus bohemicus sp. nov., Novel Macrococci from human clinical material with virulence potential and suspected uptake of foreign DNA by natural transformation. Front. Microbiol. 2018, 9, 1178. [Google Scholar] [CrossRef]
- Schöbitz, R.; González, C.; Villarreal, K.; Horzella, M.; Nahuelquín, Y.; Fuentes, R. A biocontroller to eliminate Listeria monocytogenes from the food processing environment. Food Control 2014, 36, 217–223. [Google Scholar] [CrossRef]
- Camargo, A.C.; Todorov, S.D.; Chihib, N.E.; Drider, D.; Nero, L.A. Lactic Acid Bacteria (LAB) and Their Bacteriocins as Alternative Biotechnological Tools to Control Listeria monocytogenes Biofilms in Food Processing Facilities. Mol. Biotechnol. 2018, 60, 712–726. [Google Scholar] [CrossRef]
- Smith, S.A.; Newman, S.J.; Harrison, C.E.; Loch, T.P. First isolation of Carnobacterium maltaromaticum from farmed Rainbow Trout in Virginia. J. Aquat. Anim. Health 2023, 35, 3–10. [Google Scholar] [CrossRef]
- Nan, Y.; Rodas-Gonzalez, A.; Stanford, K.; Nadon, C.; Yang, X.; McAllister, T.; Narváez-Bravo, C. Lactic acid bacteria and spoilage bacteria: Their interactions in Escherichia coli O157:H7 biofilms on food contact surfaces and implications for beef contamination. J. Food Saf. 2024, 44, e13101. [Google Scholar] [CrossRef]
- Fagerlund, A.; Møretrø, T.; Heir, E.; Briandet, R.; Langsrud, S. Cleaning and Disinfection of Biofilms Composed of Listeria monocytogenes and Background Microbiota from Meat Processing Surfaces. Appl. Environ. Microbiol. 2017, 83, e01046-17. [Google Scholar] [CrossRef]
- Pátek, M.; Grulich, M.; Nešvera, J. Stress response in Rhodococcus strains. Biotechnol. Adv. 2021, 53, 107698. [Google Scholar] [CrossRef]
- Calliauw, F.; Horemans, B.; Broekaert, K.; Michiels, C.; Heyndrickx, M. Spoilage potential of Vagococcus salmoninarum in preservative-free, MAP-stored brown shrimp and differentiation from Brochothrix thermosphacta on streptomycin thallous acetate actidione agar. J. Appl. Microbiol. 2016, 120, 1302–1312. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Saá-Ibusquiza, P.; Mosquera-Fernández, M.; López-Cabo, M. Listeria monocytogenes-carrying consortia in food industry. Composition, subtyping and numerical characterisation of mono-species biofilm dynamics on stainless steel. Int. J. Food Microbiol. 2015, 206, 84–95. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; Alexa, E.A.; Crispie, F.; Prieto, M.; López, M.; Cotter, P.D.; Alvarez-Ordóñez, A. Sequencing-based analysis of the microbiomes of Spanish food processing facilities reveals environment-specific variation in the dominant taxa and antibiotic resistance genes. Food Res. Int. 2023, 173, 113442. [Google Scholar] [CrossRef]
- Shintani, H. Validation Studies for Microbial Contamination and Control of Contaminants. Biocontrol. Sci. 2015, 20, 161–170. [Google Scholar] [CrossRef]
- Stiles, M.E.; Holzapfel, W.H. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 1997, 36, 1–29. [Google Scholar] [CrossRef]
- Fidan, H.; Esatbeyoglu, T.; Simat, V.; Trif, M.; Tabanelli, G.; Kostka, T.; Montanari, C.; Ibrahim, S.A.; Özogul, F. Recent developments of lactic acid bacteria and their metabolites on foodborne pathogens and spoilage bacteria: Facts and gaps. Food Biosci. 2022, 47, 101741. [Google Scholar] [CrossRef]
- Pang, X.; Song, X.; Chen, M.; Tian, S.; Lu, Z.; Sun, J.; Li, X.; Lu, Y.; Yuk, H. Combating biofilms of foodborne pathogens with bacteriocins by lactic acid bacteria in the food industry. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1657–1676. [Google Scholar] [CrossRef]
- Ghanbari, M.; Jami, M.; Domig, K.J.; Kneifel, W. Seafood biopreservation by lactic acid bacteria—A review. LWT-Food Sci. Technol. 2013, 54, 315–324. [Google Scholar] [CrossRef]
- Castellano, P.; Ibarreche, M.P.; Massani, M.B.; Fontana, C.; Vignolo, G.M. Strategies for Pathogen Biocontrol Using Lactic Acid Bacteria and Their Metabolites: A Focus on Meat Ecosystems and Industrial Environments. Microorganisms 2017, 5, 38. [Google Scholar] [CrossRef]
- Hossain, M.I.; Mizan, F.R.; Ashrafudoulla; Nahar, S.; Joo, H.-J.; Jahid, I.K.; Park, S.H.; Kim, K.-S.; Ha, S.-D. Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBECTM biofilm device. LWT 2020, 118, 108864. [Google Scholar] [CrossRef]
- Toushik, S.H.; Kim, K.; Ashrafudoulla; Mizan, F.R.; Roy, P.K.; Nahar, S.; Kim, Y.; Ha, S.-D. Korean kimchi-derived lactic acid bacteria inhibit foodborne pathogenic biofilm growth on seafood and food processing surface materials. Food Control 2021, 129, 108276. [Google Scholar] [CrossRef]
- Stupar, J.; Holøymoen, I.G.; Hoel, S.; Lerfall, J.; Jakobsen, A.N.; Rustad, T. Diversity and Antimicrobial Activity towards Listeria spp. and Escherichia coli among Lactic Acid Bacteria Isolated from Ready-to-Eat Seafood. Foods 2021, 10, 271. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, J.; Chen, Q.; Wang, H.; Kong, B. Physiological, Morphological and Antioxidant Responses of Pediococcus pentosaceus R1 and Lactobacillus fermentum R6 Isolated from Harbin Dry Sausages to Oxidative Stress. Foods 2021, 10, 1203. [Google Scholar] [CrossRef]
- Banerji, R.; Karkee, A.; Saroj, S.D. Bacteriocins against Foodborne Pathogens (Review). Appl. Biochem. Microbiol. 2022, 58, 518–539. [Google Scholar] [CrossRef]
- Alara, J.A.; Alara, O.R. Antimicrobial and anti-biofilm potentials of biosurfactants. In Industrial Applications of Biosurfactants and Microorganisms: Green Technology Avenues from Lab to Commercialization; Academic Press: Cambridge, MA, USA, 2024; pp. 307–339. [Google Scholar] [CrossRef]
- Schmidt, R.H.; Piotter, H.M. The Hygienic/Sanitary Design of Food and Beverage Processing Equipment. In Food Engineering Series; Springer: Cham, Switzerland, 2020; pp. 267–332. [Google Scholar] [CrossRef]
- de Oliveira, C.A.F.; da Cruz, A.G.; Tavolaro, P.; Corassin, C.H. Food Safety: Good Manufacturing Practices, Sanitation Standard Operating Procedures, Hazard Analysis, and Critical Control Point. In Antimicrobial Food Packaging, 2nd ed.; Academic Press: Cambridge, MA, USA, 2025; pp. 163–173. [Google Scholar] [CrossRef]
- Quinto, E.J.; Caro, I.; Villalobos-Delgado, L.H.; Mateo, J.; De-Mateo-silleras, B.; Redondo-Del-río, M.P. Food Safety through Natural Antimicrobials. Antibiotics 2019, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Nag, M.; Lahiri, D.; Dey, A.; Sarkar, T.; Pati, S.; Joshi, S.; Bunawan, H.; Mohammed, A.; Edinur, H.A.; Ghosh, S.; et al. Seafood Discards: A Potent Source of Enzymes and Biomacromolecules with Nutritional and Nutraceutical Significance. Front. Nutr. 2022, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, E.; de Niederhäusern, S.; Messi, P.; Sabia, C.; Iseppi, R.; Anacarso, I.; Bondi, M. Use of lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes in a small-scale model. Food Control 2009, 20, 861–865. [Google Scholar] [CrossRef]

| Seafood Factories | Year Est. | Years of FSMS Adoption | No. of Workers | FSMS Status | Market | Production Shift | Annual Returns |
|---|---|---|---|---|---|---|---|
| Uncertified | |||||||
| A | 1995 | 0 | 14 | None | Domestic | Morning and afternoon | >RM300 k |
| B | 2018 | 0 | 15 | None | Domestic | Morning | RM300 k |
| D | 2010 | 1 | 9 | None | Domestic | Morning and afternoon | >RM1.5 m |
| Certified | |||||||
| C | 2007 | 7 | 45 | MeSTI | Asia | Morning | >RM1.5 m |
| E | 1985 | 12 | 58 | HACCP, FSSC22000, BRCGS, BAP | Asia, Europe, North America, Australia | Morning | >RM1.5 m |
| F | 1999 | 15 | 20 | HACCP, GMP, VHM | Asia, South America | Morning | >RM1.5 m |
| Factory | “Uncertified” | “Certified” | ||||
|---|---|---|---|---|---|---|
| Product |  Dim Sum |  Fish ball |  Fish |  Dim Sum |  Fish ball |  Fish |
| Sampling Site | A | B | D | C | F | E |
| 1 Direct food contact | Tray | Tray | Tank | Mixer blade | Mixer bowl | Conveyor belt |
| 2 Direct food contact | Rack | Tray | Net scoop | Tray | Hopper bowl | Degutting table |
| 3 Adjacent food contact | Table | Table | Table | Table | Forming machine | Skinning machine |
| 4 Floor | Floor | Floor | Floor | Floor | Floor | Floor |
| 5 Drain | Drain | Drain | Drain | Drain | Drain | Drain |
| ID | Sites | Condition and Material | Serovar (Pre-Cleaning) | Serovar (Post-Cleaning) |
|---|---|---|---|---|
| “Uncertified” seafood factories | ||||
| A | 1: Prawn-paste tray | Unclean, uneven aluminum surface | Escherichia coli † | Listeria monocytogenes |
| 2: Prawn pressing rack | Unclean, HDPE | Escherichia coli †, Salmonella Hindmarsh | Escherichia coli † | |
| 3: Processing table | Unclean, SUS and rope | Escherichia coli †, Salmonella typhimurium | N/A | |
| 4: Floor | Cracked and porous tiles | N/A | Listeria monocytogenes | |
| 5: Drain | Uncovered cement/half-covered SUS | Salmonella Weltevreden | N/A | |
| B | 1: Fish-cake short-forming tray | Unclean, SUS | N/A | N/A |
| 2: Fish-cake long-forming tray | Unclean, SUS | N/A | Bacillus cereus | |
| 3: Forming table | Unclean, SUS | N/A | N/A | |
| 4: Floor | Cracked and porous tiles | Salmonella Bareilly | Bacillus cereus Salmonella Braenderup Escherichia coli † | |
| 5: Drain | Uncovered, porous and cracked cement surface | Salmonella Bareilly, Bacillus cereus | Bacillus cereus, Salmonella Bareilly, Escherichia coli †, | |
| D | 1. Brine tank | Polycarbonate | N/A | N/A |
| 2. Net scoop | SUS and rope | N/A | N/A | |
| 3. Salting table | Unclean, SUS | N/A | N/A | |
| 4: Floor | Epoxy | N/A | N/A | |
| 5. Drain | Half-covered, SUS | N/A | N/A | |
| “Certified” seafood factories | ||||
| C | 1. Prawn-paste mixer blade | SUS | N/A | N/A |
| 2. Prawn-paste holding tray | PP | N/A | N/A | |
| 3. Hopper bowl | SUS | N/A | N/A | |
| 4. Floor | Tiles | N/A | N/A | |
| 5. Drain | Fully/half-covered, SUS | N/A | N/A | |
| E | 1. Descaler conveyor belt | PP | N/A | N/A |
| 2. Degutting and filleting table | SUS | N/A | N/A | |
| 3. Water-jet skinning machine | Rubber and SUS | N/A | N/A | |
| 4. Floor | Epoxy and cement | N/A | N/A | |
| 5. Drain | Half-covered, SUS | N/A | N/A | |
| F | 1. Fish-paste mixer bowl | Unclean, SUS | N/A | N/A |
| 2. Hopper bowl | SUS | N/A | N/A | |
| 3. Forming machine | Iron | N/A | N/A | |
| 4. Floor | Epoxy | N/A | N/A | |
| 5. Drain | Half-covered, SUS | N/A | N/A | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuan, S.; Chin, N.L.; Tee, T.P.; Hasnan, N.Z.N. Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia. Foods 2025, 14, 1517. https://doi.org/10.3390/foods14091517
Kuan S, Chin NL, Tee TP, Hasnan NZN. Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia. Foods. 2025; 14(9):1517. https://doi.org/10.3390/foods14091517
Chicago/Turabian StyleKuan, Shuping, Nyuk Ling Chin, Tuan Poy Tee, and Noor Zafira Noor Hasnan. 2025. "Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia" Foods 14, no. 9: 1517. https://doi.org/10.3390/foods14091517
APA StyleKuan, S., Chin, N. L., Tee, T. P., & Hasnan, N. Z. N. (2025). Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia. Foods, 14(9), 1517. https://doi.org/10.3390/foods14091517






