Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention
Abstract
1. Introduction
2. Materials and Methods
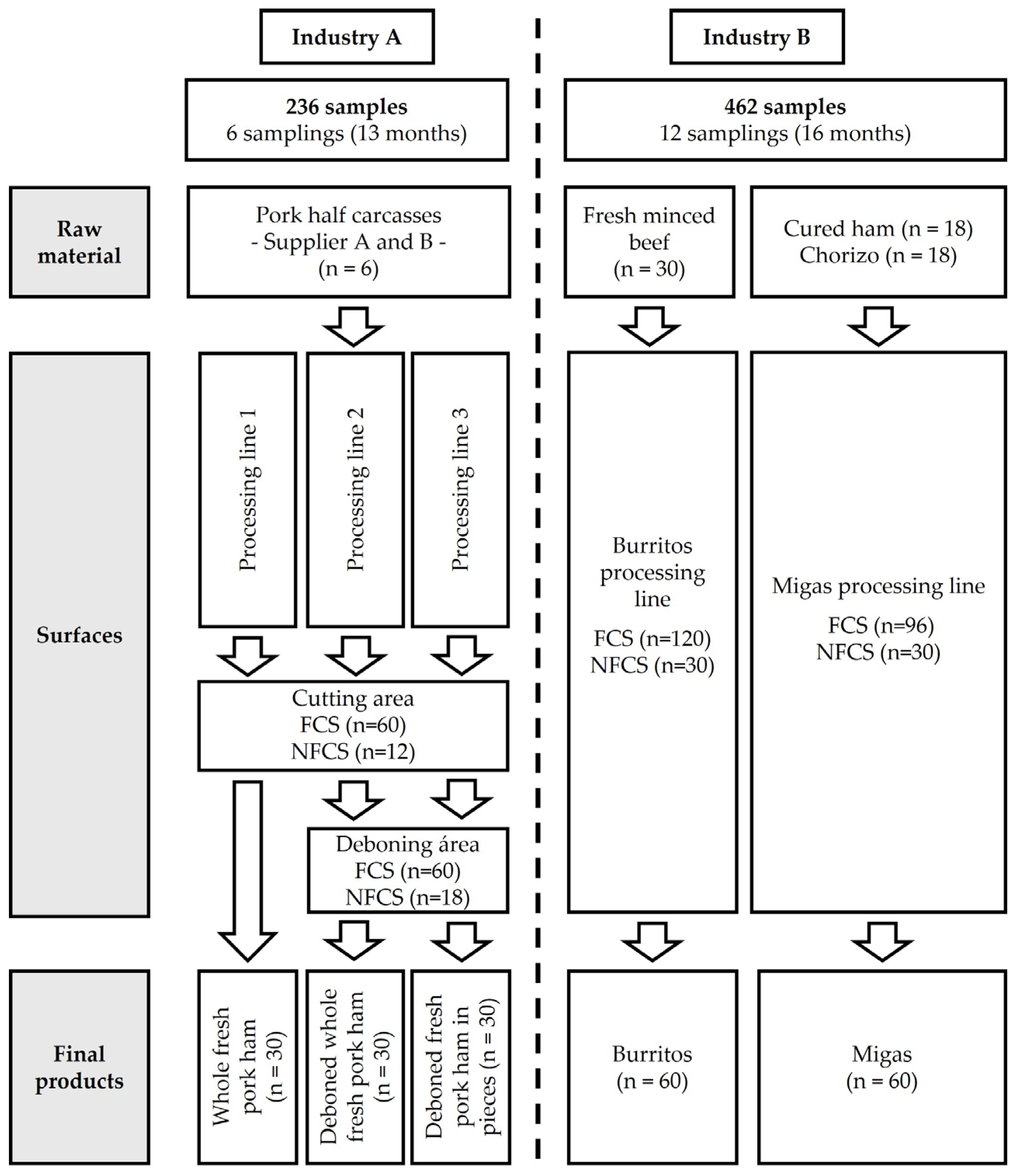
2.1. Study Design
2.2. Sampling Procedure
2.3. Detection of Listeria spp.
2.4. Isolation and Identification
2.5. L. monocytogenes Typing
2.5.1. Serotype Identification
2.5.2. Pulsed-Field Gel Electrophoresis Typing
2.6. Statistical Analysis
3. Results
3.1. Prevalence of Listeria spp. and L. monocytogenes
3.1.1. Industry A
3.1.2. Industry B
3.2. Serotype Identification
3.2.1. Industry A
3.2.2. Industry B
3.3. Pulsed-Field Gel Electrophoresis Typing
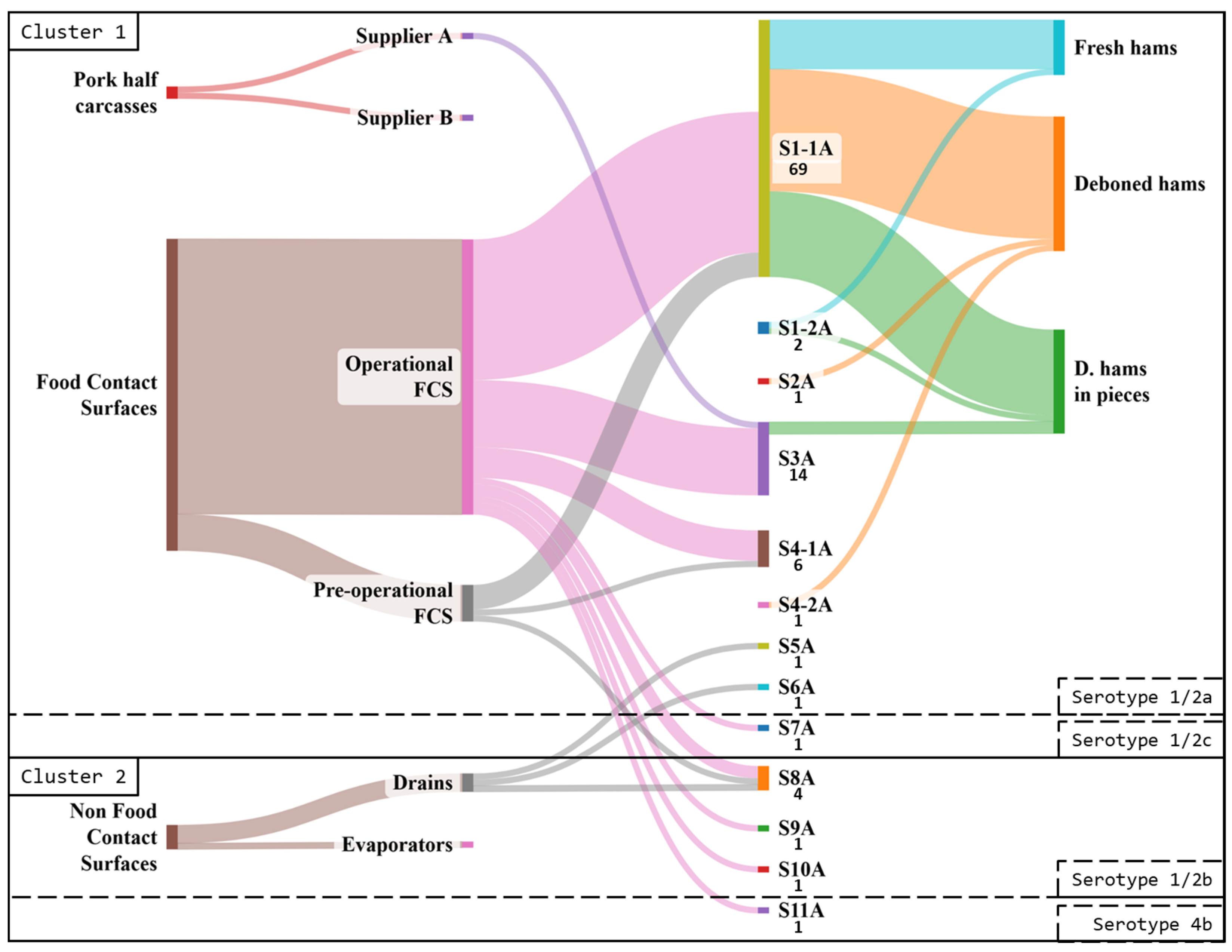
3.3.1. Industry A
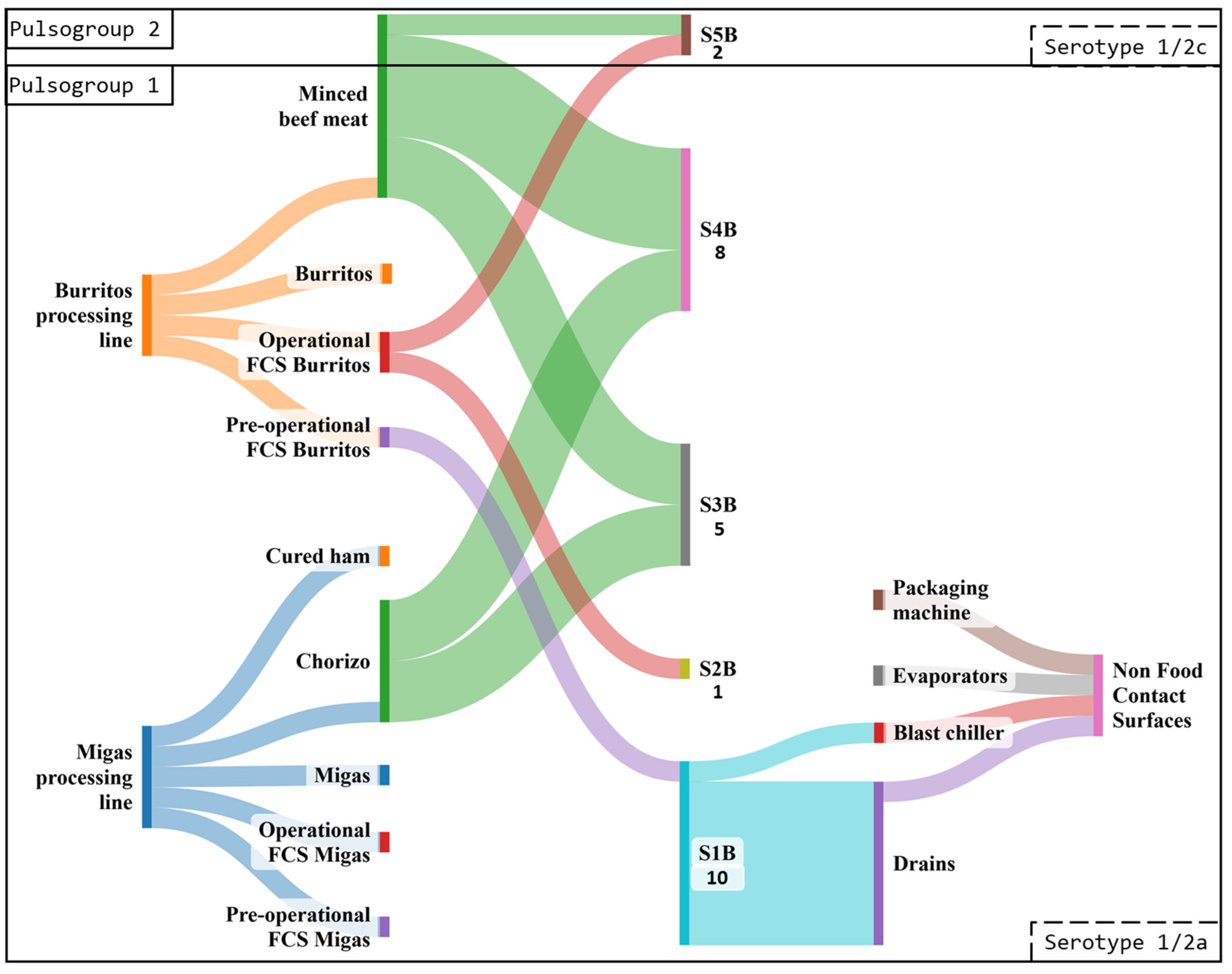
3.3.2. Industry B
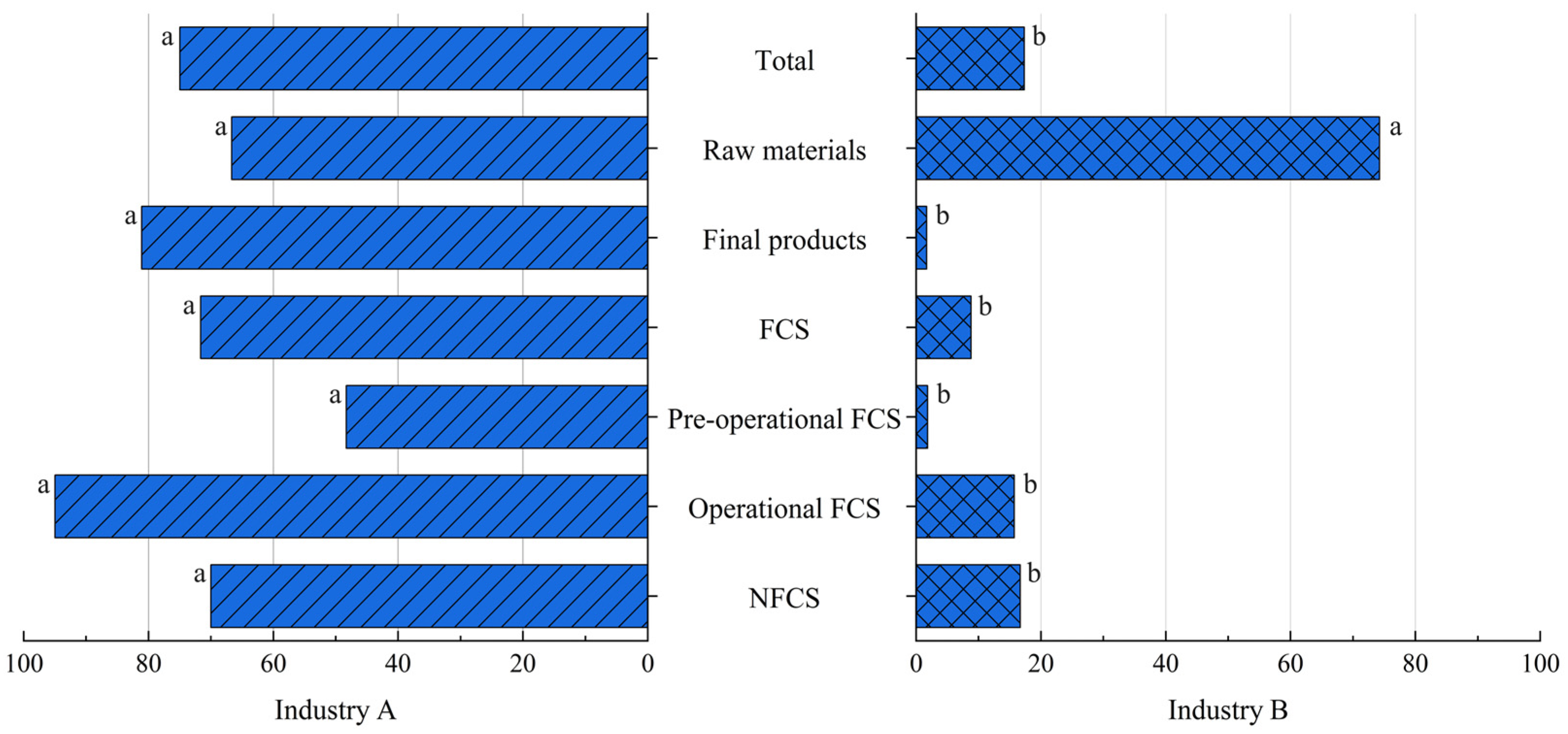
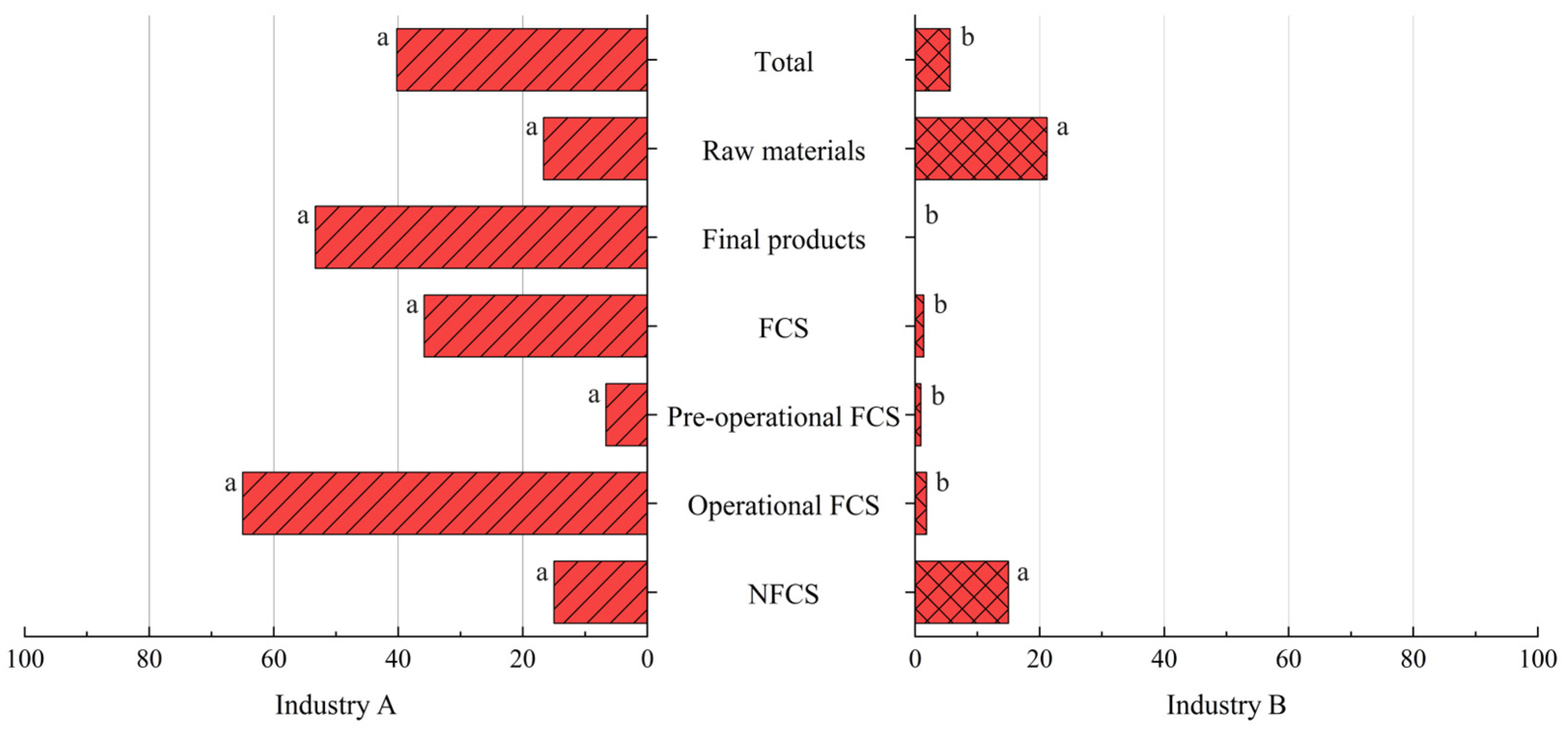
3.4. Comparison Between Industries A and B
3.4.1. Prevalence of Listeria spp. and L. monocytogenes
3.4.2. Pulsed-Field Gel Electrophoresis Typing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EU | European Union |
| RTE | Ready-To-Eat |
| GDP | Spanish Gross Domestic Product |
| C&D | Cleaning and Disinfection |
| FCS | Food Contact Surfaces |
| NFCS | Non-Food Contact Surfaces |
| OB | One Broth Listeria |
| OCLA | Oxoid Chromogenic Listeria Agar |
| TSA | Tryptone Soy Agar |
| TSB | Tryptone Soya Broth |
| PFGE | Pulsed-Field Gel Electrophoresis |
| FH | Whole Fresh Pork Hams |
| DFH | Deboned Whole Fresh Pork Hams |
| DFHP | Deboned Fresh Pork Hams In Pieces |
| OP | Operational |
| PO | Pre-Operational |
| PT | Pulsotype |
References
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses Report. EFSA J. 2024, 22, e9106. [Google Scholar]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Kérouanton, A.; Marault, M.; Petit, L.; Grout, J.; Dao, T.T.; Brisabois, A. Evaluation of a Multiplex PCR Assay as an Alternative Method for Listeria monocytogenes Serotyping. J. Microbiol. Methods 2010, 80, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, H.P.R.; Höhne, K. Chapter II Serotyping of Listeria monocytogenes and Related Species. In Methods in Microbiology; Bergan, T., Norris, J.R., Eds.; Academic Press: Cambridge, MA, USA, 1979; Volume 13, pp. 31–49. [Google Scholar]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A Hybrid Sub-Lineage of Listeria monocytogenes Comprising Hypervirulent Isolates. Nat. Commun. 2019, 10, 4283. [Google Scholar] [CrossRef]
- Borucki, M.K.; Call, D.R. Listeria monocytogenes Serotype Identification by PCR. J. Clin. Microbiol. 2003, 41, 5537–5540. [Google Scholar] [CrossRef]
- Bergis, H.; Bonanno, L.; Asséré, A.; Lombard, B. EURL Lm Technical Guidance Document on Challenge Tests and Durability Studies for Assessing Shelf-Life of Ready-to-Eat Foods Related to Listeria monocytogenes. EURL Lm 2021, 4, 196. [Google Scholar]
- Rugna, G.; Carra, E.; Bergamini, F.; Franzini, G.; Faccini, S.; Gattuso, A.; Morganti, M.; Baldi, D.; Naldi, S.; Serraino, A.; et al. Distribution, Virulence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes Isolated over One-Year Monitoring from Two Pig Slaughterhouses and Processing Plants and Their Fresh Hams. Int. J. Food Microbiol. 2021, 336, 108912. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in Biofilm Formation among Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef]
- Dygico, L.K.; Gahan, C.G.M.; Grogan, H.; Burgess, C.M. The Ability of Listeria monocytogenes to Form Biofilm on Surfaces Relevant to the Mushroom Production Environment. Int. J. Food Microbiol. 2020, 317, 108385. [Google Scholar] [CrossRef]
- Burdová, A.; Véghová, A.; Minarovičová, J.; Drahovská, H.; Kaclíková, E. The Relationship between Biofilm Phenotypes and Biofilm-Associated Genes in Food-Related Listeria monocytogenes Strains. Microorganisms 2024, 12, 1297. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Listeria monocytogenes Contamination of Ready-to-Eat Foods and the Risk for Human Health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef] [PubMed]
- Tsaloumi, S.; Aspridou, Z.; Tsigarida, E.; Gaitis, F.; Garofalakis, G.; Barberis, K.; Tzoumanika, F.; Dandoulaki, M.; Skiadas, R.; Koutsoumanis, K. Quantitative Risk Assessment of Listeria monocytogenes in Ready-to-Eat (RTE) Cooked Meat Products Sliced at Retail Stores in Greece. Food Microbiol. 2021, 99, 103800. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 338, 1–26. [Google Scholar]
- Asociación Nacional de Industrias de la Carne de España (ANICE) Memoria de Actividades. 2023. Available online: https://www.anice.es/industrias/memoria-anice/memoria-de-actividades-de-anice-2023_36573_201_48093_0_1_in.html (accessed on 18 July 2024).
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria monocytogenes—How This Pathogen Survives in Food-Production Environments? Front. Microbiol. 2022, 13, 866462. [Google Scholar] [CrossRef]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernades Júnior, A.; Araújo Júnior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of Biofilm by Listeria monocytogenes in Different Materials and Temperatures. Food Control 2014, 35, 386–391. [Google Scholar] [CrossRef]
- Conficoni, D.; Losasso, C.; Cortini, E.; Di Cesare, A.; Cibin, V.; Giaccone, V.; Corno, G.; Ricci, A. Resistance to Biocides in Listeria monocytogenes Collected in Meat-Processing Environments. Front. Microbiol. 2016, 7, 1627. [Google Scholar] [CrossRef]
- Ortiz, S.; López, V.; Villatoro, D.; López, P.; Dávila, J.C.; Martínez-Suárez, J.N.V. A 3-Year Surveillance of the Genetic Diversity and Persistence of Listeria monocytogenes in an Iberian Pig Slaughterhouse and Processing Plant. Foodborne Pathog. Dis. 2010, 7, 1177–1184. [Google Scholar] [CrossRef]
- Li, H.; Wang, P.; Lan, R.; Luo, L.; Cao, X.; Wang, Y.; Wang, Y.; Li, H.; Zhang, L.; Ji, S.; et al. Risk Factors and Level of Listeria monocytogenes Contamination of Raw Pork in Retail Markets in China. Front. Microbiol. 2018, 9, 1090. [Google Scholar] [CrossRef]
- Sereno, M.J.; Viana, C.; Pegoraro, K.; da Silva, D.A.L.; Yamatogi, R.S.; Nero, L.A.; Bersot, L. dos S. Distribution, Adhesion, Virulence and Antibiotic Resistance of Persistent Listeria monocytogenes in a Pig Slaughterhouse in Brazil. Food Microbiol. 2019, 84, 103234. [Google Scholar] [CrossRef] [PubMed]
- D’Arrigo, M.; Mateo-Vivaracho, L.; Guillamón, E.; Fernández-León, M.F.; Bravo, D.; Peirotén, Á.; Medina, M.; García-Lafuente, A. Characterization of Persistent Listeria monocytogenes Strains from Ten Dry-Cured Ham Processing Facilities. Food Microbiol. 2020, 92, 103581. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Persistence of Microbiological Hazards in Food and Feed Production and Processing Environments. EFSA J. 2024, 22, e8521. [Google Scholar] [CrossRef]
- Ortiz, S.; López-Alonso, V.; Rodríguez, P.; Martínez-Suárez, J.V. The Connection between Persistent, Disinfectant-Resistant Listeria monocytogenes Strains from Two Geographically Separate Iberian Pork Processing Plants: Evidence from Comparative Genome Analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef]
- Pérez-Baltar, A.; Pérez-Boto, D.; Medina, M.; Montiel, R. Genomic Diversity and Characterization of Listeria monocytogenes from Dry-Cured Ham Processing Plants. Food Microbiol. 2021, 99, 103779. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission Regulation (EU) 2024/2895 of 20 November 2024 Amending Regulation (EC) No 2073/2005 as Regards Listeria monocytogenes. Off. J. Eur. Union 2024, 2895, 1–3. [Google Scholar]
- Larivière-Gauthier, G.; Letellier, A.; Kérouanton, A.; Bekal, S.; Quessy, S.; Fournaise, S.; Fravalo, P. Analysis of Listeria monocytogenes Strain Distribution in a Pork Slaughter and Cutting Plant in the Province of Quebec. J. Food Prot. 2014, 77, 2121–2128. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp. Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2017.
- ISO 18593:2018; Microbiology of the Food Chain—Horizontal Methods for Surface Sampling. International Organization for Standardization: Geneva, Switzerland, 2018.
- Labrador, M.; Rota, M.C.; Pérez-Arquillué, C.; Herrera, A.; Bayarri, S. Comparative Evaluation of Impedanciometry Combined with Chromogenic Agars or RNA Hybridization and Real-Time PCR Methods for the Detection of L. monocytogenes in Dry-Cured Ham. Food Control 2018, 94, 108–115. [Google Scholar] [CrossRef]
- Labrador, M. Desarrollo de Métodos Rápidos Para el Análisis de Listeria monocytogenes y su Aplicación al Proceso de Evaluación del Riesgo en la Industria Cárnica. Ph.D. Thesis, Universidad de Zaragoza, Zaragoza, Spain, 2018. [Google Scholar]
- PulseNet. Standard Operating Procedure for Pulsenet PFGE of Listeria monocytogenes. Available online: https://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL04_ListeriaPFGEProtocol.pdf (accessed on 18 September 2024).
- Graves, L.M.; Swaminathan, B. PulseNet Standardized Protocol for Subtyping Listeria monocytogenes by Macrorestriction and Pulsed-Field Gel Electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- Martin, P.; Jacquet, C.; Goulet, V.; Vaillant, V.; De Valk, H. Participants in the PulseNet Europe Feasibility Study Pulsed-Field Gel Electrophoresis of Listeria monocytogenes Strains: The PulseNet Europe Feasibility Study. Foodborne Pathog. Dis. 2006, 3, 303–308. [Google Scholar] [CrossRef]
- Meloni, D.; Consolati, S.G.; Mazza, R.; Mureddu, A.; Fois, F.; Piras, F.; Mazzette, R. Presence and Molecular Characterization of the Major Serovars of Listeria monocytogenes in Ten Sardinian Fermented Sausage Processing Plants. Meat Sci. 2014, 97, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Demaître, N.; Rasschaert, G.; De Zutter, L.; Geeraerd, A.; De Reu, K. Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens 2021, 10, 717. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Prencipe, V.A.; Rizzi, V.; Acciari, V.; Iannetti, L.; Giovannini, A.; Serraino, A.; Calderone, D.; Rossi, A.; Morelli, D.; Marino, L.; et al. Listeria monocytogenes Prevalence, Contamination Levels and Strains Characterization throughout the Parma Ham Processing Chain. Food Control 2012, 25, 150–158. [Google Scholar] [CrossRef]
- Sala, C.; Morar, A.; Tîrziu, E.; Nichita, I.; Imre, M.; Imre, K. Environmental Occurrence and Antibiotic Susceptibility Profile of Listeria monocytogenes at a Slaughterhouse Raw Processing Plant in Romania. J. Food Prot. 2016, 79, 1794–1797. [Google Scholar] [CrossRef]
- Thévenot, D.; Delignette-Muller, M.-L.; Christieans, S.; Leroy, S.; Kodjo, A.; Vernozy-Rozand, C. Serological and Molecular Ecology of Listeria monocytogenes Isolates Collected from 13 French Pork Meat Salting–Curing Plants and Their Products. Int. J. Food Microbiol. 2006, 112, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Belias, A.; Sullivan, G.; Wiedmann, M.; Ivanek, R. Factors That Contribute to Persistent Listeria in Food Processing Facilities and Relevant Interventions: A Rapid Review. Food Control 2022, 133, 108579. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from a Pork Processing Plant and Its Respective Meat Markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef]
- Knabel, S.J.; Reimer, A.; Verghese, B.; Lok, M.; Ziegler, J.; Farber, J.; Pagotto, F.; Graham, M.; Nadon, C.A.; Gilmour, M.W. Sequence Typing Confirms That a Predominant Listeria monocytogenes Clone Caused Human Listeriosis Cases and Outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 2012, 50, 1748–1751. [Google Scholar] [CrossRef]
- Arslan, S.; Baytur, S. Prevalence and Antimicrobial Resistance of Listeria Species and Subtyping and Virulence Factors of Listeria monocytogenesfrom Retail Meat. J. Food Saf. 2019, 39, e12578. [Google Scholar] [CrossRef]
- Paduro, C.; Montero, D.A.; Chamorro, N.; Carreño, L.J.; Vidal, M.; Vidal, R. Ten Years of Molecular Epidemiology Surveillance of Listeria monocytogenes in Chile 2008–2017. Food Microbiol. 2020, 85, 103280. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequences (5′–3′) | Product Size (pb) | Reference |
|---|---|---|---|
| lmo1118 | F: AGG GGT CTT AAA TCC TGG AA R: CGG CTT GTT CGG CAT ACT TA | 906 | [2] |
| lmo0737 | F: AGG GCT TCA AGG ACT TAC CC R: ACG ATT TCT GCT TGC CAT TC | 691 | [2] |
| ORF2110 | F: AGT GGA CAA TTG ATT GGT GAA R: CAT CCA TCC CTT ACT TTG GAC | 597 | [2] |
| ORF2819 | F: AGC AAA ATG CCA AAA CTC GT R: CAT CAC TAA AGC CTC CCA TTG | 471 | [2] |
| prs | F: GCT GAA GAG ATT GCG AAA GAA G R: CAA AGA AAC CTT GGA TTT GCG G | 370 | [2] |
| prfA | F: GAT ACA GAA ACA TCG GTT GGC R: GTG TAA TCT TGA TGC CAT CAG G | 274 | [3] |
| flaA | F: TTA CTA GAT CAA ACT GCT CC R: AAG AAA AGC CCC TCG TCC | 538 | [6] |
| Sample Category | No. of Samples | Listeria Species No. of Positive Samples (%) | |
|---|---|---|---|
| Listeria spp. | L. monocytogenes | ||
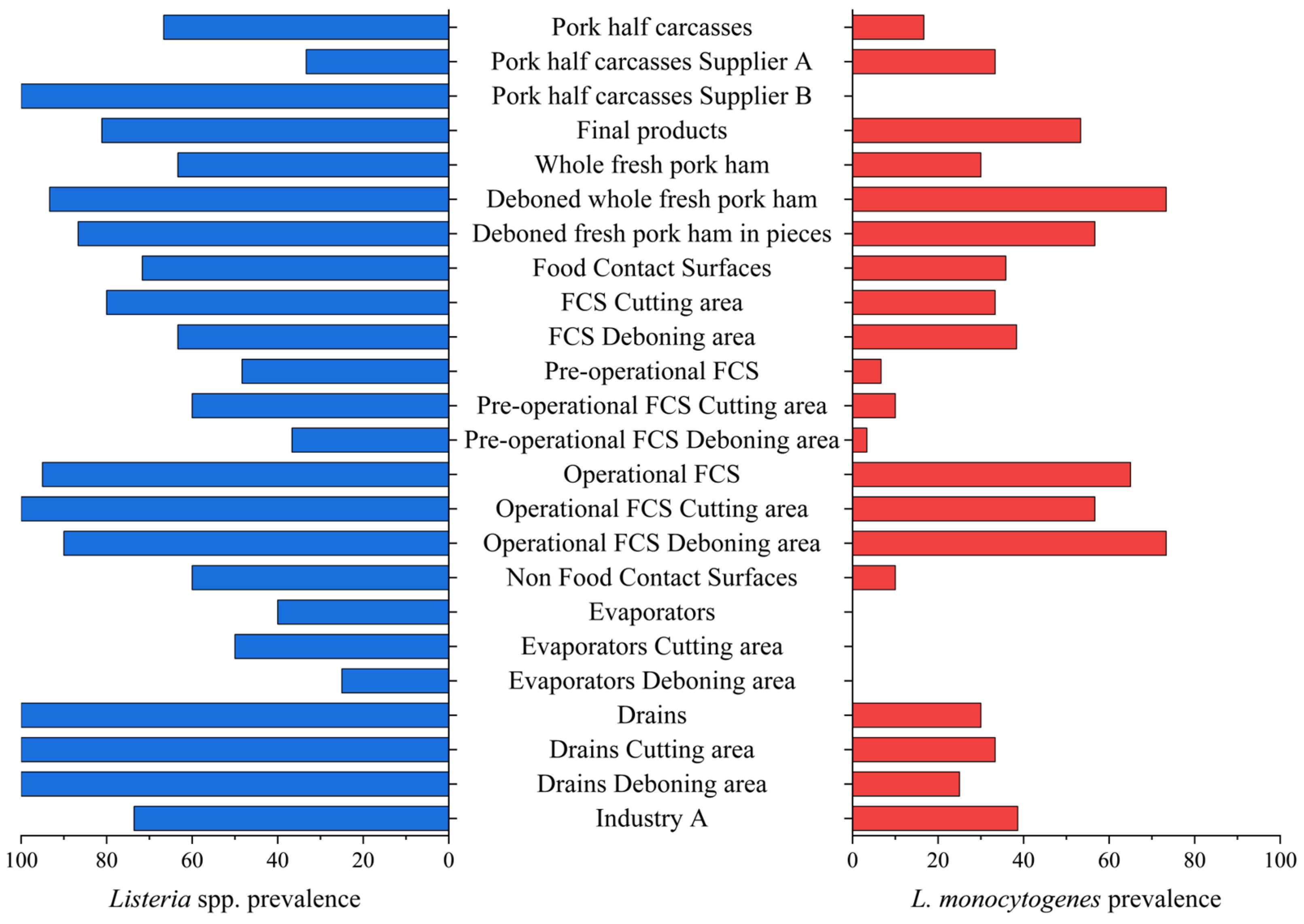
| Pork half carcasses | 6 | 4 (66.7) | 1 (16.7) |
| Pork half carcasses, Supplier A | 3 | 1 (33.3) | 1 (33.3) |
| Pork half carcasses, Supplier B | 3 | 3 (100.0) | 0 (0.0) |
| Final products | 90 | 73 (81.1) | 48 (53.3) |
| Whole fresh pork ham | 30 | 19 (63.3) a | 9 (30.0) a |
| Deboned whole fresh pork ham | 30 | 28 (93.3) b | 22 (73.3) b |
| Deboned fresh pork ham in pieces | 30 | 26 (86.7) ab | 17 (56.7) b |
| Total FCSs | 120 | 86 (71.7) | 43 (35.8) |
| FCSs cutting area | 60 | 48 (80.0) a | 20 (33.3) a |
| FCSs deboning area | 60 | 38 (63.3) b | 23 (38.3) a |
| Pre-operational FCSs | 60 | 29 (48.3) | 4 (6.7) |
| PO FCSs cutting area | 30 | 18 (60.0) a | 3 (10.0) a |
| PO FCSs deboning area | 30 | 11 (36.7) a | 1 (3.3) a |
| Operational FCSs | 60 | 57 (95.0) | 39 (65.0) |
| OP FCSs cutting area | 30 | 30 (100.0) a | 17 (56.7) a |
| OP FCSs deboning area | 30 | 27 (90.0) a | 22 (73.3) a |
| Total NFCSs | 20 | 14 (70.0) | 3 (15.0) |
| Evaporators | 10 | 4 (40.0) | 0 (0.0) |
| Evaporators cutting area | 6 | 3 (50.0) | 0 (0.0) |
| Evaporators deboning area | 4 | 1 (25.0) | 0 (0.0) |
| Drains | 10 | 10 (100.0) | 3 (30.0) |
| Drains cutting area | 6 | 6 (100.0) | 2 (33.3) |
| Drains deboning area | 4 | 4 (100.0) | 1 (25.0) |
| Industry A | 236 | 177 (75.0) | 95 (40.3) |
| Sample Category | No. of Samples | Listeria Species No. of Positive Samples (%) | |
|---|---|---|---|
| Listeria spp. | L. monocytogenes | ||
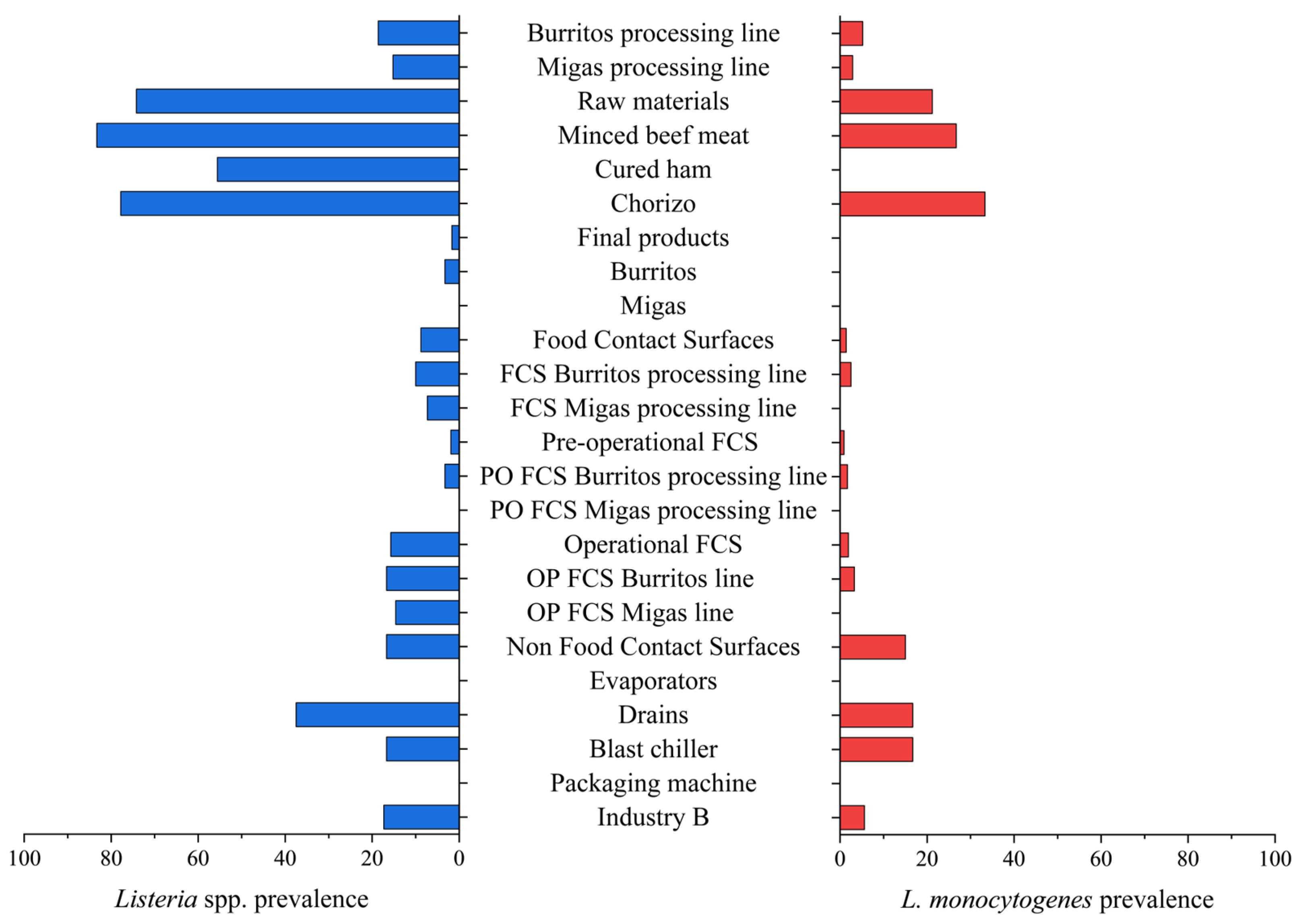
| Burritos processing line | 210 | 39 (18.6) a | 11 (5.2) a |
| Migas processing line | 192 | 31 (16.1) a | 6 (3.1) a |
| Raw materials | 66 | 49 (74.2) | 14 (21.2) |
| Minced beef meat | 30 | 25 (83.3) a | 8 (26.7) a |
| Cured ham | 18 | 10 (55.6) b | 0 (0.0) b |
| Chorizo | 18 | 14 (77.8) ab | 6 (33.3) a |
| Final products | 120 | 2 (1.7) | 0 (0.0) |
| Burritos | 60 | 2 (3.3) | 0 (0.0) |
| Migas | 60 | 0 (0.0) | 0 (0.0) |
| Total FCSs | 216 | 19 (8.8) | 3 (1.4) |
| FCSs Burritos processing line | 120 | 12 (10.0) a | 3 (2.5) a |
| FCSs Migas processing line | 96 | 7 (7.3) a | 0 (0.0) a |
| Pre-operational FCSs | 108 | 2 (1.9) | 1 (0.9) |
| PO FCSs Burritos processing line | 60 | 2 (3.3) a | 1 (1.7) a |
| PO FCSs Migas processing line | 48 | 0 (0.0) a | 0 (0.0) a |
| Operational FCSs | 108 | 17 (15.7) | 2 (1.9) |
| OP FCSs Burritos processing line | 60 | 10 (16.7) a | 2 (3.3) a |
| OP FCSs Migas processing line | 48 | 7 (14.6) a | 0 (0.0) a |
| Total NFCSs | 60 | 10 (16.7) | 9 (15.0) |
| Evaporators | 24 | 0 (0.0) | 0 (0.0) |
| Drains | 24 | 9 (37.5) | 8 (33.3) |
| Blast chiller | 6 | 1 (16.7) | 1 (16.7) |
| Packaging machine | 6 | 0 (0.0) | 0 (0.0) |
| Industry B | 462 | 80 (17.3) | 26 (5.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero de Castilla López, B.; Gómez Lozano, D.; Herrera Marteache, A.; Conchello Moreno, P.; Rota García, C. Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods 2025, 14, 1519. https://doi.org/10.3390/foods14091519
Romero de Castilla López B, Gómez Lozano D, Herrera Marteache A, Conchello Moreno P, Rota García C. Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods. 2025; 14(9):1519. https://doi.org/10.3390/foods14091519
Chicago/Turabian StyleRomero de Castilla López, Belén, Diego Gómez Lozano, Antonio Herrera Marteache, Pilar Conchello Moreno, and Carmen Rota García. 2025. "Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention" Foods 14, no. 9: 1519. https://doi.org/10.3390/foods14091519
APA StyleRomero de Castilla López, B., Gómez Lozano, D., Herrera Marteache, A., Conchello Moreno, P., & Rota García, C. (2025). Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods, 14(9), 1519. https://doi.org/10.3390/foods14091519





