Abstract
High quality stands as a pivotal competitive edge in the rice industry. Optimizing amylose content (AC) and the physicochemical properties of endosperm starch by regulating the Wx gene is crucial for enhancing rice grain quality. In this study, we created a novel Wxb-d25 allele by deleting a 25 bp segment (−26 to −2) within the Wx core promoter using CRISPR/Cas9. Compared with the wild type and the previously reported Wxb-i1, Wxb-d25 exhibited no significant changes in agronomic traits. However, its grains displayed temperature-dependent variations in AC and altered transparency and viscosity characteristics, holding the potential to synergistically improve both the eating and cooking quality (ECQ) and appearance quality (AQ) of rice. Further studies demonstrated that this promoter modification, by partially disrupting the transcription initiator, significantly downregulated the original Wx-01 transcript and generated a novel Wx transcript (ONT.7395.1) in Wxb-d25 grains. Despite its low expression abundance, the ONT.7395.1 transcript could be completely processed into mature Wx mRNA. The combined effects of the dual transcripts resulted in significantly increased Wx gene expression and AC in Wxb-d25 grains under conventional cultivation conditions. These findings provide a genetic resource and a theoretical foundation for utilizing the Wxb-d25 allele to improve rice grain quality.
1. Introduction
High-quality rice is a central objective in genetic improvement programs, with starch composition and structure playing a decisive role in overall grain quality [1,2]. Among starch-related characteristics, the amylose content (AC), which reflects the ratio of amylose to amylopectin in the endosperm, is the primary determinant of rice physicochemical properties [3,4,5]. Research has shown that AC profoundly influences eating and cooking quality (ECQ), appearance quality (AQ), nutrition quality (NQ), and milling quality (MQ) [5,6,7,8,9]. For ECQ, AC correlates positively with rice hardness and inversely with stickiness [10]. Modulating AC of rice exerts a significant influence on its physicochemical attributes, including hardness and stickiness, which in turn significantly enhances the palatability of cooked rice [3]. Globally, consumer preferences for rice with varying AC levels are highly diverse [11,12]. In East Asian countries such as China and Japan, rice with low AC (typically <16%) enjoys widespread popularity due to its softer texture and higher stickiness [11,12]. Conversely, in regions like India and Pakistan, consumer preference leans toward rice varieties characterized by higher hardness and lower stickiness, which generally correspond to medium-to-high AC levels (exceeding 20%) [11,12]. Furthermore, glutinous rice, which lacks amylose synthesis, has gained widespread application in the processing of rice-based foods, thanks to its distinctive physicochemical attributes [11]. However, its use as a staple food on a long-term basis remains relatively limited. These divergent preferences likely stem from the distinct dietary habits and culinary traditions that prevail in different regions. Concerning AQ, AC correlates positively with grain transparency. When AC is lower than about 12%, transparency declines significantly, rendering the grains increasingly translucent or even opaque [13,14]. In addition, medium- to high-AC varieties are more susceptible to chalkiness, while suppressing amylose synthesis can significantly mitigate chalkiness formation [15,16]. From a nutritional perspective, higher AC promotes resistant starch, thereby reducing the rate of digestion [17,18,19]. As for MQ, AC is intricately linked to both rice grain fissure resistance and head rice yield [20]. Lowering AC significantly boosts head rice yield. In summary, while there are clear differences in consumer preferences for rice AC, global research on rice domestication and artificial selection has consistently revealed a trend toward selecting for lower AC levels [3,21]. However, the degree of AC reduction in rice varies across regions due to differing consumer preferences. Therefore, adjusting rice AC in a region-specific manner to meet consumer preferences—without considering digestive characteristics—emerges as a fundamental strategy for enhancing rice quality.
Amylose synthesis in the rice endosperm is controlled by the Waxy (Wx) gene, which encodes granule-bound starch synthase I (GBSSI) [22]. The precise genetic regulation of the Wx gene and its encoded GBSSI protein represents a critical avenue for rice quality improvement. Multiple regulatory pathways, both direct and indirect, modulate the expression and activity of Wx, with transcriptional and post-transcriptional mechanisms playing a key role. For example, a G/T mutation at the first base of the first intron disrupts the processing of pre-mRNA into mature mRNA, accounting for the markedly lower AC observed in rice carrying the Wxb allele compared to that with Wxa [23,24]. Surrounding this variation site, several Dull genes and quantitative trait loci (QTLs) have been identified that modulate Wx splicing efficiency in the Wxb background, including Du1, Du2, Du3/OsCBP20, LowAC1, LAC6/TL1/Du13, qAC2, and qSAC3 [25,26,27,28,29,30,31,32]. Recent studies have identified multiple transcription factors involved in Wx regulation, including OsbZIP58, OsbZIP33/REB, OsNAC20, OsNAC26, NF-YB1, NF-YC12, bHLH144, OsMADS7, OsMADS14, RSR1, SLRL2/LCG1, OsMYB73, and OsDOF18. These factors regulate AC by either directly binding to the Wx promoter or indirectly interacting with other transcription factors that influence Wx expression [16,33,34,35,36,37,38,39,40,41,42]. Additionally, DNA methylation has been implicated in the transcriptional regulation of Wx. High levels of CpG methylation in the Wx promoter are closely associated with reduced AC, with two adjacent CpG islands identified in this region [43]. Promoter editing studies have further refined our understanding of Wx gene regulation, uncovering additional expression-modulating elements beyond known transcription factor binding sites. For example, our previous research identified and validated an S7 site within the Wx core promoter that effectively fine-tunes Wx expression [44]. Similarly, Zeng et al. identified multiple cis-acting elements in the Wx promoter, including the Endosperm-box, A-box, and CAAT-box [45]. These findings highlight the complexity of Wx transcriptional regulation and provide valuable insights into strategies for targeted modulation to enhance rice quality.
The advent and advancement of CRISPR/Cas technology have significantly streamlined research into gene functions and the development of superior alleles and novel germplasms [46]. Presently, CRISPR/Cas technology has been extensively utilized for gene editing in a variety of crops, with the Wx gene emerging as one of the most frequently targeted genes. To date, successful editing of the Wx gene using CRISPR/Cas technology has been reported in several major crops, including rice, maize, wheat, barley, and cassava [47,48,49,50,51]. These studies have not only expanded the potential for creating superior Wx alleles with diverse AC profiles through gene editing but also paved the way for a deeper exploration of the regulatory mechanisms governing the Wx gene.
In this study, we focused on Chinese rice consumers who favor low AC. We tackled the challenge that the popular low-AC soft rice varieties (typically <12%) in breeding and consumer markets, while offering good palatability due to their low AC, often suffer from poor grain transparency, thus failing to achieve a balance between excellent ECQ and desirable AQ. By leveraging our previously established method of editing the core promoter of the Wx gene to fine-tune its expression and regulate rice AC, we successfully developed a novel Wxb-d25 allele featuring a 25 bp deletion within the Wx core promoter. We compared the AC and transparency of rice carrying this new allele with those of the representative soft rice variety Nangeng 46 and conducted a comprehensive evaluation of the physicochemical properties of the relevant materials. Furthermore, we elucidated the underlying mechanism by which this allele modulates Wx gene expression and rice AC through alterations in the transcriptional pattern of the Wx gene. These findings not only introduce a new gene and germplasm resource but also provide a robust theoretical foundation for improving the quality of soft rice.
2. Materials and Methods
2.1. Plant Materials
The japonica rice variety Nipponbare, which carries the Wxb allele, served as the recipient for generating the promoter-edited lines. The elite soft rice cultivar Nangeng 46 (NG46), which carries the Wxmp allele, was used as a control due to its low AC and excellent ECQ [52]. All rice lines were cultivated under identical climatic and management conditions in a paddy field in the summer in Yangzhou, Jiangsu Province (119° E, 32° N) or winter in Lingshui, Hainan Province (110° E, 18° N).
2.2. Construction of CRISPR/Cas9 Vector and Screening of Homozygous Edited Lines
The CRISPR/Cas9 system used in this study includes two parts: the intermediate vector SK-gRNA and the final vector pC1300-Cas9 [53]. The target site S7 was designed as a primer and inserted into the gRNA region of SK-gRNA. Afterward, the gRNA was cleaved and assembled into the pC1300-Cas9 binary vector using an isocaudamer system. The final CRISPR/Cas9 construct was transformed into rice via Agrobacterium-mediated transformation. To determine mutation type, the targeted promoter region was amplified by PCR and sequenced using the Sanger method. The sequencing data were analyzed via DSDecodeM (http://skl.scau.edu.cn/dsdecode/) (accessed on 2 August 2024) or manually decoded [54,55]. A list of the PCR primers used is provided in Table S1.
2.3. Determination of Key Agronomic Traits
Wild-type (WT) and S7-edited rice plants were dug up from the field at 15–25 days after flowering (DAF) and photographed to compare overall plant morphology. Once the rice reached maturity, 10 individual plants were randomly selected to measure plant height and tiller number, while 10 main panicles were sampled for evaluating seed setting rate. After drying, more than 50 rice grains were scanned using an SC-E rice appearance quality detector (Wanshen Testing Technology Co., Hangzhou, Zhejiang, China) to measure grain length, grain width, and 1000-grain weight. Grain thickness was measured on 15–20 grains using a vernier caliper.
2.4. Determination of Physicochemical Properties of Rice Grains
Mature seeds were air-dried, dehulled (SY88-TH, Sangyong, Seoul, Republic of Korea), polished (Pearlest, Kett, Tokyo, Japan) and ground into powder (Cyclotec CT1093, FOSS, Alingsås, Sweden), then passed through a 100-mesh sieve (pore size 0.15 mm). Total starch content (TSC) was measured using a K-TSTA total starch assay kit (Megazyme, Bray, County Wicklow, Ireland). The measurement of apparent amylose content (AAC) and gel consistency (GC) was conducted in accordance with previously published methods [56]. Gelatinization temperature (GT) was measured using a 200 F3 differential scanning calorimeter (DSC, Netzsch, Selb, Germany), providing onset (To), peak (Tp), and end (Te) temperatures, along with the enthalpy of gelatinization (ΔH). Pasting properties were assessed using a Techmaster rapid visco-analyzer (RVA) (Newport Scientific, Warriewood, NSW, Australia). The RVA parameters include peak viscosity (PKV), hot paste viscosity (HPV), breakdown value (BDV), cool paste viscosity (CPV), setback value (SBV), peak time (PeT), and pasting temperature (PaT). All measurements were performed in triplicate.
2.5. Grain Transparency Calculation
Rice grains from various lines were air-dried, dehulled (SY88-TH, Sangyong, Korea), polished (Pearlest, Kett, Japan), and then placed on a scanner coupled with the SC-E rice appearance quality detector (Wanshen Testing Technology Co.) to obtain high-resolution photos. Using ImageJ 1.53t software, the images were converted to grayscales, inverted, and then measured for grayscale values. Dividing these values by 255 yielded the grain transparency [27,56].
2.6. Scanning Electron Microscopy Analysis
Starch was purified from milled rice via a reported neutral protease method [57] with slight modifications. The resulting starch samples were suspended in ethanol, mounted on aluminum stubs using carbon double-sided conductive tape, gold-coated using a sputter coater, and then examined using a Hitachi S-4800II environmental scanning electron microscope (SEM, Hitachi, Tokyo, Japan) for imaging. Starch grain sizes were measured using ImageJ software, with at least 40 individual granules analyzed for size distribution.
2.7. Gel Permeation Chromatography Analysis
Purified rice starch (as described in Section 2.6) was enzymatically debranched using isoamylase (EC 3.2.1.68, E-ISAMY, Megazyme, Bray, County Wicklow, Ireland), dissolved in dimethyl sulfoxide (DMSO, Merck KGaA, Darmstadt, Germany), and analyzed for relative molecular weight distribution via gel permeation chromatography (GPC) using a PL-GPC 220 integrated GPC system (Agilent Technologies, Lexington, MA, USA). The resulting GPC chromatograms resolved three distinct fractions, corresponding to A and B1 chains of amylopectin (Ap1), long B chains of amylopectin (Ap2), and amylose (Am), respectively.
2.8. Total RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA was extracted from developing rice caryopses at 5, 10, 15, 20, and 25 DAF using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, Jiangsu, China). First-strand cDNA synthesis was performed using the HiScript III RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, China). Quantitative real-time PCR (RT-qPCR) was conducted on a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using ChamQ Universal SYBR qPCR Master Mix (Vazyme). The Actin01 gene was used as an internal reference for normalization, and all experiments were performed with three biological replicates. The primers used for RT-qPCR are listed in Table S1.
2.9. Western Blot and Enzyme Activity Assay
Mature rice flour was weighed and suspended in total protein extraction buffer (125 mM Tris-HCl (pH 6.8), 4 M urea, 4% SDS, and 5% β-mercaptoethanol] at 37 °C for 3 h (h) at a ratio of 1:15 (1 mg flour to 15 µL buffer). The extracted total proteins were mixed with 5× protein loading buffer, denatured at 99 °C for 10 min, separated via SDS-PAGE, and subsequently transferred onto a PVDF membrane. GBSSI accumulation was examined using a specific GBSSI antibody, while SDS-PAGE gels were stained with Coomassie Brilliant Blue as a loading control [58]. GBSSI enzyme activity was assessed using a GBSSI enzyme activity assay kit (Keming, Suzhou, Jiangsu, China) with developing rice caryopses collected at 10 DAF.
2.10. Transient Assay of Promoter Activity
The 2.7 Kb promoter sequences of S7-edited lines and their WT counterparts were amplified and cloned into the pGreenII 0800-LUC vector. The resulting reporter constructs were introduced into Agrobacterium tumefaciens and subsequently infiltrated into Nicotiana benthamiana leaves. Firefly luciferase (LUC) and Renilla luciferase (REN) activities were measured using a Dual Luciferase Reporter Assay Kit (Vazyme). Promoter activity was quantified as the LUC/REN ratio. Three plants at the same developmental stage were used for each assay, with all experiments conducted in triplicate.
2.11. Full-Length Transcriptome Analysis
Full-length transcriptome sequencing of rice caryopses collected 10 DAF from both WT and S7-edited lines was performed and analyzed by Biomarker Technology Company (Beijing, China) using the PromethION platform (Oxford Nanopore Technologies, Oxford, UK).
2.12. Statistical Analysis
All data are presented as means ± standard deviations (SD). A one-way analysis of variance (ANOVA) was conducted using GraphPad Prism 9.3.1 software to assess statistical significance, with p < 0.05 (*) and p < 0.01 (**) indicating significant differences.
3. Results
3.1. Creation of a Novel Wx Allele (Wxb-d25) via Promoter Editing
To develop novel Wx alleles with enhanced regulatory properties, we generated a series of S7-edited lines, building on our previous findings that editing the S7 site within the Wx gene promoter significantly regulates Wx gene expression and AC in the rice endosperm. Screening for mutation types in the regenerated edited lines led to the identification of a novel Wx allele, Wxb-d25, characterized by a 25 bp deletion (−26 to −2) near the S7 site (Figure 1). The Wxb-d25 allele retained the integrity of the TATA box (−35 to −29) located upstream of the S7 site but partially disrupted the transcription initiator (−2 to +4) of the Wx gene. To further investigate the genetic effects of Wxb-d25 and evaluate its potential for breeding applications, we conducted a comprehensive analysis comparing Wxb-d25 with its recipient parent, Nipponbare (Wxb), and the previously reported Wxb-i1 allele, which harbors an adenine insertion [44].
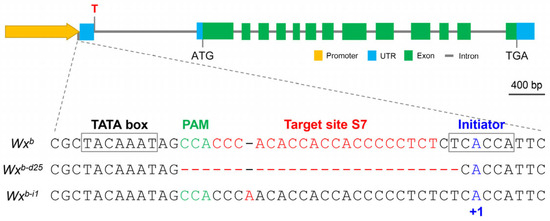
Figure 1.
Schematic representation of sequence alterations at the S7 target site within the Wx promoter in the wild-type (Wxb) and the edited lines (Wxb-d25 and Wxb-i1). The TATA box, PAM site, target region (S7), and transcriptional initiator are highlighted. The overall gene structure, including promoter, untranslated region (UTR), exons, introns, and termination codon (TGA), is indicated for reference.
We first measured the agronomic traits of the candidate lines. The results showed that, in comparison to the WT, both Wxb-d25 and Wxb-i1 exhibited no significant alterations in key vegetative and reproductive traits (Figure S1). A detailed analysis of agronomic data revealed that, aside from a modest reduction in grain length and 1000-grain weight, the edited lines were largely comparable to the WT (Table S2). These findings corroborate our previous report and reinforce the understanding that the Wx gene predominantly governs amylose synthesis in the rice endosperm [44].
3.2. Wxb-d25 Allele Synergistically Improves Amylose Content and Grain Transparency in Rice
To elucidate the regulatory effect of the Wxb-d25 allele on rice AC, and considering our previous findings that the S7 site of the Wx promoter influences Wx gene expression and AC in a temperature-dependent manner, we measured the AAC of rice grains harvested under summer planting conditions in Yangzhou and winter conditions in Lingshui. The average summer temperature in Yangzhou is approximately 2.5–3.5 °C higher than the winter temperature in Lingshui. Under the warmer summer conditions in Yangzhou, the AAC of Wxb-d25 rice significantly increased from 16.5% in the WT to 18.0%, whereas Wxb-i1 showed no significant change (Figure 2A). In contrast, under the cooler winter conditions in Lingshui, the AAC of both Wxb-d25 and Wxb-i1 was significantly reduced, from 15.8% in the WT to 12.4% and 12.5%, respectively, reaching levels characteristic of high-quality soft rice (Figure 2B). These results indicate that the Wxb-d25 allele effectively modulates rice AC, but its regulatory effect, similarly to other S7-edited Wx alleles, is temperature-dependent.
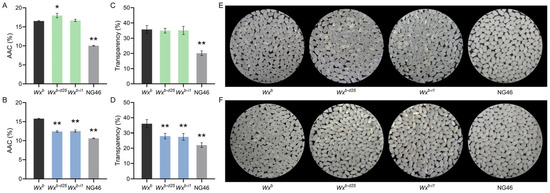
Figure 2.
Apparent amylose content, transparency, and appearance of polished rice grains from Wx promoter-edited lines and the wild-type. (A,C,E) Data from rice grains grown under summer conditions in Yangzhou. (B,D,F) Data from rice grown under winter conditions in Lingshui. (A,B) Apparent amylose content (AAC); (C,D) Grain transparency values; (E,F) Representative images of polished rice grains. “*” and “**” indicate statistically significant differences compared to the wild-type at p < 0.05 and p < 0.01, respectively. n = 3 for AAC measurements (A,B) and transparency values (C,D); n = 40 for grain transparency imaging (E,F).
AC is also a key determinant of rice grain transparency. To assess the impact of the Wxb-d25 allele on grain transparency following AC modification, we examined the grain appearance of milled rice under both planting conditions and compared it with the high-quality soft rice cultivar NG46, which carries the Wxmp allele. Under the warmer summer conditions in Yangzhou, both Wxb-d25 and Wxb-i1 exhibited a transparent grain phenotype, consistent with their respective AAC profiles, which showed either no significant change or a notable increase (Figure 2C,D). In contrast, under the cooler winter conditions in Lingshui, the transparency of Wxb-d25 and Wxb-i1 grains was significantly reduced, aligning with their low AC phenotypes. However, in terms of measured transparency values, the polished rice from both lines exhibited significantly greater transparency than the high-quality soft rice control NG46 (Figure 2E,F). These results indicate that the Wxb-d25 allele simultaneously regulates both AC and grain transparency, offering a promising strategy to enhance both ECQ and AQ in rice.
3.3. Wxb-d25 Allele Enhances Rice Grain Quality
To clarify the specific effects of Wxb-d25 and Wxb-i1 on rice grain quality, we assessed key physicochemical parameters, including TSC, GC, GT, and RVA profiles. Under summer planting conditions in Yangzhou, Wxb-d25 and Wxb-i1 exhibited no significant differences in TSC, GC, and GT compared to the WT control. Similarly, under winter planting conditions in Lingshui, both TSC and GT remained unchanged; however, GC length increased slightly, with Wxb-d25 showing a statistically significant increase (Figure 3A–D, Table S3). The RVA profile of Wxb-i1 rice flour from Yangzhou closely resembled that of the WT, except for a significant increase in PeT. In contrast, under winter planting conditions in Lingshui, Wxb-i1 displayed a general decline in its RVA profile, with significant reductions in PKV, BDV, CPV, and SBV, while PeT was significantly increased. These changes were consistent with its significantly lower AC. Unlike Wxb-i1, Wxb-d25 exhibited distinct RVA profile modifications despite differing trends and magnitudes of AC variation under the two planting conditions. In both environments, Wxb-d25 showed an overall reduction in its RVA profile, with significant decreases in HPV, CPV, SBV, and PaT, while BDV was significantly increased (Figure 3E,F, Table 1). Notably, the combination of significantly increased BDV and significantly decreased SBV is consistent with the previously reported RVA characteristics of premium-tasting rice [59,60,61], indicating that Wxb-d25 contributes to ECQ enhancement across different planting conditions.
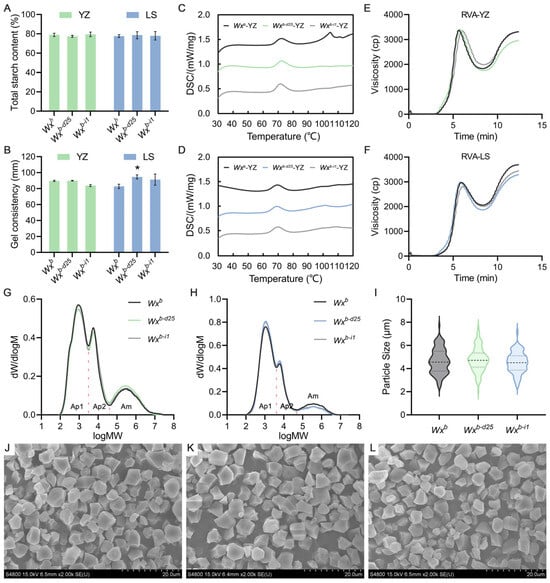
Figure 3.
Physicochemical properties of rice grains from Wx promoter-edited lines and the wild-type. (A) Total starch content. (B) Gel consistency. (C,D) Differential scanning calorimetry (DSC) curves showing gelatinization temperatures of rice flour from Yangzhou (C) and Lingshui (D). (E,F) Rapid visco analyzer (RVA) profiles of rice flours from Yangzhou (E) and Lingshui (F). (G,H) Molecular weight distribution of debranched starch from grains grown in Yangzhou (G) and Lingshui (H). (I) Starch granule particle size from rice grains grown in Yangzhou. (J,L) Scanning electron microscopy (SEM) images illustrating starch granule morphology of Wxb (J), Wxb-d25 (K), and Wxb-i1 (L). YZ, Yangzhou; LS, Lingshui. An asterisk indicates a statistically significant difference at p < 0.05; n = 3.

Table 1.
Rapid visco analyzer (RVA) parameters of rice flour.
In addition, we extracted starch from the tested rice grains and conducted a detailed analysis of its fine structure. GPC and SEM results revealed that the increase, stability, or reduction in AAC in Wxb-d25 and Wxb-i1 under different planting conditions correspondingly altered the proportion of amylopectin fractions with varying molecular weights (Figure 3G,H). However, these changes had no significant impact on the morphology or size of starch granules (Figure 3I–L). This result aligns with the previously observed AAC trends and is consistent with existing reports on the Wx gene’s role in starch composition.
3.4. Expression and Promoter Activity Analysis of Wxb-d25
To clarify the impact of promoter editing on Wx gene transcription and translation, we first conducted Western blot analysis to analyze the expression of GBSSI protein in mature grains under two distinct planting conditions. Under summer planting conditions in Yangzhou, the accumulation of GBSSI protein in Wxb-d25 exhibited a significant increase, whereas Wxb-i1 showed no significant change (Figure 4A). Conversely, under winter planting conditions in Lingshui, GBSSI protein accumulation was significantly reduced in both Wxb-d25 and Wxb-i1 (Figure 4B), aligning with their previously observed AC phenotypes. To further validate promoter activity in vitro, we employed a dual-luciferase reporter gene assay to assess transient expression levels of Wxb-d25 and Wxb-i1 promoters in tobacco. The results revealed an upward trend in the promoter activity of Wxb-d25, though statistical significance was not achieved. Meanwhile, the promoter activity of Wxb-i1 remained comparable to that of the WT (Figure 4C,D). Additionally, developing seeds at 5–25 DAF under summer planting conditions in Yangzhou were analyzed for GBSSI enzyme activity and Wx gene expression. Enzyme activity assays showed a significant increase in GBSSI enzyme activity and Wx gene expression. Enzyme activity assays indicated a significant increase in GBSSI activity in 10 DAF in Wxb-d25, while Wxb-i1 displayed no discernible change (Figure 4E). Given previous studies noting that the G/T mutation at the first base of intron 1 in the Wxb allele disrupts normal splicing by causing intron 1 retention and a significant reduction in splicing efficiency [58], we employed RT-qPCR with three primer pairs to investigate Wx gene expression in developing grains from 5 to 25 DAF. With a few exceptions, Wxb-i1 exhibited no significant differences in precursor mRNA (Wx-Pre), mature mRNA (Wx-Mat), or total mRNA (Wx-Tot) expression compared to WT. This was consistent with the absence of significant changes in its AC, promoter activity, GBSSI enzyme activity, and protein expression. However, in Wxb-d25, the expression of all three transcript variants was significantly reduced, contradicting the significant increases observed in AC, promoter activity, and GBSSI protein accumulation (Figure 4F–H). Given that the 25 bp deletion in the core promoter region of Wxb-d25 likely disrupted the transcriptional initiator of the Wx gene, we speculate that this discrepancy between phenotype and Wx gene transcriptional levels may stem from alterations in mRNA splicing mechanisms.
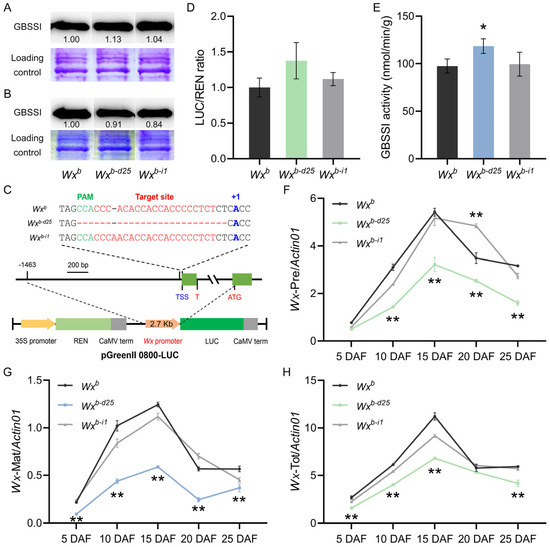
Figure 4.
Expression and activity analyses of Wx promoter-edited lines and wild-type. (A,B) GBSSI protein accumulation in grains from each line grown in Yangzhou (A) and Lingshui (B). (C) Schematic representation of the pGreenII 0800-LUC vector construction. (D) Transient assay of Wx allele promoter activity. (E) GBSSI activity in developing rice caryopses at 10 DAF under Yangzhou field conditions. (F–H) Relative expression levels of Wx gene precursor mRNA (F), mature mRNA (G), and total mRNA (H) in developing caryopses grown in Yangzhou. Asterisks (* and **) indicate statistically significant differences at p < 0.05 and p < 0.01, respectively (n = 3).
3.5. Full-Length Transcriptome Sequencing Reveals a Novel Transcription Mechanism in Wxb-d25
To verify this supposition, we utilized Nanopore third-generation full-length transcriptome sequencing to investigate Wx gene transcripts in Wxb-d25, Wxb-i1, and their corresponding WT. Data analyses revealed that, in addition to the two canonical transcripts Os06t0133000-01 (hereafter Wx-01, predominantly expressed isoform) and Os06t0133000-02 (Wx-02), several novel Wx transcripts were identified. Among these, the transcript designated ONT.7395.1 closely resembles Wx-01 in overall structure, retaining all 14 exons without altering the protein-coding sequence. However, it exhibits distinct modifications in the sequence and length of exon 1, exon 2, and exon 14 (Figure 5A). Specifically, exon 1 of the ONT.7395.1 transcript contains a promoter sequence (−64 bp to −26 bp) along with a partial fragment of exon 1 (+1 bp to +46 bp) from the original Wx-01 transcript. The absence of a 25 bp segment (−26 bp to −2 bp), which was deleted through gene editing, indicates that this transcript is uniquely associated with Wxb-d25. Meanwhile, exon 2 of ONT.7395.1 is nearly identical to that of Wx-01, except for a 5 bp (TGCAG) insertion derived from the terminal region of intron 1 in the original Wx-01 transcript, which is added at the exon’s 5′ end. It is particularly noteworthy that the sequence alterations in exon 1 and exon 2 of ONT.7395.1 effectively bypass the G/T mutation at the 5′ splicing site of intron 1 in Wxb, a mutation known to severely impair splicing efficiency. Additionally, exon 14 of ONT.7395.1 extends 60 bp beyond the original Wx-01 transcript (Figure 5B). These findings suggest that the 25 bp deletion in Wxb-d25 alters the splicing pattern of the Wx gene, leading to the generation of a novel transcript. This transcript retains the full protein-coding sequence but undergoes structural modifications that may influence its stability, expression, or regulatory interactions.
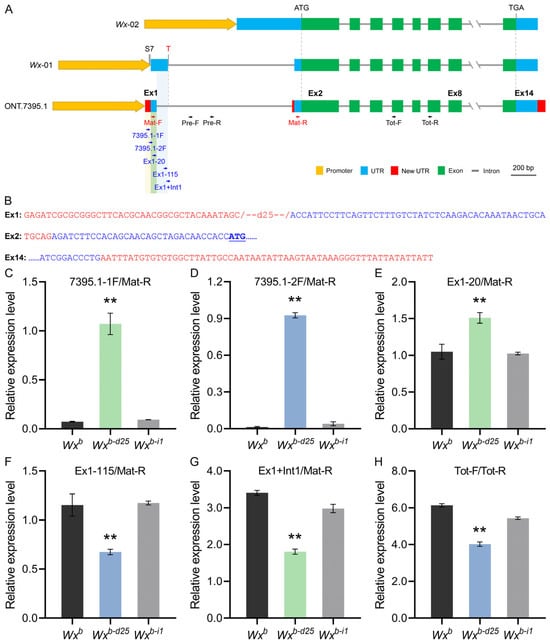
Figure 5.
Transcript identification and validation of Wxb-d25 allele. (A) Schematic illustration of the three transcripts derived from the Wxb-d25 allele. (B) Sequence changes in exons 1, 2, and 14 of the ONT.7395.1 transcript. Text in red and blue represents altered and unchanged sequences relative to Wx-01, respectively. (C–H) Relative expression levels measured with primer sets 7395.1-1F/Mat-R (C), 7395.1-2F/Mat-R (D), Ex1-20/Mat-R (E), Ex1-115/Mat-R (F), Ex1+Int1/Mat-R (G), and 7395.1-1F/Mat-R (H). The primer locations are indicated in (A). Double asterisks (**) indicate statistically significant differences at p < 0.01 (n = 3).
To further validate the authenticity and expression levels of the ONT.7395.1 transcript and investigate the transcriptional mechanism of Wxb-d25, we designed multiple RT-qPCR primers targeting distinct Wx transcripts. This was particularly necessary given that the original Mat-F primers (+4 bp to +29 bp), used for detecting mature Wx mRNA expression, are located near the transcription initiator of the Wx gene. Among the designed primers, 7395.1-1F and 7395.1-2F primers specifically detected the ONT.7395.1 transcript, while the Ex1-20 primer identified both the Wx-01 and ONT.7395.1 transcripts. The Ex1-115 primer was selective for Wx-01, and the Ex1 + Int1 primer specifically detected the pre-mRNA containing unspliced intron 1 of Wx-01. Expression analysis of developing grains at 10 DAF confirmed that the ONT.7395.1 transcript was uniquely present and actively expressed in Wxb-d25 (Figure 5C,D). Additionally, expression levels of precursor mRNA, mature mRNA, and total mRNA of the Wx-01 transcript were significantly reduced compared to the WT, indicating that although Wx-01 remained transcribed, its transcription was significantly reduced by approximately 50% (Figure 5F–H).
In terms of specific expression levels, the ONT.7395.1 transcript in developing grains of Wxb-d25 at 10 DAF exhibited an expression level of approximately 0.9–1.1 relative to the internal reference Actin01 gene (Figure 5C,D). Despite this relatively low expression, ONT.7395.1 was fully translated into functional GBSSI protein without being affected by compromised splicing efficiency. Meanwhile, the Wx-01 transcript in Wxb-d25 exhibited an overall expression level of 2.5 (sum of precursor mRNA and mature mRNA) relative to the internal reference Actin01 gene (Figure 5F–G), representing approximately 55% of WT (Wxb) expression. However, due to the G/T variation at the intron 1 splice site, only about 0.7 of this fraction was successfully processed into mature Wx mRNA (Figure 5F). The combined expression of these two transcripts resulted in a significant increase in the mature Wx mRNA encoding GBSSI protein in Wxb-d25 compared to the WT (Figure 5E). This, in turn, led to the elevated AC and physicochemical changes of starch in Wxb-d25 under summer planting conditions.
4. Discussion
4.1. Development of an Elite Wx Allele
AC is a crucial factor in determining rice grain quality, with the Wx gene serving as the primary genetic regulator. Modulating the expression and activity of the Wx gene and its encoded GBSSI protein to optimize endosperm amylose synthesis is a critical approach to improving rice grain quality. Existing strategies for regulating Wx and GBSSI expression primarily include (1) utilization of superior Wx alleles with low AC, such as Wxb (AC~16%), Wxmw/Wxla (AC~13%), and Wxmp (AC~10.5%) [3,14,21]; (2) transcriptional regulation of Wx through specific transcription factors; (3) regulation of Wx splicing efficiency by Dull genes and QTLs; (4) indirect modification of AC via other starch synthesis-related genes, particularly those involved in amylopectin synthesis (e.g., SBEIIb, SSIIIa) and non-enzymatic proteins that facilitate GBSSI localization to the starch granule surface (e.g., OsGBP, OsLESV, OsESV1) [62,63,64,65,66]; and (5) regulation of genes that influence starch synthesis indirectly, including those controlling floury endosperm formation [67,68], chalkiness [69], hormone synthesis and transport [70], and grain filling [71]. However, most of these genetic modifications not only alter AC but also lead to compromised yield and some undesirable quality traits, such as floury endosperm and high chalkiness. Consequently, despite the extensive identification of genes directly or indirectly involved in amylose biosynthesis, few have been proven suitable for rice breeding programs. Instead, the incorporation of superior alleles of key starch synthesis genes, particularly Wx, remains the predominant strategy for breeding high-quality rice varieties [72]. For rice consumers in East Asian regions such as China and Japan, rice with low AC is widely popular due to its lower hardness and higher stickiness [12]. In current low-AC rice breeding programs, the superior Wx alleles primarily utilized are Wxb (AC around 16%) and Wxmp (AC about 10.5%), respectively aligning with conventional rice cultivars and soft rice varieties [3]. Rice carrying the Wxb allele generally exhibits inferior ECQ compared to soft rice varieties carrying the Wxmp allele, which have even lower AC [52]. However, grains carrying the Wxb allele maintain high transparency. Although soft rice varieties carrying Wxmp are more favored by Chinese consumers, their grains have low transparency and poor AQ due to the extremely low AC [14,27,52].
Grain transparency is a key component of rice AQ and is positively correlated with AC. High transparency forms the foundation of excellent AQ [52]. AC and RVA are key physicochemical indicators for assessing rice ECQ [73]. Early studies found that RVA profiles are significantly correlated with rice AC and texture (hardness and stickiness). Rice varieties with higher AC typically exhibit lower BDV and higher SBV [59]. For rice texture, changes in rice hardness and stickiness are significantly correlated with changes in BDV and SBV in the RVA profile. In rice production, high-quality varieties with good taste are generally recognized to have larger BDV and smaller SBV. Conversely, varieties with poor taste often have smaller BDV and larger SBV [60,61].
In this study, we developed a novel Wxb-d25 allele by further editing the S7 site in the core promoter region of the Wx gene, as previously reported [44]. Compared with the wild type (Wxb) and the reported Wxb-i1 control, Wxb-d25 showed no significant changes in agronomic traits, consistent with other reports on Wx gene editing [45]. Physicochemical quality analysis revealed that under summer planting conditions in Yangzhou, the AC of Wxb-d25 rice increased slightly compared with the wild type, while its grain transparency remained high. In contrast, under winter planting conditions in Lingshui, both the AC and transparency of Wxb-d25 rice significantly decreased. The AC approached that of the high-ECQ soft rice control NG46, but the transparency remained significantly higher than that of the latter. Further RVA analysis showed that under both planting conditions, Wxb-d25 rice exhibited characteristics similar to those of high-ECQ rice, with a significant increase in BDV and a significant decrease in SBV, suggesting that its rice texture and final ECQ might be improved under both conditions. We speculate that under Yangzhou summer planting conditions, the significant increase in AC of Wxb-d25 rice, yet exhibiting RVA characteristics similar to those of elite low-AC rice, might be due to the relatively small increase in AC combined with significant decreases in HPV, CPV, and PaT. These results indicate that Wxb-d25 has the potential to enhance rice ECQ by optimizing texture and maintaining high transparency under conventional summer conditions, building on the high AQ of Wxb rice. Under slightly lower winter planting conditions, it can moderately reduce rice AC and optimize rice texture, thereby bringing its ECQ closer to that of high-quality soft rice carrying the Wxmp allele while maintaining significantly higher grain transparency than the latter. Therefore, despite significant differences in AC variation under different planting conditions, Wxb-d25 has the potential to simultaneously improve both rice ECQ and AQ. Among these, the enhancement of rice AQ through grain transparency by Wxb-d25 is readily observable. However, the impact of Wxb-d25 on rice ECQ, given the complexity of its definition and the diverse dietary preferences of consumers, still requires further validation, such as through human sensory evaluation.
4.2. Revelation of a Novel Transcriptional Pattern of the Wx Gene
The advancement of gene editing technologies has facilitated the creation of superior Wx alleles devoid of foreign T-DNA insertions, making them highly applicable for rice breeding [74]. Currently, various gene editing approaches, including Wx gene knockout, promoter editing, upstream open reading frame (uORF) modification, single-base editing, and splice-site editing, have been successfully implemented in rice [13,44,45,48,75,76,77,78,79]. However, these studies have primarily focused on modifying key regulatory sites previously identified as influencing Wx gene expression, GBSSI activity, and splicing efficiency. While promoter editing has revealed novel regulatory elements within the Wx promoter, it has not been shown to induce significant alterations in the transcriptional pattern of the Wx gene [45,79].
In this study, the S7 site was identified within the core promoter region of the Wx gene. The 25-base deletion in this region partially disrupted the transcription initiator, not only affecting Wx gene expression but also altering its transcriptional pattern, leading to the emergence of a novel transcript, ONT.7395.1. Interestingly, despite substantial sequence modifications in ONT.7395.1, Western blot analysis confirmed that the GBSSI protein it encodes remained unchanged. While the original Wx-01 transcript persisted in Wxb-d25, its expression was significantly reduced due to the partial disruption of the transcription initiator. In contrast, ONT.7395.1, though expressed at a lower level, retained full splicing efficiency and was completely processed into mature Wx mRNA. The co-existence of both the original and novel transcripts resulted in an overall increase in Wx gene expression, leading to a higher GBSSI protein accumulation and subsequent changes in AC and physicochemical properties in Wxb-d25 rice under Yangzhou summer planting conditions. These results not only uncover a novel transcriptional pattern of the Wx gene but also provide new insights into promoter editing as a tool for modulating gene expression. However, since the Wxb-d25 allele only partially disrupts the transcription initiator, further research is needed to explore the effects of its complete disruption on Wx gene transcription.
The core promoter, a minimal DNA sequence essential for initiating transcription in eukaryotes, serves as a platform for the RNA polymerase system to recognize and bind to the gene promoter [80,81]. Typical eukaryotic gene core promoters usually contain cis-acting elements recognized and bound by general transcription factors (GTFs), such as the initiator and the TATA box [82,83,84,85]. In Wxb-d25, the deletion of 25 bases did not disrupt the TATA box on the Wx core promoter but partially disrupted the transcription initiator and may have affected other unknown cis-acting elements (Figure 1). This likely altered the binding efficiency of one or more GTFs, thereby changing the transcription of the downstream Wx gene. The AC phenotype of Wxb-d25 is highly temperature-sensitive, showing opposite trends in AC changes under Yangzhou and Lingshui planting conditions, yet maintaining an AC above 12%. This suggests that while GTFs can still recognize and bind to the modified Wx core promoter, their recognition or binding to the edited region is significantly less thermally stable. This highlights the need for further investigation into their regulatory mechanisms under varying environmental conditions. Due to the absence of RNA samples from Lingshui, we are currently unable to determine the specific temperature response of the two transcripts.
5. Conclusions
In conclusion, we successfully developed the novel Wxb-d25 allele by editing the core promoter of the Wx gene. Compared with the wild-type control and Wxb-i1, Wxb-d25 exhibited no significant changes in agronomic traits. Through comprehensive physicochemical quality analyses, we demonstrated that this allele exhibits a temperature-responsive pattern of AC regulation under different planting conditions and has the potential to synergistically enhance both the ECQ and AQ of rice by modulating AC, grain transparency, and texture. Moreover, we uncovered a unique transcriptional pattern in Wxb-d25, characterized by the co-regulation of Wx gene expression and rice AC through dual transcripts (Wx-01 and ONT.7395.1) under different planting conditions. These findings not only provide valuable insights for leveraging gene editing technology to precisely regulate target gene expression via promoter editing but also lay a solid theoretical foundation for using the Wxb-d25 allele to optimize endosperm starch composition and structure, ultimately improving rice grain quality.
Building on these research findings, the Wxb-d25 allele can be introduced into cultivated rice varieties to systematically evaluate its potential for breeding applications. Additionally, further editing of the core promoter of the Wx gene, including the creation of new Wx alleles with complete deletion of the initiator, can be carried out. This approach will not only aid in further elucidating the transcriptional regulation mechanisms of the Wx gene but also address the increasing demands for variations in rice AC and physicochemical quality driven by consumers’ diverse dietary preferences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14081330/s1, Figure S1: Morphology of rice plants and grains from Wxb-d25 and the wild-type; Table S1: Primers used in this study; Table S2: Main agronomic traits of Wx promoter-edited lines and the wild-type; Table S3: DSC parameters of rice flour; Table S4: The physiological parameters, methods, reagents and equipment involved in this study [3,5,9,27,52,56,58].
Author Contributions
Conceptualization, L.H. and Q.L. (Qiaoquan Liu); Formal analysis, X.F., Q.L. (Qianfeng Li) and C.Z.; Methodology, J.Y. (Jiali Yan), J.Y. (Jiawen Yu), H.S., L.Z. and Z.C.; Writing—original draft, J.Y. (Jiali Yan) and J.Y. (Jiawen Yu); Writing—review and editing, Q.L. (Qing Liu), L.H. and Q.L. (Qiaoquan Liu). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32201740, 32472071 and 32230074), the Programs from Government of Jiangsu Province (BK20220567, BE2021301 and PAPD), the Jiangsu Youth Science & Technology Talent Support Project (JSTJ-2023-XH028), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX23_1975 and KYCX21_3236).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gilbert, R.G. Starch molecular structure: The basis for an improved understanding of cooked rice texture. Carbohydr. Polym. 2018, 195, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhu, J.; Chen, S.; Fan, X.; Li, Q.; Lu, Y.; Wang, M.; Yu, H.; Yi, C.; Tang, S.; et al. Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant 2019, 12, 1157–1166. [Google Scholar] [CrossRef]
- Seung, D. Amylose in starch: Towards an understanding of biosynthesis, structure and function. New Phytol. 2020, 228, 1490–1504. [Google Scholar] [CrossRef]
- Tan, Y.; Yu, S.; Xing, Y.; Xu, C.; Zhang, Q.; Li, J. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef]
- Zaman, S.A.; Sarbini, S.R. The potential of resistant starch as a prebiotic. Crit. Rev. Biotechnol. 2016, 36, 578–584. [Google Scholar] [CrossRef]
- Mikami, I.; Aikawa, M.; Hirano, H.Y.; Sano, Y. Altered tissue-specific expression at the Wx gene of the opaque mutants in rice. Euphytica 1999, 105, 91–97. [Google Scholar] [CrossRef]
- Sasaki, T.; Yasui, T.; Matsuki, J. Effect of amylose content on gelatinization, retrogradation, and pasting properties of starches from waxy and nonwaxy wheat and their F1 seeds. Cereal Chem. 2000, 77, 58–63. [Google Scholar] [CrossRef]
- Tian, Z.; Qian, Q.; Liu, Q.; Yan, M.; Liu, X.; Yan, C.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef]
- Li, H.; Prakash, S.; Nicholson, T.M.; Fitzgerald, M.A.; Gilbert, R.G. The importance of amylose and amylopectin fine structure for textural properties of cooked rice grains. Food Chem. 2016, 196, 702–711. [Google Scholar] [CrossRef]
- Mestres, C.; Briffaz, A.; Valentin, D. Rice cooking and sensory quality. In Rice, 4th ed.; Bao, J., Ed.; AACC International Press: Saint Paul, MN, USA, 2019; pp. 385–426. [Google Scholar]
- Calingacion, M.; Laborte, A.; Nelson, A.; Resurreccion, A.; Concepcion, J.C.; Daygon, V.D.; Mumm, R.; Reinke, R.; Dipti, S.; Bassinello, P.Z.; et al. Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS ONE 2014, 9, e85106. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Zhang, Y.; He, L.; Li, C.; Liang, W.; Chen, T.; Zhao, Q.; Zhu, Z.; Zhao, L.; Zhao, C.; et al. Adjusting the amylose content of semi-glutinous japonica rice by genome editing of uORF6 in the Wx gene. Crop J. 2024, 12, 1806–1811. [Google Scholar] [CrossRef]
- Zhou, H.; Xia, D.; Zhao, D.; Li, Y.; Li, P.; Wu, B.; Gao, G.; Zhang, Q.; Wang, G.; Xiao, J.; et al. The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 2021, 63, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, C.; Luo, X.; Zhang, T.; Zhang, X.; Liu, P.; Yang, W.; Lei, Y.; Tang, S.; Kang, L.; et al. Fine mapping of the grain chalkiness quantitative trait locus qCGP6 reveals the involvement of Wx in grain chalkiness formation. J. Exp. Bot. 2023, 74, 3544–3559. [Google Scholar] [CrossRef]
- Tu, B.; Zhang, T.; Liu, P.; Yang, W.; Zheng, L.; Dai, Y.; Wang, H.; Lin, S.; Zhang, Z.; Zheng, X.; et al. The LCG1-OsBP5/OsEBP89-Wx module regulates the grain chalkiness and taste quality in rice. Plant Biotechnol. J. 2025, 23, 36–50. [Google Scholar] [CrossRef]
- Huang, L.; Sreenivasulu, N.; Liu, Q. Waxy editing: Old meets new. Trends Plant Sci. 2020, 25, 963–966. [Google Scholar] [CrossRef]
- Wang, A.; Jing, Y.; Cheng, Q.; Zhou, H.; Wang, L.; Gong, W.; Kou, L.; Liu, G.; Meng, X.; Chen, M.; et al. Loss of function of SSIIIa and SSIIIb coordinately confers high RS content in cooked rice. Proc. Natl. Acad. Sci. USA 2023, 120, e2220622120. [Google Scholar] [CrossRef]
- Shen, L.; Li, J.; Li, Y. Resistant starch formation in rice: Genetic regulation and beyond. Plant Commun. 2022, 3, 100329. [Google Scholar] [CrossRef]
- Deng, Z.; Liu, Y.; Gong, C.; Chen, B.; Wang, T. Waxy is an important factor for grain fissure resistance and head rice yield as revealed by a genome-wide association study. J. Exp. Bot. 2022, 73, 6942–6954. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, Z.; Liu, J.; Yu, L.; Wang, Z.; Zhu, S.; Shi, W.; Pan, C.; Wu, Y.; Li, Y.; et al. Genetic improvement of eating and cooking quality of rice cultivars in southern China. Plant Biotechnol. J. 2025, 23, 518–531. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Z.; Xing, Y.; Zheng, F.; Guo, X.; Zhang, W.; Hong, M. Nucleotide sequence of rice Waxy gene. Nucleic Acids Res. 1990, 18, 5898. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, Z.; Xing, Y.; Zhang, J.; Hong, M. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of Waxy gene in rice cultivars of intermediate amylose content. Plant J. 1998, 14, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.Y.; Eiguchi, M.; Sano, Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 1998, 15, 978–987. [Google Scholar] [CrossRef]
- Zeng, D.; Yan, M.; Wang, Y.; Liu, X.; Qian, Q.; Li, J. Du1, encoding a novel Prp1 protein, regulates starch biosynthesis through affecting the splicing of Wxb pre-mRNAs in rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 501–509. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, W.; Fu, Y.; Shan, Z.; Xu, J.; Wang, P.; Kong, F.; Jin, J.; Yan, H.; Ge, X.; et al. Du13 encodes a C2H2 zinc-finger protein that regulates Wxb pre-mRNA splicing and microRNA biogenesis in rice endosperm. Plant Biotechnol. J. 2022, 20, 1387–1401. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, T.; Shi, Y.; Lu, Y.; Li, Q.; Fan, X.; Huang, L.; Chen, L.; Song, X.; Liu, Q.; et al. Introgression of lac6/tl1/du13 improves the palatability of japonica rice. Crop J. 2024, 12, 1259–1265. [Google Scholar] [CrossRef]
- Zhao, G.; Xie, S.; Zong, S.; Wang, T.; Mao, C.; Shi, J.; Li, J. Mutation of TL1, encoding a novel C2H2 zinc finger protein, improves grains eating and cooking quality in rice. Theor. Appl. Genet. 2022, 135, 3531–3543. [Google Scholar] [CrossRef]
- Dung, L.V.; Mikami, I.; Amano, E.; Sano, Y. Study on the response of dull endosperm 2-2, du2-2, to two Wx alleles in rice. Breed. Sci. 2000, 50, 215–219. [Google Scholar] [CrossRef]
- Isshiki, M.; Takasaki, M.A.; Wong, H.L.; Satoh, H.; Shimamoto, K. Du3, a mRNA cap-binding protein gene, regulates amylose content in japonica rice seeds. Plant Biotechnol. 2008, 25, 483–487. [Google Scholar] [CrossRef]
- Takemoto-Kuno, Y.; Mitsueda, H.; Suzuki, K.; Hirabayashi, H.; Ideta, O.; Aoki, N.; Umemoto, T.; Ishii, T.; Ando, I.; Kato, H.; et al. qAC2, a novel QTL that interacts with Wx and controls the low amylose content in rice (Oryza sativa L.). Theor. Appl. Genet. 2015, 128, 563–573. [Google Scholar] [CrossRef]
- Igarashi, H.; Ito, H.; Shimada, T.; Kang, D.-J.; Hamada, S. A novel rice dull gene, LowAC1, encodes an RNA recognition motif protein affecting Waxyb pre-mRNA splicing. Plant Physiol. Biochem. 2021, 162, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Xie, D.; Wang, Z.; Hong, M. Interaction of rice bZIP protein REB with the 5′-upstream region of both rice SBE1 gene and Waxy gene. Chin. Sci. Bull. 2002, 47, 310–314. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Z.; Zhang, Q.; Meng, S.; Wei, C. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020, 184, 1775–1791. [Google Scholar] [CrossRef]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef]
- Xiong, Y.; Ren, Y.; Li, W.; Wu, F.; Yang, W.; Huang, X.; Yao, J. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Wang, L.; Li, L.; Liu, Y.; Chong, K.; Theißen, G.; Meng, Z. OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm. New Phytol. 2022, 234, 77–92. [Google Scholar] [CrossRef]
- Wang, J.-D.; Wang, J.; Huang, L.-C.; Kan, L.-J.; Wang, C.-X.; Xiong, M.; Zhou, P.; Zhou, L.-H.; Chen, C.; Zhao, D.-S.; et al. ABA-mediated regulation of rice grain quality and seed dormancy via the NF-YB1-SLRL2-bHLH144 Module. Nat. Commun. 2024, 15, 4493. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, H.; Feng, M.; Zhu, Y. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol. J. 2018, 16, 18–26. [Google Scholar] [CrossRef]
- Fu, F.; Xue, H. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Jiao, G.; Yu, J.; Zhao, R.; Lu, A.; Zhou, W.; Cao, N.; Wu, J.; Hu, S.; et al. The elite eating quality alleles Wx and ALK are regulated by OsDOF18 and coordinately improve head rice yield. Plant Biotechnol. J. 2024, 22, 1582–1595. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Mawia, A.M.; Wei, X.; Cao, R.; Jiao, G.; Wu, Y.; Zhang, J.; Xie, L.; Sheng, Z.; et al. A novel transcription factor OsMYB73 affects grain size and chalkiness by regulating endosperm storage substances’ accumulation-mediated auxin biosynthesis signalling pathway in rice. Plant Biotechnol. J. 2025, 23, 1021–1038. [Google Scholar] [CrossRef] [PubMed]
- Anacleto, R.; Badoni, S.; Parween, S.; Butardo, V.M., Jr.; Misra, G.; Cuevas, R.P.; Kuhlmann, M.; Trinidad, T.P.; Mallillin, A.C.; Acuin, C.; et al. Integrating a genome-wide association study with a large-scale transcriptome analysis to predict genetic regions influencing the glycaemic index and texture in rice. Plant Biotechnol. J. 2019, 17, 1261–1275. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, Q.; Zhang, C.; Chu, R.; Gu, Z.; Tan, H.; Zhao, D.; Fan, X.; Liu, Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 2164–2166. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Liu, T.; Ma, X.; Wang, B.; Zheng, Z.; Zhang, Y.; Xie, X.; Yang, B.; Zhao, Z.; Zhu, Q.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Zhong, Y.; Blennow, A.; Kofoed-Enevoldsen, O.; Jiang, D.; Hebelstrup, K.H. Protein Targeting to Starch 1 is essential for starchy endosperm development in barley. J. Exp. Bot. 2018, 70, 485–496. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Bull, S.E.; Seung, D.; Chanez, C.; Mehta, D.; Kuon, J.E.; Truernit, E.; Hochmuth, A.; Zurkirchen, I.; Zeeman, S.C.; Gruissem, W.; et al. Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 2018, 4, eaat6086. [Google Scholar] [CrossRef]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, R.; Song, G.; Gao, J.; Li, W.; Han, X.; Chen, M.; Li, Y.; Li, G. Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol. 2018, 18, 302. [Google Scholar] [CrossRef]
- Li, Q.F.; Huang, L.C.; Chu, R.; Li, J.; Jiang, M.Y.; Zhang, C.Q.; Fan, X.L.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. Down-regulation of SSSII-2 gene expression results in novel low-amylose rice with soft, transparent grains. J. Agric. Food Chem. 2018, 66, 9750–9760. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shen, L.; Fu, Y.; Yan, C.; Wang, K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genom. 2015, 42, 703–706. [Google Scholar] [CrossRef]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, L.; Feng, L.; Jiang, J.; Huang, L.; Liu, Q.; Zhang, Y.; Zhang, C.; Liu, Q. Deciphering the role of Waxy gene mutations in enhancing rice grain quality. Foods 2024, 13, 1624. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.J. Comparison of protease digestion at neutral pH with alkaline steeping method for rice starch isolation. Cereal Chem. 2001, 78, 690–692. [Google Scholar] [CrossRef]
- Huang, L.; Gu, Z.; Chen, Z.; Yu, J.; Chu, R.; Tan, H.; Zhao, D.; Fan, X.; Zhang, C.; Li, Q.; et al. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 2021, 106, 419–432. [Google Scholar] [CrossRef]
- Xiang, X.; Kang, C.; Xu, S.; Yang, B. Combined effects of Wx and SSIIa haplotypes on rice starch physicochemical properties. J. Sci. Food Agric. 2017, 97, 1229–1234. [Google Scholar] [CrossRef]
- Shu, Q.; Wu, D.; Xia, Y.; Gao, M.; McClung, A. Relationship between RVA profile character and eating quality in Oryza sativa L. Sci. Agric. Sin. 1998, 34, 25–29. [Google Scholar]
- Jia, L.; Ding, X.; Wang, P.; Deng, X. RVA profile characteristics and correlation with the physical/chemical quality. Acta Agron. Sin. 2008, 34, 790–794. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, L.; Liu, G.; Meng, X.; Jing, Y.; Shu, X.; Kong, X.; Sun, J.; Yu, H.; Smith, S.M.; et al. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 12844–12849. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, X.; Jiao, G.; Chen, W.; Wu, Y.; Sheng, Z.; Hu, S.; Xie, L.; Wang, J.; Tang, S.; et al. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. J. Integr. Plant Biol. 2020, 62, 948–966. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Jiao, G.; Cao, R.; Li, S.; Zhao, S.; Duan, Y.; Ma, L.; Li, X.; Lu, F.; Wang, H.; et al. OsLESV and OsESV1 promote transitory and storage starch biosynthesis to determine rice grain quality and yield. Plant Commun. 2024, 5, 100893. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, W.; Wang, Y.; Jin, J.; Xu, H.; Fu, Y.; Shan, Z.; Wang, X.; Teng, X.; Li, X.; et al. Rice LIKE EARLY STARVATION1 cooperates with FLOURY ENDOSPERM6 to modulate starch biosynthesis and endosperm development. Plant Cell 2024, 36, 1892–1912. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef]
- Yan, M.; Pan, T.; Zhu, Y.; Jiang, X.; Yu, M.; Wang, R.; Zhang, F.; Luo, S.; Bao, X.; Chen, Y.; et al. FLOURY ENDOSPERM20 encoding SHMT4 is required for rice endosperm development. Plant Biotechnol. J. 2022, 20, 1438–1440. [Google Scholar] [CrossRef]
- Wu, H.; Ren, Y.; Dong, H.; Xie, C.; Zhao, L.; Wang, X.; Zhang, F.; Zhang, B.; Jiang, X.; Huang, Y.; et al. FLOURY ENDOSPERM24, a heat shock protein 101 (HSP101), is required for starch biosynthesis and endosperm development in rice. New Phytol. 2024, 242, 2635–2651. [Google Scholar] [CrossRef]
- Wu, B.; Yun, P.; Zhou, H.; Xia, D.; Gu, Y.; Li, P.; Yao, J.; Zhou, Z.; Chen, J.; Liu, R.; et al. Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 2022, 34, 1912–1932. [Google Scholar] [CrossRef]
- Qin, P.; Zhang, G.; Hu, B.; Wu, J.; Chen, W.; Ren, Z.; Liu, Y.; Xie, J.; Yuan, H.; Tu, B.; et al. Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 2021, 7, eabc8873. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, L.; Gao, Q.; Wang, J.; Li, X.; Wang, H.; Liu, Y.; Lin, H.; Liu, J.; Wang, X.; et al. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 2021, 53, 906–915. [Google Scholar] [CrossRef]
- Fujita, N.; Miura, S.; Crofts, N. Effects of various allelic combinations of starch biosynthetic genes on the properties of endosperm sarch in rice. Rice 2022, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Tan, H.; Zhang, C.; Li, Q.; Liu, Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 100237. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J.K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 2018, 60, 369–375. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Q.; Li, X.; Wang, F.; Chen, Z.; Wang, J.; Li, W.; Fan, F.; Tao, Y.; Jiang, Y.; et al. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 2021, 19, 11–13. [Google Scholar] [CrossRef]
- Huang, X.; Su, F.; Huang, S.; Mei, F.; Niu, X.; Ma, C.; Zhang, H.; Zhu, X.; Zhu, J.-K.; Zhang, J. Novel Wx alleles generated by base editing for improvement of rice grain quality. J. Integr. Plant Biol. 2021, 63, 1632–1638. [Google Scholar] [CrossRef]
- Liu, X.; Ding, Q.; Wang, W.; Pan, Y.; Tan, C.; Qiu, Y.; Chen, Y.; Li, H.; Li, Y.; Ye, N.; et al. Targeted deletion of the first intron of the Wxb allele via CRISPR/Cas9 significantly increases grain amylose content in rice. Rice 2022, 15, 1. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, G.; Zhao, Y.; Zhang, R.; Tang, X.; Li, L.; Jia, X.; Guo, Y.; Wu, Y.; Han, Y.; et al. An efficient CRISPR–Cas12a promoter editing system for crop improvement. Nat. Plants 2023, 9, 588–604. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000, 34, 77–137. [Google Scholar] [CrossRef]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Juven, G.T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Sandelin, A.; Carninci, P.; Lenhard, B.; Ponjavic, J.; Hayashizaki, Y.; Hume, D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet. 2007, 8, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.M.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).