Abstract
HMGR is a crucial enzyme in the biosynthesis of terpenoids. We cloned FaHMGR and found that FaHMGR expression in fruit was significantly higher than other tissues, especially during the coloring stage. Suppression of FaHMGR (FaHMGRR) promoted coloration by increasing anthocyanin content and produced five new components. In contrast, FaHMGR overexpression (FaHMGROE) downregulated most anthocyanin genes and reduced hexanoic acid methyl ester and linalool contents, thereby inhibiting coloring. Transcriptomic and metabolomic analyses showed that DEGs in HMGROE vs. HMGRC (pCAMBIA1302 empty vector transformant serving as a control) were significantly enriched in phenylpropanoid biosynthesis pathway and pathways related to terpenoid metabolism and MeJA, suggesting MeJA as a potential mediator of HMGR’s influence on terpenoid pathways. Additionally, DEGs in HMGRR vs. HMGRC were enriched in anthocyanin biosynthesis, particularly keracyanin and pelargonidin, which may explain the promoted coloration observed in HMGRR. WGCNA analysis identified five module genes with distinct expression patterns in HMGRR and HMGROE, including ERF118 and WRKY12, which may impact fruit quality by regulating HMGR activity.
1. Introduction
As the most abundant class of secondary metabolites in plants, terpenoids play an important role in plant growth and development, environmental adaptation, and fruit quality attributes. As a globally popular berry crop, strawberries (Fragaria ananassa (Weston) Duch.) are well-recognized for their distinctive flavor and nutritional benefits. In the French strawberry cultivar Falandi, nine sesquiterpenes and three triterpenes have been identified. Terpenes are important components of volatile organic compounds (VOCs). Although they constitute only 0.001–0.01% of strawberry weight, these compounds are essential to the fruit’s favorable flavor, with small changes greatly impacting taste [1]. Strawberry fruits produce a favorable aroma, with terpenoids significantly contributing to their aroma profile. Key terpenoids in strawberries include linalool, nerol, menthol, α-pinene, β-myrcene, α-terpineol, and β-phellandrene, which vary across varieties and developmental stages [2]. Among them, linalool and nerol are two major aroma components in strawberries; however, their contents differ significantly between wild (Fragaria × vesca) and cultivated strawberries (Fragaria × ananassa). The terpenoid synthase gene FaNES1, which is highly expressed in cultivated strawberries, catalyzes the conversion of geranyl diphosphate (GPP) and farnesyl diphosphate (FPP) into linalool and nerol [3]. A study by Fan et al. [4] identified grape flavor genes and their regulatory elements using multi-omics. This research implies that the distal region of strawberry Chromosome 3C may be related to the production of multiple terpenes, with FaNES1 significantly correlated with the biosynthesis of linalool, β-myrcene, α-pinene, (E)-β-farnesene, and nerol.
Studies on the disease resistance mechanisms in strawberries suggest terpenes as one of the three crucial metabolites involved in strawberry defense [5]. Xu et al. [6] demonstrated that UV-C exposure induces monoterpene production in strawberry leaves, which interacts with abscisic acid (ABA) to enhance pathogen resistance. Additionally, terpinen-4-ol improves resistance against Botrytis cinerea and soft rot by activating the phenylpropanoid metabolic pathway in strawberry fruits [7]. Triterpene accumulation has also been linked to differential resistance across strawberry varieties exposed to nanoplastics [8]. Moreover, rapid induction of terpenoid synthesis may enhance anthracnose resistance in certain wild strawberry species [9].
Terpenoids are primarily synthesized in plants through two pathways: the mevalonate (MVA) pathway in the cytoplasm and the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway in the plastids [10]. In the MVA pathway, 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) is the first rate-limiting enzyme and acts as a central regulator in the biosynthesis of cytoplasmic terpenoids [11]. Studies have demonstrated a positive correlation between HMGR activity and the production of diverse terpenoids across virtually all plant species [12,13,14,15]. Upregulation of HMGR and TPS genes has been shown to enhance terpene levels in roses [16]. Terpenoids influenced by HMGR include beneficial compounds such as sterols, rubber, resin, and saponins [17,18,19,20], which improve plant adaptability to adverse environmental conditions [21,22]. HMGR also directly affects the synthesis of sesquiterpene and triterpene aromas and accelerates the accumulation of tetraterpenoid carotenoids in Escherichia coli (E. coli). [23,24,25]. In addition, HMGR also participates in the regulation of anthocyanin accumulation in plants by affecting hormone accumulation and signal transduction. Downregulation of the HMGR gene can positively induce the expression of a variety of hormones and affect the accumulation of anthocyanins [19,24,26]. However, the association between HMGR and quality traits, such as fruit aroma and color, remains poorly understood [26], with no reported studies in strawberries. Given the significance of HMGR in strawberry fruit development and ripening, we cloned FaHMGR in strawberries and examined its expression across various tissues and developmental stages. Moreover, by integrating transcriptomic and metabolomic analyses with transient gene expression, we assessed the effect of FaHMGR on strawberry fruit quality attributes.
2. Materials and Methods
2.1. Plant Materials
Experiments were conducted at the Yangdu Experimental Vineyard, the Zhejiang Academy of Agricultural Science (E 120°24′, N 30°26′), Jiaxing, China. The strawberry plants used in this study were Fragaria × ananassa cv. Benihoppe. Young roots, stems, leaves, petals, and pistils were collected during flowering period, and the fruits were collected at different developmental stages, individually rapidly frozen in liquid nitrogen, and stored at −80 °C for the determination of HMGR gene expression.
2.2. RNA Extraction, cDNA Synthesis, and qRT-PCR
Total RNA was extracted from 0.1 g of each tissue material using the cetyltrimethylammonium bromide (CTAB) method with the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). cDNA was synthesized using a HifairII® 1st Strand cDNA Synthesis SuperMix (Yeasen, Shanghai, China). The qRT-PCR reaction consisted of 5 μL SYBR Premix Ex Taq™ (Takara, Beijing, China), 0.3 μL of each primer (10 μM), 2 μL cDNA, and 2.4 μL RNase-free water, for a total volume of 10 μL. Reactions were performed on a LightCycler 1.5 instrument (Roche, Baden, Germany), starting with the preliminary step at 95 °C for 30 s, followed by 35 cycles of 95 °C for 5 s and 58 °C for 35 s [27]. The relative gene expression was calculated using the 2−ΔCt method [10]. Experiments were conducted with three biological and three technical replicates. Gene-specific primers are listed in Table S1.
2.3. Determination of Hormone Contents
The contents of the hormones auxin indole-3-acetic acid (IAA), ABA, gibberellin (GA3), zeatin riboside (ZR), and brassinosteroids (BRs) were determined using an enzyme-linked immunosorbent assay (ELISA), following the instruction by Wang et al. [28]. For hormone extraction, 0.5 g of strawberry fruits were ground in an ice bath with 2 mL of extract solution (80% methanol containing 1 mmol·L−1 BHT). The resulting mixture was diluted to 10 mL, incubated at 4 °C for 4 h, and then centrifuged at 3500 rpm for 8 min. The supernatant was collected and extracted by C-18 solid phase extraction. The eluted sample was transferred to a 10 mL centrifuge tube and freeze-dried before being dissolved in 2 mL of buffer.
2.4. Transient Expression Levels of FaHMGR and FaHMGRi in Strawberries
Full-length coding sequences of FaHMGR and FaHMGRi (see Table S2) were amplified from the cDNA of ‘Benihoppe’ strawberries and cloned into the pCAMBIA1302 vector carrying a 35S promoter (abbreviated as p35S) to generate constructs p35S-FaHMGR-GFP and p35S-FaHMGRi-GFP. For transient expression, these two plasmids and pCAMBIA1302 empty vector were individually mobilized into Agrobacteria (strain EHA105), which were then used to transform strawberries at the green stage [29], generating the transgenic lines FaHMGROE and FaHMGRR, with the pCAMBIA1302 empty vector transformant serving as a control (CK, FaHMGRC). Following confirmation of the expression levels of FaHMGR-GFP and FaHMGRi-GFP by qRT-PCR (gene specific primers are listed in Table S2), the anthocyanin content and aroma components were determined.
2.5. Determination of Anthocyanin and Chlorophyll Content
Chlorophyll content was determined using spectrophotometry. Total anthocyanins were extracted using a methanol–HCl method. Samples of 0.1 g were submerged and incubated overnight in 5 mL of methanol containing 0.1% (v/v) HCl in the dark at room temperature. Anthocyanin content was measured using the PH differential method, with absorbance at 520 and 700 nm recorded using a UV-2550 spectrophotometer (Shimadzu, Kyoto, Japan). The anthocyanin components were determined using liquid chromatography-mass spectrometer (LC-MS), and the peak area was measured to quantify each component [30].
2.6. Determination of the Aroma Components
Aroma components in transgenic strawberries FaHMGROE, FaHMGRR, and FaHMGRC were analyzed using gas chromatography-mass spectrometry (GC-MS). According to the method described by Zheng et al. [30], samples (3 g) were ground, transferred to a 20 mL headspace bottle, and mixed with three milliliters of saturated NaCl solution and 2 μL 3-nonanone (internal standard). The samples were analyzed with a gas chromatograph (TRACE 1310, Thermo Scientific, Shanghai, China) coupled to a triple quadrupole mass spectrometer (TSQ 9000, Thermo Scientific, Shanghai, China). The column temperature was programmed as follows: the initial temperature was set at 50 °C for 6 min and then increased to 250 °C at a rate of 6 °C min−1, and held for 3 min. The MS conditions were as follows: EI mode at voltage 70 eV; ion source temperature at 230 °C; a scanning rate of 2.88 scan·s−1; and a detection range of 29–540 m·z−1, with helium as the carrier gas at a flow rate of 1.0 mL min−1.
2.7. Transcriptomic Analysis
Library preparation and transcriptome sequencing of FaHMGROE, FaHMGRR, and FaHMGRC strawberries were conducted by the Beijing Novogene Technology Corporation (Beijing, China). Differentially expressed gene (DEG) analysis was performed using the DESeq R package (1.18.0) with the following criteria: false discovery rate (FDR) < 0.05, |fold change| ≥ 2, and adjusted p-value < 0.05 [31]. Principal component analysis (PCA) was performed with the gmodels R package. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed with the GOseq R 3.12 software package and KOBAS 3.0 software, respectively [32]. A weighted gene correlation network (WGCNA) was constructed based on a hierarchical clustering tree. All treatments included three biological replicates. To validate transcriptomic data, ten DEGs were selected for confirmation via qRT-PCR.
2.8. Metabolomic Profile Detection and Analysis
For metabolome sequencing, fruits of strawberries FaHMGROE, FaHMGRR, and FaHMGRC were individually grounded in liquid nitrogen, and each homogenate was resuspended in pre-chilled 80% methanol and 0.1% formic acid and well vortexed before incubation. The samples were then centrifuged, diluted, and filtered before injection into the LC-MS/MS system. LC-MS/MS analysis was performed using a Vanquish UHPLC system (Thermo Scientific, Shanghai, China) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Raw data were processed using Compound Finder 3.0 (CD 3.0, Thermo Fisher). Peaks were matched with the mzCloud (https://www.mzcloud.org/, accessed on 20 March 2022) and ChemSpider (http://www.chemspider.com/, accessed on 20 March 2022) databases to obtain accurate qualitative results, with areas measured for quantitative analysis. The statistical analyses were conducted using R (R version R-3.4.3), Python (Python 2.7.6 version), and CentOS (CentOS release 6.6). Positive ion mode (POS) and negative ion mode (NEG) were both used to detect metabolites. The quality control (QC) sample was included to assess system stability, and a blank sample was used to eliminate background ions.
2.9. Statistical Analysis
All data (with at least three replications, n = 3) are presented as means with standard errors of the means (SEMs). Mean ± SEM values for each treatment were calculated using Microsoft Excel (Microsoft Corporation, Albuquerque, NM, USA). Statistical analysis of variance (ANOVA) and Duncan’s multiple range test (p < 0.05) were performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Figures were generated using Origin Pro 9 (Origin Inc., Northampton, MA, USA).
3. Results
3.1. Expression Characteristics of FaHMGR
To clarify the patterns of FaHMGR accumulation in strawberries, we examined its expression across various tissues and observed significant variation, with the highest level found in fruit tissue, followed by root, flower, stem, pistil, and leaf in descending order (Figure 1A). During fruit development, FaHMGR expression gradually decreased during the fruit enlargement phase (fruit 1 to fruit 3), then began to increase steadily during the fruit whitening phase (fruit 4 to fruit 6), peaking at the onset of fruit coloring (fruit7). The transcription of FaHMGR then decreased as the fruits turned red, reaching its lowest level at fruit mature stage (fruit 10).
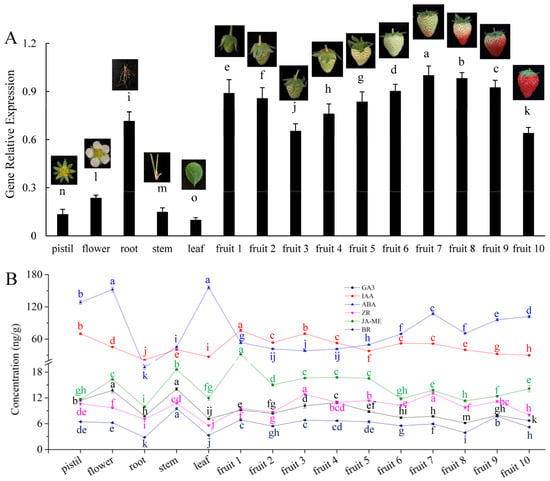
Figure 1.
Expression of FaHMGR (A) and hormone levels (B) in various tissues and fruits at different developmental stages. Different letters represent significant differences between groups.
Analyses of six hormones across different tissues and at various stages of fruit development revealed that IAA and ABA were significantly more abundant than the other hormones, including GA3, ZR, JA-ME, and BRs. JA-ME levels were slightly lower than those of IAA and ABA, while BR levels were the lowest across examined tissues (Figure 1B). Notably, hormone levels in roots were generally lower than in other tissues, contrasting with the high expression of HMGR in this tissue. At the onset of fruit coloring (fruit 7), levels of GA3, ABA, ZR, JA-ME, and BRs all increased to varying extents and then decreased as fruit coloration progressed (fruit 8).
3.2. Effects of Transient FaHMGR and FaHMGRi Expression on Strawberry Coloration and Aroma
Overexpression of HMGR in strawberries inhibited fruit coloration and reduced anthocyanin content, where opposite phenomena were observed when HMGR was suppressed, with a 3.33 mg·g−1 increase in anthocyanin content compared to the control (Figure 2A). Analysis of anthocyanin components revealed that cyanidin 3-O-(6″-malonyl-3″-glucosyl-glucoside), cyanidin 3-O-glucoside, cyanidin 3-O-rutinoside, cyanidin 3-O-sophoroside, and delphinidin 3-O-rutinoside were present in FaHMGRR strawberries but absent in FaHMGRC and FaHMGROE (Figure 2B). Additionally, the concentrations of other anthocyanin components in FaHMGRR were significantly higher than in both FaHMGRC and FaHMGROE strawberries. Cyanidin 3-O-(6″-p-coumaroyl-glucoside) was only detected in FaHMGROE. Furthermore, the content of ABA in FaHMGRR was significantly higher than in both FaHMGROE and FaHMGRC (Figure 2C). Conversely, the concentrations of BRs, ZR, and GA3 were significantly elevated in FaHMGROE plants compared to FaHMGRR strawberries.
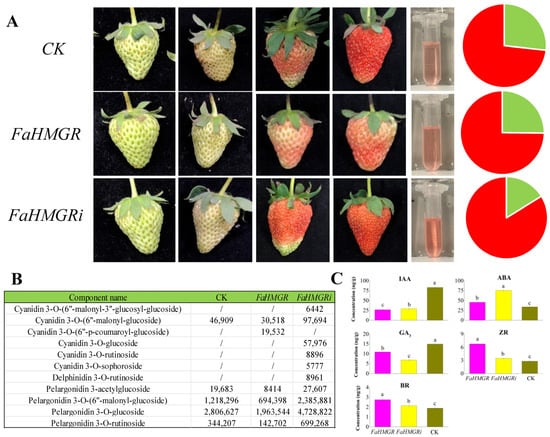
Figure 2.
Influence of FaHMGR expression on strawberry fruit development. (A) Fruit development in strawberries transiently overexpressing FaHMGR (FaHMGROE) and FaHMGRi (FaHMGRR), using the pCAMBIA1302 empty vector transformant as a control (CK). The pie chart illustrates the relative chlorophyll (green) and anthocyanin (red) contents in strawberries. (B) Composition of anthocyanins in strawberries with varying FaHMGR levels. Data in the table represent the peak area. (C) Impact of FaHMGR overexpression and suppression on phytohormone levels in strawberries treated as in (A). Different letters represent significant differences between groups.
The type and content of aroma compounds are crucial in determining strawberry fruit flavors and distinguishing strawberry varieties. In this study, a total of 93, 84, and 120 aroma compounds—including esters, ketones, aldehydes, alcohols, terpenes, and minor amounts of furans, phenols, and acids—were detected in FaHMGROE, FaHMGRR, and FaHMGRC strawberries, respectively (Figure 3, Table 1). Esters were found to be the dominant aroma components in wild-type strawberries, with hexanoic acid and its methyl ester accounting for 18.96%. Their concentrations decreased in FaHMGROE and FaHMGRR strawberries. Another major component, (Z)-hex-2-enyl acetate, decreased in FaHMGRR strawberries but remained unchanged in FaHMGROE. Linalool, the primary terpene aroma in strawberries, had a content of 0.62% in CK, which decreased to 0.13% and 0.37% in FaHMGROE and FaHMGRR strawberries, respectively. Additionally, other detected terpenes included D-limonene (0.16%) in CK and α-pinene (0.03%), β-pinene (0.05%), α-terpineol (0.01%), and caryophyllene (0.01%) in FaHMGRR strawberries.
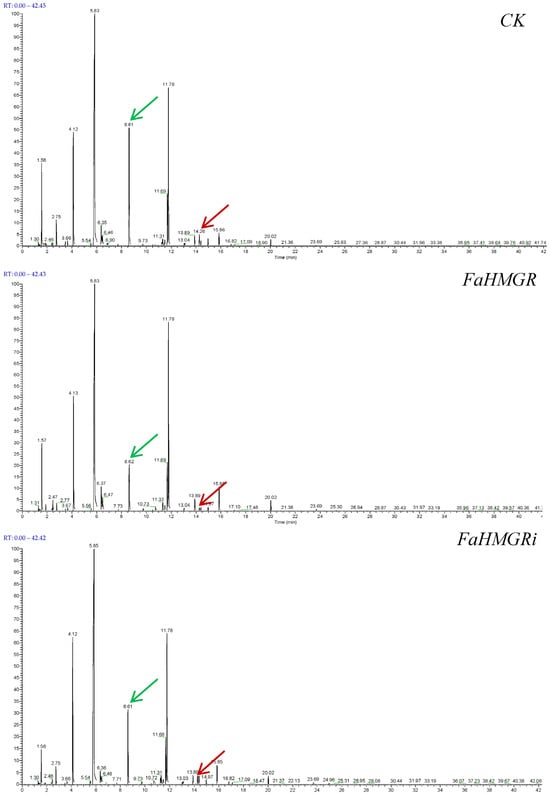
Figure 3.
Impact of FaHMGR overexpression and suppression on fruit aroma components in strawberries. The red arrow indicates linalool, which decreased after both FaHMGR overexpression and suppression; and the green indicates methyl hexanoate, another main aroma component, which also showed a decrease in response to changes in FaHMGR expression. (The specific data are presented in Table 1).

Table 1.
Aroma components in the fruits of FaHMGR overexpressing and suppressing strawberry.
The expression levels of genes involved in anthocyanin and aroma formation were assessed in FaHMGROE, FaHMGRR, and FaHMGRC strawberries. The results showed that the expression of key anthocyanin metabolic pathway genes, including FaC4H, FaCHI, FaCHS, FaF3H, FaANS, FaUFGT, FaMYB10, FabHLH143, FaWD40, and FaPAL2, was lowest in FaHMGROE strawberries, whereas Fa4CL, Fa4CL2, and FaDFR increased. In contrast, in FaHMGRR fruits, levels of FaC4H, FaCHS, FaANS, FaUFGT, FaMYB10, FabHLH143, FaWD40, and FaPAL2 were higher than in both CK and FaHMGROE strawberries (Figure 4). This result suggests a negative correlation between FaHMGR level and the anthocyanin accumulation. For aroma synthesis genes, FaCNL, FaPIN, and FaNCED3 exhibited the highest expression in FaHMGROE fruits, while FaDAHPS1, FaDAHPS2, FaNES1, FaOMT, FaCHP1, FaAAT, FaQR, and FaNCED1 showed the lowest expression. When FaHMGR was inhibited, genes FaNES1, FaOMT, FaCHP1, FaAAT, FaQR, and FaNCED1 were significantly upregulated.
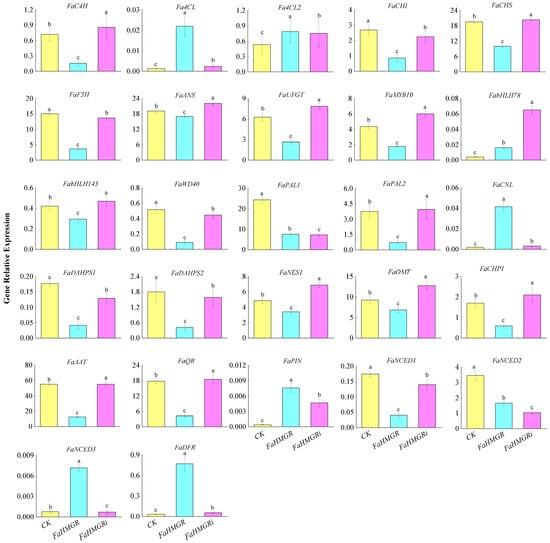
Figure 4.
Effects of changes in FaHMGR levels on the expression of genes associated with anthocyanin and aroma metabolic pathways following transient overexpression of FaHMGR or suppression via FaHMGRi in strawberries. The pCAMBIA1302 empty vector was used as a control. Different letters represent significant differences between groups.
3.3. Manipulating FaHMGR Expression Caused Changes in Strawberry Transcription and Metabolism
To determine the transcriptional and metabolic regulatory effects of FaHMGR on strawberries, RNA sequencing (RNA-Seq) and metabolic profiling were performed on the strawberries with transient overexpression or suppression of FaHMGR (HMGROE, HMGRR), with the empty vector transformed plants as the control (HMGRC). RNA sequencing generated approximately 6.6 GB of data and an average of 44,377,805 clean reads (Table S3). Mapping to the strawberry genome database yielded a high coverage of 94.77%, suggesting effective detection of transcriptional changes due to overexpression alteration of FaHMGR in strawberries. To validate RNA-Seq reliability, 10 DEGs were randomly selected for qRT-PCR, which revealed consistent expression patterns of these genes with the sequencing results (Figure S1).
The PCA of the transcriptome data (Figure 5A) revealed that all samples were positioned along the positive axis of PC1, which explained 32.95% of the variance, indicating a strong association among HMGROE, HMGRR, and HMGRC. HMGROE and HMGRR overlapped along PC1, while minimal differences were observed among the three samples along PC2 (17.74%), with HMGRC positioned in the positive axis. As shown in Figure 5B, the correlation among triplicates for each treatment sample was high, exceeding 0.923. T1 showed slightly lower correlations (below 0.97) with the other two replicates. A total of 3811 DEGs were identified across all samples, with 2713 unique to the comparison pair HMGRR vs. HMGRC and 236 unique to HMGROE vs. HMGRC. There were more downregulated genes than upregulated ones in both comparison pairs (Figure 5C). Of these DEGs, 842 were common between HMGRR vs. HMGRC and HMGROE vs. HMGRC. All DEGs showed four distinct expression patterns and were thereby clustered into four groups comprising 1705, 2049, 54, and 3 genes. Most genes exhibited no significant differences between HMGROE and HMGRR but differed from HMGRC. In contrast, genes in cluster 4, including cold-regulated 413 plasma membrane protein 1 (AtCOR413-PM1), were expressed significantly differently between HMGROE and HMGRR (Figure 5D).
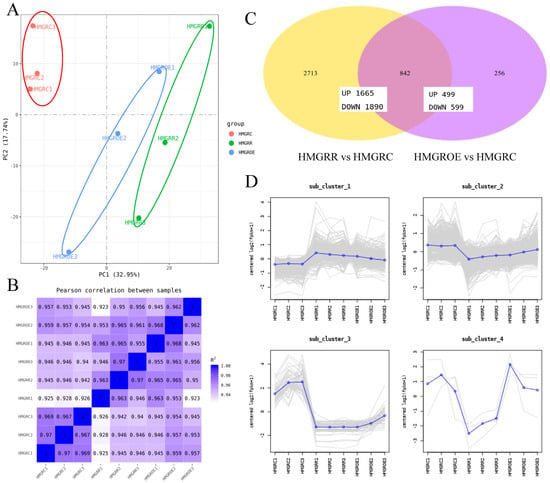
Figure 5.
Differentially expressed genes (DEGs) in strawberries after the overexpression and suppression of FaHMGR in strawberry fruits. (A). Principal component analysis (PCA) scatter plot of transcriptomic profiles from different samples. (B). Correlation coefficient graph showing the relationships among all samples, demonstrating the degree of similarity in gene expression profiles. (C). Venn diagram illustrating the number of DEGs in comparison pairs HMGROE vs. HMGRC and HMGRR vs. HMGRC. (D). Expression patterns of DEGs. The Y-axis represents the p-value and the X-axis represents samples.
GO and KEGG enrichment analyses of the DEGs in HMGRR vs. HMGRC and HMGROE vs. HMGRC were performed to identify the functional roles of the DEGs. GO analysis revealed that these DEGs in both the HMGROE vs. HMGRC and HMGRR vs. HMGRC comparisons showed significant enrichment in the item nutrient reservoir activity (Figure 6A, Table S4). KEGG analysis showed significant enrichment of the DEGs in the phenylpropanoid biosynthesis pathway for HMGROE vs. HMGRC and in the ribosome, DNA replication, and pyruvate metabolism pathways for the HMGRR vs. HMGRC comparison (Figure 6B, Table S4).
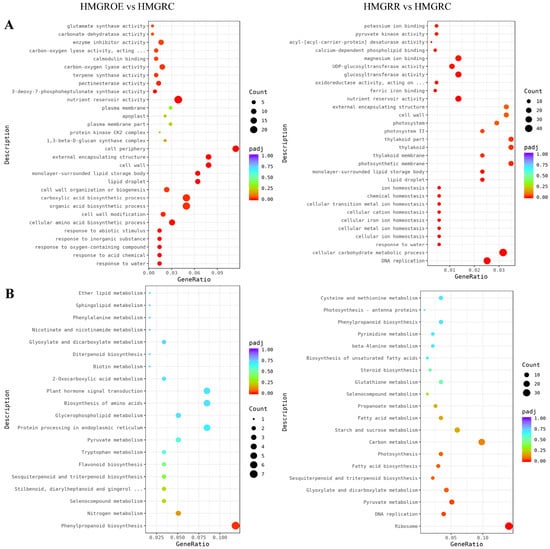
Figure 6.
Gene Ontology (GO) categories (A) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (B) of significantly enriched DEGs in the comparisons HMGRR vs. HMGRC and HMGROE vs. HMGRC.
Furthermore, WGCNA divided the DEGs into 63 modules (Figure S2). Module–sample relationship analysis identified five modules indicated in Table S5 by the colors dark magenta, honeydew, tan, violet, and white, containing 82, 40, 269, 88, and 102 genes, respectively (Table S5). Genes in each module displayed distinct expression patterns in HMGRR and HMGROE strawberries (Table S5). In the violet module, gene expression was low in HMGRR and high in HMGROE, while the other four modules exhibited the opposite pattern. Notably, two abscisic acid receptors, PYL11 and PYL9, were identified in the dark magenta and honeydew modules. The tan, violet, and white modules contained several enzymes involved in sugar and acid synthesis pathways. Additionally, transcription factors ERF118 and WRKY12 were found in the white module, suggesting that they may play an important role in FaHMGR-mediated effects on fruit quality.
By integrating database comparisons, we conducted qualitative and quantitative analyses of metabolites, identifying a total of 501 components in positive ion mode and 312 in negative ion mode (Table S6). Principal component analysis (PCA) was employed to evaluate the overall metabolic differences between samples (Figure 7A,D), revealing that HMGROE was more closely related to HMGRC than to HMGRR. In positive ion mode, 63 differential metabolites were common to both HMGROE vs. HMGRC and HMGRR vs. HMGRC. Additionally, 70 and 128 unique metabolites were identified in each pairwise comparison, respectively (Figure 7B,E). In the negative ion mode, 30 differential metabolites were shared in the two pairs, with 34 and 70 metabolites uniquely present in each. Further analysis showed that in HMGROE vs. HMGRC, 20 differential metabolites were upregulated and 52 were downregulated in negative ion mode, while 133 were upregulated and 131 were downregulated in positive ion mode. For HMGRR vs. HMGRC, 116 differential metabolites were upregulated and 115 were downregulated in negative ion mode, while 191 were upregulated and 189 were downregulated in positive ion mode. KEGG pathway analysis indicated that most metabolites mapped to the item “Global and overview maps”, with four specific substances associated with the “Metabolism of terpenoids and polyketides” pathway in both negative and positive ion modes (Figure 7C,F).
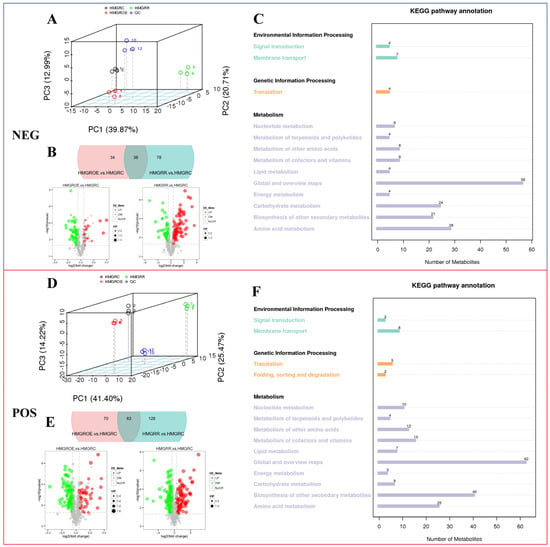
Figure 7.
Differentially accumulated metabolites (DAMs) in negative and positive ion modes (NEG, POS) in strawberries after the overexpression and suppression of FaHMGR in fruits. (A,D). PCA analyses in NEG and POS. Quality control (QC) was an equivalent standard to evaluate the stability and signal response strength of the instrument during metabolite detection. (B,E). Venn diagrams and volcano maps illustrating the differential metabolites and their quantities. (C,F). Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation of identified metabolites in samples.
KEGG analysis of differentially accumulated metabolites (DAMs) identified 30 enriched pathways in HMGROE vs. HMGRC, with significant enrichment in the phenylpropanoid biosynthesis pathway (p value = 0.029) (Table S7). It is worth noting that the DEGs were enriched in the diterpenoid biosynthesis pathway (p value = 0.228), indicating that HMGR overexpression caused DAMs to decrease and affected terpenoid metabolism. For HMGRR vs. HMGRC, 56 pathways were enriched, including ubiquinone and other terpenoid–quinone biosynthesis pathways (p value = 0.500), with specific metabolites such as shikonin linked to the HMGR metabolic pathway. This group was also enriched in the anthocyanin biosynthesis pathway (p value = 0.126), involving metabolites like keracyanin and pelargonidin, which is likely one reason for the enhanced coloration in strawberries under the HMGRR condition. Notably, both HMGROE vs. HMGRC and HMGRR vs. HMGRC were enriched in methyl jasmonate-related pathways, including alpha-linolenic acid metabolism and plant hormone signal transduction (p value = 0.500).
4. Discussion
4.1. Correlation Between HMGR Expression Levels and Strawberry Fruit Quality
Research on HMGR and its influence on fruit quality related to terpenoids remains limited, particularly concerning its effects on fruit color and aroma. The two terpenoid biosynthesis pathways MVA and MEP are not completely isolated, as transporters in the plastid membrane allow metabolite exchange, facilitating a ‘metabolic cross-talk’ [1]. HMGR, which typically catalyzes the conversion of 3-hydroxy-3-methyl-glutaryl-CoA to mevalonic acid in the MVA pathway, has been shown to influence color formation by affecting the MEP pathway and anthocyanin synthesis [10]; it also encodes functional proteins that accelerate carotenoid biosynthesis [23,25]. Consistently, Zheng et al. [10] found that the expression of VvHMGRs in yellow grape varieties was generally higher than in red varieties. HMGR positively regulates terpenoid metabolism, thereby affecting hormone synthesis and indirectly regulating anthocyanin accumulation [24]. Moreover, a truncated form of HMGR (tHMG) was found to increase endogenous sesquiterpenes level by 37-fold and phytoene by 100-fold [33]. Negative regulation of HMGR by phyB and HY5 via interaction with PIF3 may further affect anthocyanin synthesis [19]. In this study, strawberries overexpressing FaHMGR exhibited reduced coloration, along with downregulated expression of most anthocyanin pathway genes. Conversely, HMGR silencing by expressing FaHMGRi promoted strawberry coloration, with anthocyanin content increasing by 3.33 mg·g−1 compared to the control, aligning with findings by Zheng et al. [26] in grape studies.
Strawberry aroma contributes significantly to fruit quality, enhancing consumer appeal and potential health benefits. Strawberry aroma results from a mixture of more than 350 volatile compounds, making it one of the most complex fruit aromas [34]. Volatiles in strawberry aroma include esters, terpenoids, alkanes, alcohols, acids, aldehydes, and ketones, with terpenes providing the main floral notes [35]. The terpenoids responsible for strawberry aroma primarily consist of monoterpenes (C10), sesquiterpenes (C15), and triterpenes (C30). Monoterpenes were the only type detected in this study. The highest diversity of terpenes, especially monoterpenes, was observed in FaHMGRi-expressing strawberries. This finding suggests that inhibiting HMGR expression promotes monoterpene synthesis via the MEP pathway. However, this contrasts with studies in grapes, which indicated a positive correlation between HMGR expression and monoterpene accumulation. This inconsistency suggests species-specific differences in how HMGR influences fruit aroma composition [36].
Beyond color and aroma, HMGR expression also affects fruit size. HMGR impacts early fruit development by regulating MVA synthesis, a role confirmed in studies of melons and tomatoes. Moreover, reduced HMGR activity has been shown to disrupt normal plant development [37,38].
4.2. Key Factors Regulating HMGR Expression
Hormones are important regulators of terpenoid synthesis, functioning by influencing HMGR activity. Several studies have demonstrated that exogenous application of hormones, including ABA and indole-3-acetic acid (IAA), regulates HMGR activity, while CTK counteracts HMGR inhibitors to support cell growth [39,40,41,42]. Zheng et al. [26] found that BR treatment in Kyoho grapes inhibited HMGR expression and promoted fruit coloration. Here, our study showed significantly higher BR levels in FaHMGROE strawberries than in FaHMGRR.
Several key proteins interact with HMGR to regulate its activity, including Protein Phosphatase 2A (PP2A), sucrose nonfermenting-1 (SNF1)-related kinase 1 (SnRK1), HIGH STEROL ESTER 1 (HISE1), Phytoene synthases 1 (PSY1), GATA24, and PHYTOCHROME INTERACTING FACTOR 3 (PIF3). PP2A, a multi-stage regulator of plant HMGR, negatively impacts HMGR activity through post-translational regulation and is involved in signal transduction for hormones such as ABA, IAA, BRs, and ETH [40]. SnRK1 helps maintain energy homeostasis by post-translationally regulating HMGR and TPS5 [43]. Protein HISE1 participates in the downregulation of HMGR to prevent sterol overproduction in the endoplasmic reticulum, as evidenced by the loss-of-function mutant hise1, which displays elevated levels of HMGR and sterol-producing activity [44]. Additionally, tHMG1 complexes with ZmPSY1 and PaCRTI to significantly boost carotenoid accumulation in rice endosperm by enhancing MVA pathway flux and creating a metabolic sink for carotenoids [45]. In Vitis vinifera, twelve amino acid residues of VvGATA24, including Pro218B, directly bind to the GATA-box cis-element of VvHMGR2 to mediate wax terpenoid biosynthesis [46].
4.3. Transcription Factors Related to Terpenoid Synthesis
Terpenoids encompass a diverse range of compounds with intricate metabolic regulation. The synthesis of plant terpenoids is a complex process involving various enzymes encoded by structural genes that catalyze the formation of multiple intermediate and final products in the terpenoid biosynthesis pathways. Regulatory genes are another crucial factor influencing terpenoid accumulation, forming complexes either individually or through interactions, and specifically binding to cis-elements in the promoters of structural genes. These regulatory complexes modulate terpenoid synthesis by upregulating or downregulating the expression of structural genes.
Transcription factors play a critical role in regulating multiple key genes involved in terpenoid metabolism [47]. Identifying these transcription factors contributes to our understanding of plant terpenoid synthetic biology. Six transcription factor families are currently known to be involved in terpenoid production, including AP2/ERF, bHLH, MYB, NAC, WRKY, and bZIP [6,46,48,49]. For example, AaERF1 and AaERF2 bind to CBF2 and RAA motifs in the promoters of ADS and CYP-71AV1 to regulate artemisinin synthesis [50]; in Citrus sinensis, citERF71 activates CitTPS16 geraniol synthase expression by binding to the CitTPS16 promoter [51]. WRKY regulates plant physiological and biochemical processes by modulating signaling pathways such as SA, ABA, JA, and ETH, thereby participating in plant growth, development, and stress responses. In Catharanthus roseus, Gossypium arboretum, and Artemisia annua, WRKY1 has been shown to regulate monoterpene and sesquiterpene synthesis. For instance, CrWRKY1 inhibits CrMYC2 transcription to suppress monoterpene synthase, thereby regulating monoterpene production [52]. GaWRKY1 promotes sesquiterpene synthesis in cotton by upregulating CAD1-A expression [53]; AaWRKY1, induced by MeJA, binds to the W-box cis-acting element of the ADS promoter to enhance artemisinin synthesis in Artemisia annua [54].
Research on HMGR-related transcription factors remains limited, and current knowledge primarily derives from Arabidopsis thaliana. Studies indicate that HY5 and PIF3 act antagonistically to regulate HMGR expression and sterol biosynthesis by physically binding to the AtHMGR2 promoter [55]. In this study, WGCNA analysis suggests that ERF118 and WRKY12 may act as transcription factors that modulate HMGR-related pathways affecting fruit quality.
5. Conclusions
In conclusion, this study offers new insights into the role of HMGR in influencing strawberry fruit coloration and aroma composition (Figure 8). HMGR regulates both the terpenoid and anthocyanin pathways, with its overexpression inhibiting fruit coloration, downregulating most anthocyanin pathway genes, and reducing aroma content. Conversely, suppression of HMGR promoted coloration and led to the identification of five new anthocyanin components. Our work also suggests MeJA as a potential mediator of HMGR’s influence on terpenoid pathways. Additionally, transcription factors ERF118 and WRKY12 potentially regulate HMGR to affect fruit quality. Overall, our findings contribute to the current understanding of terpenoid metabolism and strawberry fruit quality regulation.
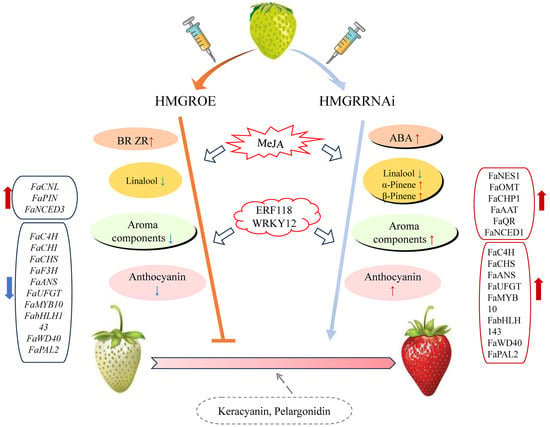
Figure 8.
Proposed model illustrating the impact of varying HMGR expression levels on transcriptional changes and metabolite profiles linked to strawberry fruit development and aroma production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods14071199/s1, Figure S1: Comparison of the gene transcription level detected by RNA-sequencing and qRT-PCR. The green columns indicate levels detected by RNA sequencing, and the black lines indicate the qRT-PCR result. Bars represent standard deviations of the means; Figure S2: Gene dendrogram from weighted gene co-expression network analysis (WGCNA). (A) Hierarchical clustering tree illustrating gene relationships. (B) Module–sample correlation showing the strength of associations between modules and samples. (C) Analysis of the expression patterns of differentially expressed genes (DEGs) from selected typical modules; Figure S3: Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of significantly enriched differentially accumulated metabolites (DAMs) in the comparisons of HMGRR vs. HMGRC and HMGROE vs. HMGRC; Table S1: Primer used for qRT-PCR on strawberry; Table S2: CDs sequence of FaHMGR and FaHMGRi; Table S3: General overview of transcriptome data; Table S4: The GO with the most significant enrichment of DEGs; Table S5: Genes in five main module; Table S6: The metabolites in positive and negative ion mode; Table S7: The KEGG pathways with the most significant enrichment of DEGs.
Author Contributions
Conceptualization, T.Z. and J.C.; methodology, L.W.; software, J.X.; validation, L.W. and J.W.; formal analysis, T.Z.; investigation, L.W.; resources, J.X.; data curation, L.W.; writing—original draft preparation, T.Z.; writing—review and editing, J.W. and J.C.; visualization, J.X.; supervision, J.C.; project administration, J.W.; funding acquisition, T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32202414).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All transcriptomic raw data reported in this paper have been deposited in the National Center for Biotechnology Information sequence read archive (transcriptome sequencing accession SRA accession number: PRJNA781262).
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Yan, J.W.; Ban, Z.J.; Lu, H.Y.; Li, D.; Poverenov, E.; Luo, Z.S.; Li, L. The aroma volatile repertoire in strawberry fruit: A review. J. Sci. Food Agric. 2018, 98, 4395–4402. [Google Scholar] [CrossRef]
- Contreras-Avilés, W.; Heuvelink, E.; Marcelis, L.F.M.; Kappers, I.F. Ménage à trois: Light, terpenoids, and quality of plants. Trends Plant Sci. 2024, 29, 572–588. [Google Scholar] [CrossRef]
- Liu, Z.C.; Liang, T.; Kang, C.Y. Molecular bases of strawberry fruit quality traits: Advances, challenges, and opportunities. Plant Physiol. 2023, 193, 900–914. [Google Scholar] [CrossRef]
- Fan, Z.; Tieman, D.M.; Knapp, S.J.; Zerbe, P.; Famula, R.; Barbey, C.R.; Folta, K.M.; Amadeu, R.R.; Lee, M.; Oh, Y.; et al. A multi-omics framework reveals strawberry flavor genes and their regulatory elements. New Phytol. 2022, 236, 1089–1107. [Google Scholar] [CrossRef]
- Badmi, R.; Gogoi, A.; Prestwich, B.D. Secondary Metabolites and Their Role in Strawberry Defense. Plants 2023, 12, 3240. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Charles, M.T.; Luo, Z.S.; Mimee, B.; Tong, Z.C.; Véronneau, P.Y.; Roussel, D.; Rolland, D. Ultraviolet-C priming of strawberry leaves against subsequent Mycosphaerella fragariae infection involves the action of reactive oxygen species, plant hormones, and terpenes. Plant Cell Environ. 2019, 42, 815–831. [Google Scholar] [CrossRef]
- Li, Z.B.; Wang, N.; Wei, Y.Y.; Zou, X.R.; Jiang, S.; Xu, F.; Wang, H.F.; Shao, X.F. Terpinen-4-ol Enhances Disease Resistance of Postharvest Strawberry Fruit More Effectively than Tea Tree Oil by Activating the Phenylpropanoid Metabolism Pathway. J. Agric. Food Chem. 2020, 68, 6739–6747. [Google Scholar] [CrossRef]
- Sun, C.D.; Yang, X.F.; Gu, Q.J.; Jiang, G.H.; Shen, L.; Zhou, J.Y.; Li, L.; Chen, H.X.; Zhang, G.F.; Zhang, Y.C. Comprehensive analysis of nanoplastic effects on growth phenotype, nanoplastic accumulation, oxidative stress response, gene expression, and metabolite accumulation in multiple strawberry cultivars. Sci. Total Environ. 2023, 897, 165432. [Google Scholar] [CrossRef]
- Mehmood, N.; Yuan, Y.; Ali, M.; Ali, M.; Iftikhar, J.; Cheng, C.; Lyu, M.; Wu, B. Early transcriptional response of terpenoid metabolism to Colletotrichum gloeosporioides in a resistant wild strawberry Fragaria nilgerrensis. Phytochemistry 2021, 181, 112590. [Google Scholar] [CrossRef]
- Zheng, T.; Guan, L.; Yu, K.; Haider, M.S.; Nasim, M.; Liu, Z.; Li, T.; Zhang, K.; Jiu, S.; Jia, H.; et al. Expressional diversity of grapevine 3-Hydroxy-3-methylglutaryl-CoA reductase (VvHMGR) in different grapes genotypes. BMC Plant Biol. 2021, 21, 279. [Google Scholar] [CrossRef]
- Friesen, J.A.; Rodwell, V.W. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004, 5, 248. [Google Scholar] [CrossRef]
- Wani, T.A.; Kaloo, Z.A.; Reshi, S.A. Molecular characterization of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) in relation to aconite biosynthesis in Aconitum heterophyllum Wall ex Royle. Gene Rep. 2022, 26, 101432. [Google Scholar]
- Wang, Y.Y.; Jing, F.Y.; Yu, S.Y.; Chen, Y.F.; Wang, T.; Liu, P.; Wang, G.F.; Sun, X.F.; Tang, K.X. Co-overexpression of the HMGR and FPS genes enhances artemisinin content in Artemisia annua L. J. Med. Plants Res. 2011, 5, 3396–3403. [Google Scholar]
- Nafis, T.; Akmal, M.; Ram, M.; Alam, P.; Ahlawat, S.; Mohd, A.; Abdin, M.Z. Enhancement of artemisinin content by constitutive expression of the HMG-CoA reductase gene in high-yielding strain of Artemisia annua L. Plant Biotechnol. Rep. 2011, 5, 53–60. [Google Scholar] [CrossRef]
- Alam, P.; Abdin, M.Z. Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep. 2011, 30, 1919–1928. [Google Scholar] [CrossRef]
- Shang, J.Z.; Feng, D.D.; Liu, H.; Niu, L.T.; Li, R.H.; Li, Y.J.; Chen, M.X.; Li, A.; Liu, Z.H.; He, Y.H.; et al. Evolution of the biosynthetic pathways of terpene scent compounds in roses. Curr. Biol. 2024, 34, 3550–3563. [Google Scholar] [CrossRef]
- Choi, D.; Ward, B.L.; Bostock, R.M. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 1992, 4, 1333–1344. [Google Scholar] [CrossRef][Green Version]
- Dong, N.; Ponciano, G.; McMahan, C.M.; Coffelt, T.A.; Johnson, L.; Creelman, R.; Whalen, M.C.; Cornish, K. Overexpression of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Parthenium argentatum (guayule). Ind. Crops Prod. 2013, 46, 15–24. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, O.R.; Oh, J.Y.; Jang, M.G.; Yang, D.C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014, 165, 373–387. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, C.; Wen, G.; Zhang, H.; Zeng, X.; Liu, Z.; Xu, S.; Lin, C. Longitudinal expression patterns of HMGR, FPS, SS, SE and DS and their correlations with saponin contents in green-purple transitional aerial stems of Panax notoginseng. Ind. Crop. Prod. 2018, 119, 132–143. [Google Scholar]
- Ma, X.C.; Luo, N.; Bai, W.P.; Wang, X.R.; Wang, C.C.; Cheng, N.N.; Liu, H. Genome-wide analysis of the PtHMGR gene family and functional validation of PtHMGR5 improving drought tolerance in Populus trichocarpa. Environ. Exp. Bot. 2023, 216, 105544. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Xu, C.; Sun, W.B.; Li, L.L.; Wang, P.; Li, D.W.; Zhuge, Q. Overexpression of PtHMGR enhances the drought and salt tolerance of poplar. Ann. Bot. 2020, 125, 785–803. [Google Scholar]
- Akhtar, N.; Gupta, P.; Sangwan, N.S.; Sangwan, R.S.; Trivedi, P.K. Cloning and functional characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Withania somnifera: An important medicinal plant. Protoplasma 2013, 250, 613–622. [Google Scholar] [CrossRef]
- Li, W.F.; Mao, J.; Yang, S.J.; Guo, Z.G.; Ma, Z.H.; Dawuda, M.M.; Zuo, C.W.; Chu, M.Y.; Chen, B.H. Anthocyanin accumulation correlates with hormones in the fruit skin of ‘Red Delicious’ and its four generation bud sport mutants. BMC Plant Biol. 2018, 18, 363. [Google Scholar] [CrossRef]
- Bhambhani, S.; Lakhwani, D.; Shukla, T.; Pandey, A.; Dhar, Y.V.; Asif, M.H.; Trivedi, P.K. Genes encoding members of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) gene family from Azadirachta indica and correlation with azadirachtin biosynthesis. Acta Physiol. Plant 2017, 39, 65. [Google Scholar] [CrossRef]
- Zheng, T.; Dong, T.; Haider, M.S.; Jin, H.; Jia, H.; Fang, J. Brassinosteroid Regulates 3-Hydroxy-3-methylglutaryl CoA Reductase to Promote Grape Fruit Development. J. Agric. Food Chem. 2020, 68, 11987–11996. [Google Scholar] [CrossRef]
- Xu, Y.; Lv, Z.; Manzoor, M.A.; Song, L.; Wang, M.; Wang, L.; Wang, S.; Zhang, C.; Jiu, S. VvD14c-VvMAX2-VvLOB/VvLBD19 module is involved in the strigolactone-mediated regulation of grapevine root architecture. Mol. Hortic. 2024, 4, 40. [Google Scholar] [CrossRef]
- Wang, Y.; Li, B.; Du, M.W.; Eneji, A.E.; Wang, B.M.; Duan, L.S.; Li, Z.H.; Tian, X.L. Mechanism of phytohormone involvement in feedback regulation of cotton leaf senescence induced by potassium deficiency. J. Exp. Bot. 2012, 63, 5887–5901. [Google Scholar]
- Li, B.J.; Shi, Y.N.; Xiao, Y.N.; Jia, H.R.; Yang, X.F.; Dai, Z.R.; Sun, Y.F.; Shou, J.H.; Jiang, G.H.; Grierson, D.; et al. AUXIN RESPONSE FACTOR 2 mediates repression of strawberry receptacle ripening via auxin-ABA interplay. Plant Physiol. 2024, 196, 2638–2653. [Google Scholar] [CrossRef]
- Zheng, T.; Lv, J.H.; Sadeghnezhad, E.; Cheng, J.H.; Jia, H.F. Transcriptomic and metabolomic profiling of strawberry during postharvest cooling and heat storage. Front. Plant Sci. 2022, 13, 1009747. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, P.; Zhang, P.; Su, L.; Jia, H.; Wei, X.; Fang, J.; Jia, H. Integrative transcriptomics and metabolomics data exploring the effect of chitosan on postharvest grape resistance to Botrytis cinerea. Postharvest Biol. Technol. 2020, 167, 111248. [Google Scholar]
- Andersen, T.B.; Llorente, B.; Morelli, L.; Torres-Montilla, S.; Bordanaba-Florit, G.; Espinosa, F.A.; Rodriguez-Goberna, M.R.; Campos, N.; Olmedilla-Alonso, B.; Llansola-Portoles, M.J.; et al. An engineered extraplastidial pathway for carotenoid biofortification of leaves. Plant Biotechnol. J. 2021, 19, 1008–1021. [Google Scholar] [CrossRef]
- Oh, Y.; Barbey, C.R.; Chandra, S.; Bai, J.; Fan, Z.; Plotto, A.; Pillet, J.; Folta, K.M.; Whitaker, V.M.; Lee, S. Genomic Characterization of the Fruity Aroma Gene, FaFAD1, Reveals a Gene Dosage Effect on γ-Decalactone Production in Strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 12, 639345. [Google Scholar] [CrossRef]
- Peng, X.; Wang, B.; Wang, X.L.; Ni, B.B.; Zuo, Z.J. Variations in aroma and specific flavor in strawberry under different colored light-quality selective plastic film. Flavour Fragr. J. 2020, 35, 350–359. [Google Scholar] [CrossRef]
- Xie, S.; Wu, G.; Ren, R.H.; Xie, R.; Yin, H.N.; Chen, H.W.; Yang, B.W.; Zhang, Z.W.; Ge, M.S. Transcriptomic and metabolic analyses reveal differences in monoterpene profiles and the underlying molecular mechanisms in six grape varieties with different flavors. Lwt-Food Sci. Technol. 2023, 174, 114442. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kato-Emori, S.; Tomita, K.; Ezura, H. Detection of 3-hydroxy-3-methylglutaryl-coenzyme A reductase protein Cm-HMGR during fruit development in melon (Cucumis melo L.). Theor. Appl. Genet. 2002, 104, 779–785. [Google Scholar] [CrossRef]
- Enfissi, E.M.; Fraser, P.D.; Lois, L.M.; Boronat, A.; Schuch, W.; Bramley, P.M. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 2005, 3, 17–27. [Google Scholar] [CrossRef]
- Crowell, D.; Salaz, S. Inhibition of growth of cultured tobacco cells at low concentrations of lovastatin is reversed by cytokinin. Plant Physiol. 1992, 100, 2090–2095. [Google Scholar]
- Leivar, P.; Antolin-Llovera, M.; Ferrero, S.; Closa, M.; Arro, M.; Ferrer, A.; Boronat, A.; Campos, N. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 2011, 23, 1494–1511. [Google Scholar] [CrossRef]
- Antolín-Llovera, M.; Leivar, P.; Arró, M.; Ferrer, A.; Boronat, A.; Campos, N. Modulation of plant HMG-CoA reductase by protein phosphatase 2A: Positive and negative control at a key node of metabolism. Plant Signal. Behav. 2011, 6, 1127–1131. [Google Scholar] [CrossRef]
- Nishi, A.; Tsuritani, I. Effect of auxin on the metabolism of mevalonic acid in suspension-cultured carrot cells. Phytochemistry 1983, 22, 399–401. [Google Scholar] [CrossRef]
- Luo, J.; Peng, F.; Zhang, S.; Xiao, Y.; Zhang, Y. The protein kinase FaSnRK1α regulates sucrose accumulation in strawberry fruits. Plant Physiol. Biochem. 2020, 151, 369–377. [Google Scholar] [CrossRef]
- Shimada, T.L.; Shimada, T.; Okazaki, Y.; Higashi, Y.; Saito, K.; Kuwata, K.; Oyama, K.; Kato, M.; Ueda, H.; Nakano, A.; et al. HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat. Plants 2019, 5, 1154–1166. [Google Scholar] [CrossRef]
- Tian, Y.S.; Wang, B.; Peng, R.H.; Xu, J.; Li, T.; Fu, X.Y.; Xiong, A.S.; Gao, J.J.; Yao, Q.H. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering. Plant Biotechnol. J. 2019, 17, 1694. [Google Scholar] [CrossRef]
- Yang, M.Y.; Xiang, Y.Z.; Luo, Z.S.; Gao, Y.Z.; Wang, L.; Hu, Q.N.; Dong, Y.Y.; Qi, M.; Li, D.; Liu, L.L.; et al. Light-responsive transcription factors VvHYH and VvGATA24 mediate wax terpenoid biosynthesis in Vitis vinifera. Plant Physiol. 2024, 196, 1546–1561. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Y.S.; Zeng, J.G.; Duan, L.X.; Xue, X.F.; Wang, H.S.; Lin, T.; Liu, Z.Q.; Zeng, K.W.; Zhong, Y.; et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants 2016, 2, 16183. [Google Scholar] [CrossRef]
- Kong, W.L.; Zhao, P.; Zhang, Q.; Yang, J.J.; Zhu, Q.F.; Zhang, Y.B.; Deng, X.M.; Chen, X.; Lin, J.K.; Zhang, X.T. Chromatin accessibility mediated transcriptome changes contribute to flavor substance alterations and jasmonic acid hyperaccumulation during oolong tea withering process. Plant J. 2024, 117, 679–693. [Google Scholar] [CrossRef]
- Cheng, L.; Tu, G.F.; Ma, H.C.; Zhang, K.Y.; Wang, X.Y.; Zhou, H.Z.; Gao, J.W.; Zhou, J.; Yu, Y.B.; Xu, Q.S. Alternative splicing of CsbHLH133 regulates geraniol biosynthesis in tea plants. Plant J. 2024, 120, 598–614. [Google Scholar] [CrossRef]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The Jasmonate-Responsive AP2/ERF Transcription Factors AaERF1 and AaERF2 Positively Regulate Artemisinin Biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.; Klee, H.; Zhang, B.; Chen, K.; Hancock, R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (1)-d-Cadinene Synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar]
- Jiang, W.; Fu, X.; Pan, Q.; Tang, Y.; Shen, Q.; Lv, Z.; Yan, T.; Shi, P.; Li, L.; Zhang, L.; et al. Overexpression of AaWRKY1 Leads to an Enhanced Content of Artemisinin in Artemisia annua. BioMed Res. Int. 2016, 2016, 7314971. [Google Scholar] [CrossRef]
- Michael, R.; Ranjan, A.; Gautam, S.; Trivedi, P.K. HY5 and PIF antagonistically regulate HMGR expression and sterol biosynthesis in Arabidopsis thaliana. Plant Sci. 2024, 346, 112168. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).