DNA Barcoding for Tracing Biodiversity in Mixed Crop Food Products: A Proof of Concept Within the BioValue Project
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant-Based Food Products
2.2. DNA Extraction
2.3. PCR Amplification
2.4. Long-Read Nanopore Sequencing of ITS and rbcL rRNA Regions
2.5. Bioinformatic Analysis
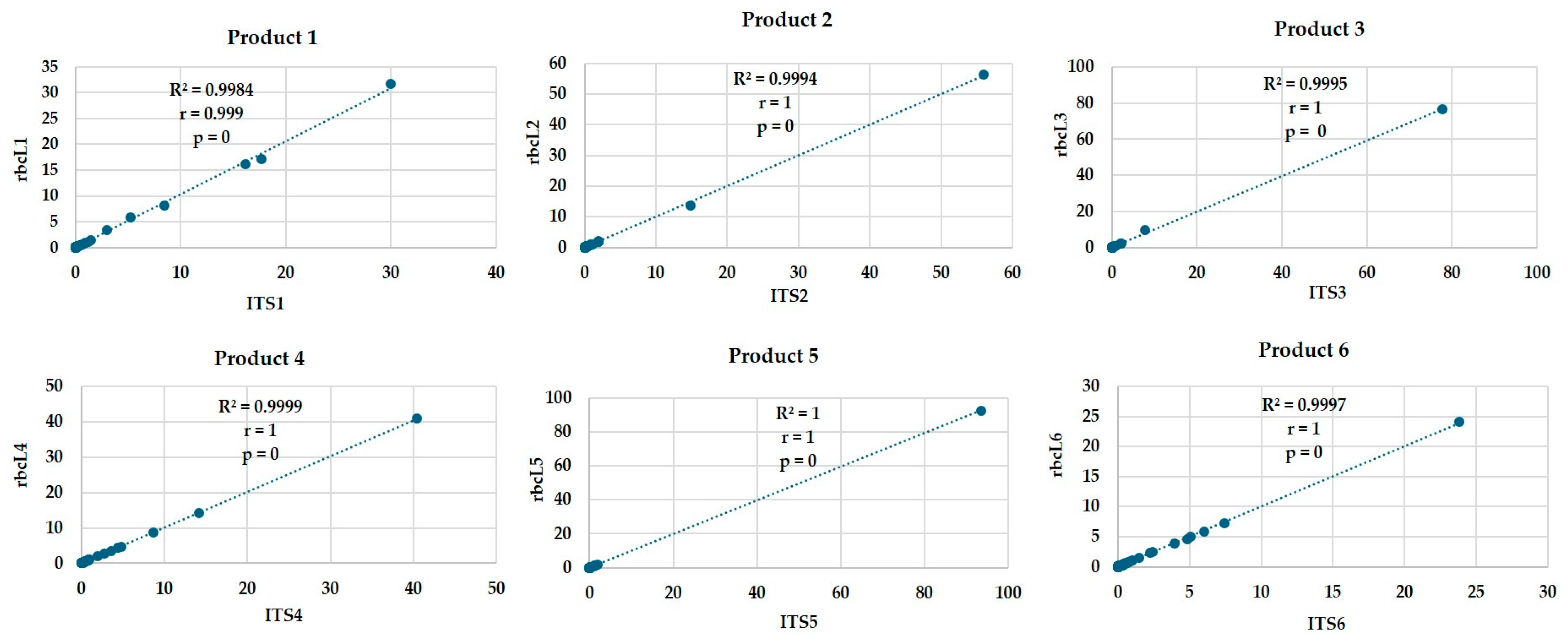
2.6. Statistical Analysis
3. Results and Discussion
3.1. DNA Extraction and PCR Performance Are Affected by Food Processing and Additives
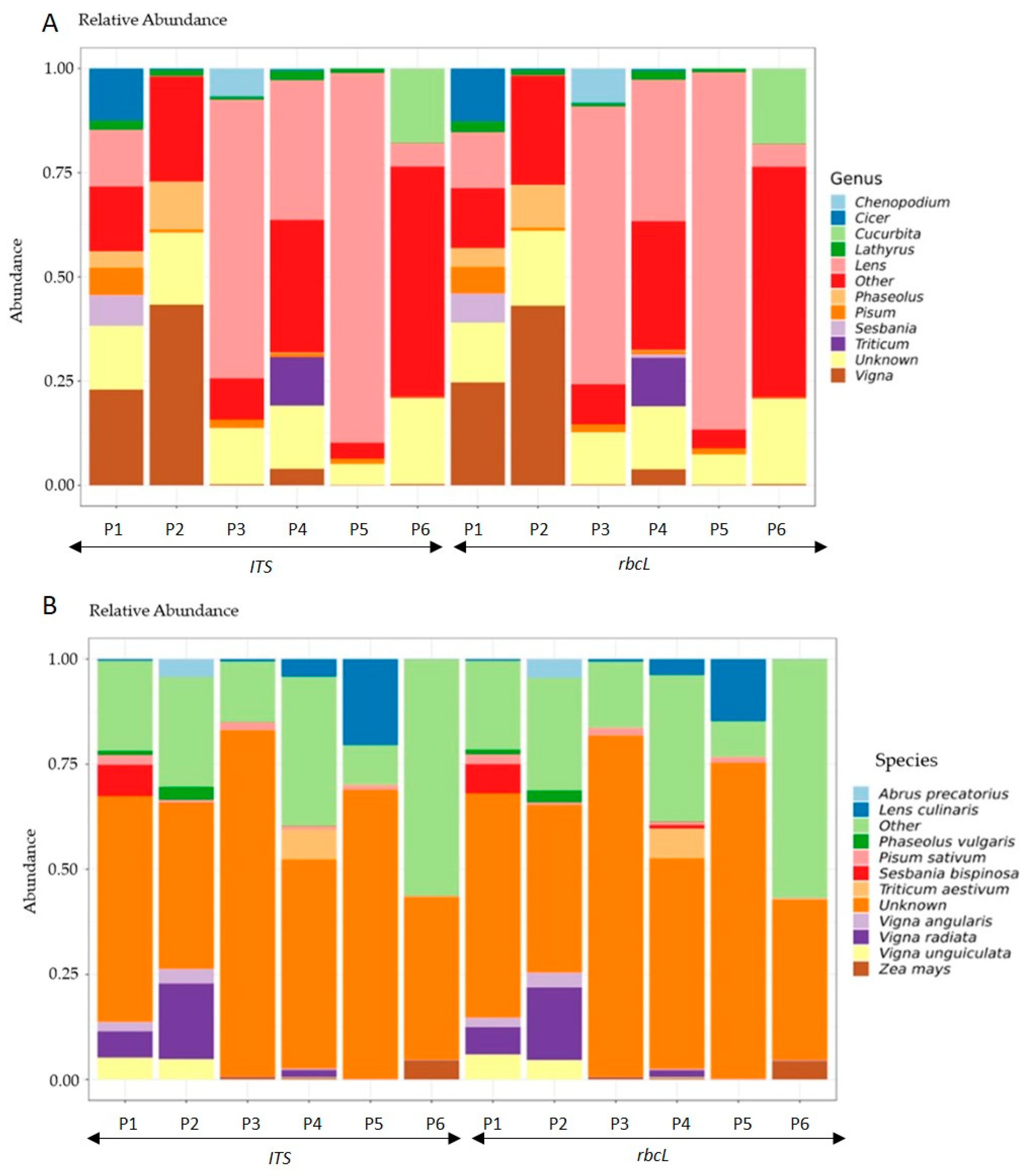
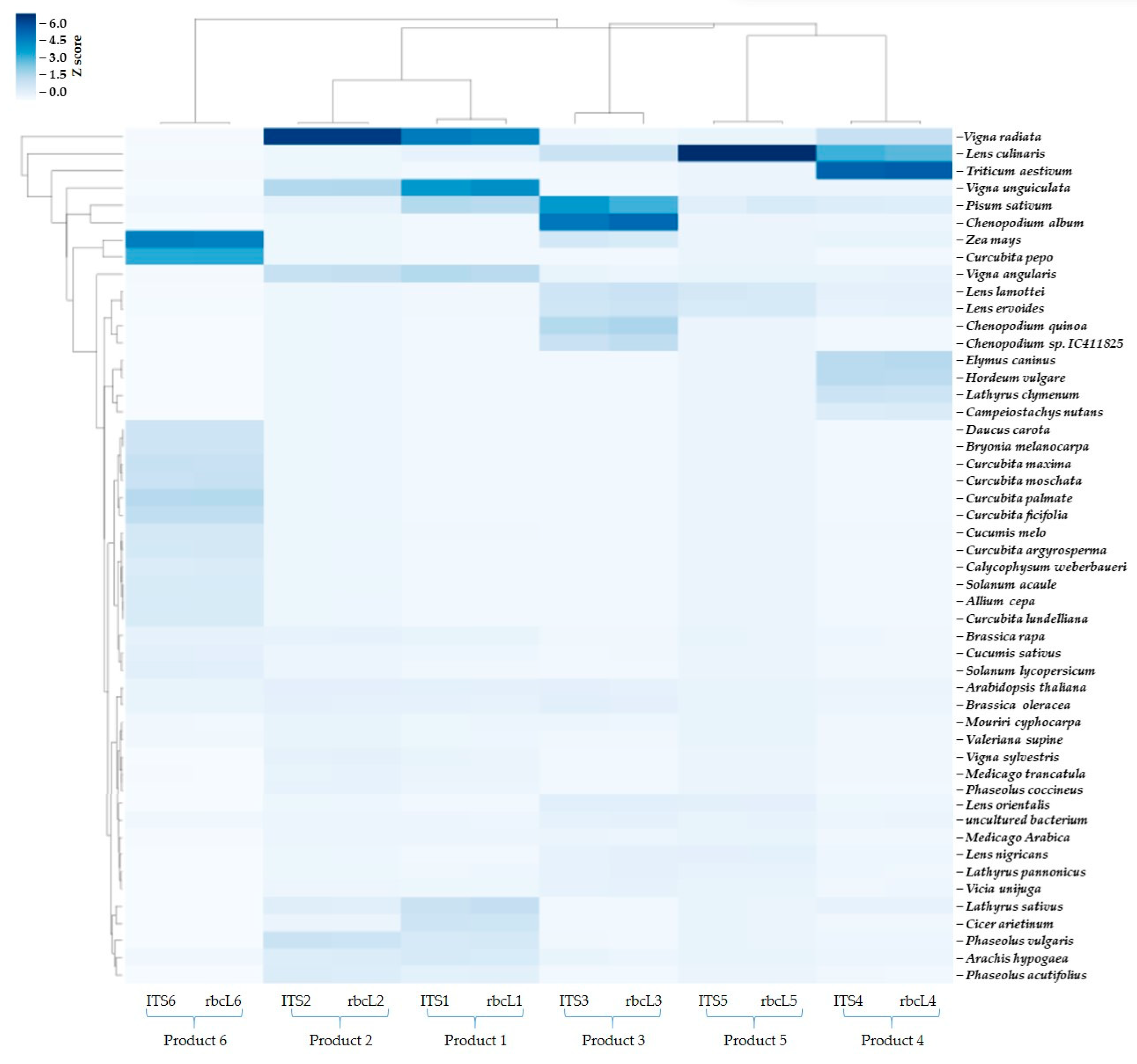
3.2. DNA Barcoding Using ITS and rbcL Genes Reveals Consistently Diverse Taxonomic Composition in Commercial Plant-Based Products
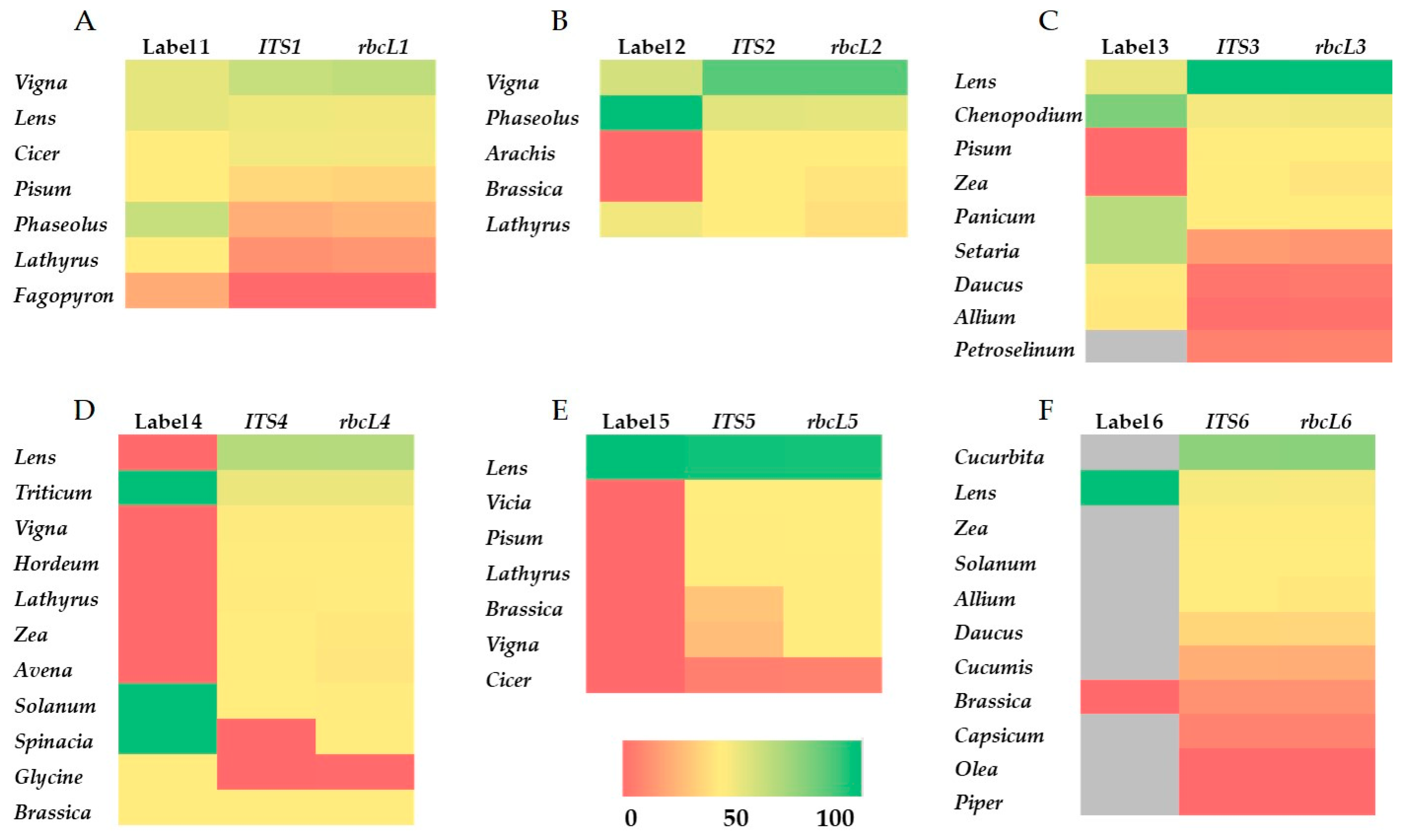
3.3. Comparison of Label Composition and Sequencing-Based Genera Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of the World’s Biodiversity for Food and Agriculture; Bélanger, J., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2019. [Google Scholar]
- Dawson, I.K.; Powell, W.; Hendre, P.; Bančič, J.; Hickey, J.M.; Kindt, R.; Hoad, S.; Hale, I.; Jamnadass, R. The role of genetics in mainstreaming the production of new and orphan crops to diversify food systems and support human nutrition. New Phytol. 2019, 224, 37–54. [Google Scholar] [CrossRef]
- Hawkins, I.W. Promoting Biodiversity in Food Systems, 1st ed.; CRC Press: Boca Raton, FL, USA, 2018; ISBN 9780367732974. [Google Scholar]
- García-Martín, M.; Huntsinger, L.; Ibarrola-Rivas, M.J.; Penker, M.; D’Ambrosio, U.; Dimopoulos, T.; Fernández-Giménez, M.E.; Kizos, T.; Muñoz-Rojas, J.; Abson, D.J.; et al. Landscape products for sustainable agricultural landscapes. Nat. Food 2022, 3, 814–821. [Google Scholar] [CrossRef]
- Martin, G.; Magne, M.A. Agricultural diversity to increase adaptive capacity and reduce vulnerability of livestock systems against weather variability—A farm-scale simulation study. Agric. Ecosyst. Environ. 2015, 199, 301–311. [Google Scholar] [CrossRef]
- Córdoba Vargas, C.A.; Hortúa Romero, S.; León Sicard, T. Key points of resilience to climate change: A necessary debate from agroecological systems. Clim. Dev. 2020, 12, 564–574. [Google Scholar] [CrossRef]
- Elouafi, I. Why biodiversity matters in agriculture and food systems. Science 2024, 386, eads8197. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.; Seddon, N.; Elliott, J. Biodiversity loss is a development issue A rapid review of evidence Issue Paper. Lancet Planet. Health 2019, 3, 1–24. [Google Scholar]
- Jarzebski, M.P.; Su, J.; Abrahamyan, A.; Lee, J.; Kawasaki, J.; Chen, B.; Andriatsitohaina, R.N.N.; Ocen, I.; Sioen, G.B.; Lambino, R.; et al. Developing biodiversity-based solutions for sustainable food systems through transdisciplinary Sustainable Development Goals Labs (SDG-Labs). Front. Sustain. Food Syst. 2023, 7, 1144506. [Google Scholar] [CrossRef]
- IPBES. Summary for Policymakers of the Global Assessment Report on Biodiversity and Ecosystem Services; IPBES: Bonn, Germany, 2019; Volume 45, ISBN 9783947851133. [Google Scholar]
- Cheng, A.; Mayes, S.; Dalle, G.; Demissew, S.; Massawe, F. Diversifying crops for food and nutrition security—A case of teff. Biol. Rev. 2017, 92, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Fardet, A.; Rock, E. Ultra-processed foods and food system sustainability: What are the links? Sustainability 2020, 12, 6280. [Google Scholar] [CrossRef]
- Leite, F.H.M.; Khandpur, N.; Andrade, G.C.; Anastasiou, K.; Baker, P.; Lawrence, M.; Monteiro, C.A. Ultra-processed foods should be central to global food systems dialogue and action on biodiversity. BMJ Glob. Health 2022, 7, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, A.; Tommasi, N.; Guzzetti, L.; Galimberti, A.; Bruni, I.; Labra, M. DNA barcoding to promote social awareness and identity of neglected, underutilized plant species having valuable nutritional properties. Food Res. Int. 2019, 115, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Casiraghi, M.; Labra, M.; Ferri, E.; Galimberti, A.; de Mattia, F. DNA barcoding: A six-question tour to improve users’ awareness about the method. Brief Bioinform. 2010, 11, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; De Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
- Hubert, N.; Hanner, R. DNA Barcoding, species delineation and taxonomy: A historical perspective. DNA Barcodes 2016, 3, 44–58. [Google Scholar] [CrossRef]
- Liberty, J.T.; Lin, H.; Kucha, C.; Sun, S.; Alsalman, F.B. Innovative approaches to food traceability with DNA barcoding: Beyond traditional labels and certifications. Ecol. Genet. Genom. 2025, 34, 100317. [Google Scholar] [CrossRef]
- Antil, S.; Abraham, J.S.; Sripoorna, S.; Maurya, S.; Dagar, J.; Makhija, S.; Bhagat, P.; Gupta, R.; Sood, U.; Lal, R.; et al. DNA barcoding, an effective tool for species identification: A review. Mol. Biol. Rep. 2023, 50, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Watson, G.M.F.; Tabita, F.R. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: A molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 1997, 146, 13–22. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid Dna Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Grigoriou, A.; Tsaniklidis, G.; Hagidimitriou, M.; Nikoloudakis, N. The cypriot indigenous grapevine germplasm is a multi-clonal varietal mixture. Plants 2020, 9, 1034. [Google Scholar] [CrossRef]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Corvalán, L.C.J.; de Melo-Ximenes, A.A.; Carvalho, L.R.; e Silva-Neto, C.D.M.; Diniz-Filho, J.A.F.; Telles, M.P.D.C.; Nunes, R. Is There a Key Primer for Amplification of Core Land Plant DNA Barcode Regions (rbcL and matK)? Ecol. Evol. 2025, 15, e70961. [Google Scholar] [CrossRef]
- Fay, M.F.; Swensen, S.M.; Chase, M.W. Taxonomic Affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull. 1997, 52, 111–120. [Google Scholar] [CrossRef]
- Robinson, J.P.; Harris, S.A.; Juniper, B.E. Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Syst. Evol. 2001, 226, 35–58. [Google Scholar] [CrossRef]
- Oshingboye, A.D.; Ogundipe, O.T. Exploring the DNA barcoding potential of Nigerian arid-land Mimosoideae using a multigene tiered approach. S. Afr. J. Bot. 2018, 117, 288–300. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, Y.; Ding, H.; Liu, X.; Cao, F.; Xu, L. From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding. Plants 2025, 14, 619. [Google Scholar] [CrossRef]
- Nikoloudakis, N.; Skaracis, G.; Katsiotis, A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Mol. Phylogenet. Evol. 2008, 46, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Stortchevoi, A.; Kamelamela, N.; Levine, S.S. SPRI beads-based size selection in the range of 2-10kb. J. Biomol. Tech. 2020, 31, 7–10. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Gorini, T.; Mezzasalma, V.; Deligia, M.; De Mattia, F.; Campone, L.; Labra, M.; Frigerio, J. Check Your Shopping Cart: DNA Barcoding and Mini-Barcoding for Food Authentication. Foods 2023, 12, 2392. [Google Scholar] [CrossRef] [PubMed]
- Mafra, I.; Silva, S.A.; Moreira, E.J.M.O.; da Silva, C.S.F.; Beatriz, M.; Oliveira, P.P. Comparative study of DNA extraction methods for soybean derived food products. Food Control 2008, 19, 1183–1190. [Google Scholar] [CrossRef]
- Ren, Y.; Ma, Y.; Li, Y.; Song, Y.; Zhao, W.; Huang, X.; Yu, D.; Li, J.; Xu, Z.; Zhao, W. Comparative evaluation of various DNA extraction methods and analysis of DNA degradation levels in commercially marketed Chestnut rose juices and beverages. BMC Biotechnol. 2025, 25, 9. [Google Scholar] [CrossRef]
- Singh, M.; Sodhi, K.K.; Paliwal, A.; Sharma, S.; Randhawa, G. Efficient DNA Extraction Procedures for Processed Food Derivatives—A Critical Step to Ensure Quality for GMO Analysis. Food Anal. Methods 2021, 14, 2249–2261. [Google Scholar] [CrossRef]
- Filipa-Silva, A.; Castro, R.; Rebelo, M.; Mota, M.J.; Almeida, A.; Valente, L.M.P.; Gomes, S. Enhancing the authenticity of animal by-products: Harmonization of DNA extraction methods from novel ingredients. Front. Chem. 2024, 12–2024, 1350433. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Spall, L.J.; Gibson, F.J.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Lorusso, L.; Shum, P.; Piredda, R.; Mottola, A.; Maiello, G.; Cartledge, E.L.; Neave, E.F.; Di Pinto, A.; Mariani, S. Mismanagement and poor transparency in the European processed seafood supply revealed by DNA metabarcoding. Food Res. Int. 2024, 194, 114901. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Kesanakurti, P.R.; Burgess, K.S.; Percy, D.M.; Graham, S.W.; Barrett, S.C.H.; Newmaster, S.G.; Hajibabaei, M.; Husband, B.C. Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol. Ecol. Resour. 2009, 9, 130–139. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; DeWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003, 270, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH-psbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. DNA barcodes: Genes, genomics, and bioinformatics. Proc. Natl. Acad. Sci. USA 2008, 105, 2761–2762. [Google Scholar] [CrossRef]
- Lanubile, A.; Stagnati, L.; Marocco, A.; Busconi, M. DNA-based techniques to check quality and authenticity of food, feed and medicinal products of plant origin: A review. Trends Food Sci. Technol. 2024, 149, 104568. [Google Scholar] [CrossRef]
- Robertson, J.M.; Damaso, N.; Meiklejohn, K.A. DNA-Based Analysis of Plant Material in Forensic Investigations; Springer: Singapore, 2021; ISBN 9789811593642. [Google Scholar]
- Singh, H.; Parveen, I.; Raghuvanshi, S.; Babbar, S.B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res. Notes 2012, 5, 42. [Google Scholar] [CrossRef]
- Cameron, K.M.; Chase, M.W.; Whitten, W.M.; Kores, P.J.; Jarrell, D.C.; Albert, V.A.; Yukawa, T.; Hills, H.G.; Goldman, D.H. A phylogenetic analysis of the Orchidaceae: Evidence from rbcL nucleotide sequences. Am. J. Bot. 1999, 86, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef]
- Nehal, N.; Choudhary, B.; Nagpure, A.; Gupta, R.K. DNA barcoding: A modern age tool for detection of adulteration in food. Crit. Rev. Biotechnol. 2021, 41, 767–791. [Google Scholar] [CrossRef]
- Brooks, C.; Parr, L.; Smith, J.M.; Buchanan, D.; Snioch, D.; Hebishy, E. A review of food fraud and food authenticity across the food supply chain, with an examination of the impact of the COVID-19 pandemic and Brexit on food industry. Food Control 2021, 130, 108171. [Google Scholar] [CrossRef]
- Raja, H.A.; Baker, T.R.; Little, J.G.; Oberlies, N.H. DNA barcoding for identification of consumer-relevant mushrooms: A partial solution for product certification? Food Chem. 2017, 214, 383–392. [Google Scholar] [CrossRef]
- Bosmali, I.; Ordoudi, S.A.; Tsimidou, M.Z.; Madesis, P. Greek PDO saffron authentication studies using species specific molecular markers. Food Res. Int. 2017, 100, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulos, I.; Bazakos, C.; Madesis, P.; Kalaitzis, P.; Tsaftaris, A. Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J. Sci. Food Agric. 2013, 93, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Intharuksa, A.; Sasaki, Y.; Ando, H.; Charoensup, W.; Suksathan, R.; Kertsawang, K.; Sirisa-ard, P.; Mikage, M. The combination of ITS2 and psbA-trnH region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J. Nat. Med. 2020, 74, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Khilare, V.; Tiknaik, A.; Prakash, B.; Ughade, B.; Korhale, G.; Nalage, D.; Ahmed, N.; Khedkar, C.; Khedkar, G. Multiple tests on saffron find new adulterant materials and reveal that Ist grade saffron is rare in the market. Food Chem. 2019, 272, 635–642. [Google Scholar] [CrossRef]
- Mosa, K.A.; Soliman, S.; El-Keblawy, A.; Ali, M.A.; Hassan, H.A.; Tamim, A.A.B.; Al-Ali, M.M. Using DNA Barcoding to Detect Adulteration in Different Herbal Plant- Based Products in the United Arab Emirates: Proof of Concept and Validation. Recent Pat. Food Nutr. Agric. 2018, 9, 55–64. [Google Scholar] [CrossRef]
- Tripodi, P. Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species. Genes 2023, 14, 1594. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Y.; Liu, J.; Xu, C.; Zou, X.; Chen, X.; Liu, Y.; Wu, P.; Yang, X.; Zhou, S. DNA barcoding of Oryza: Conventional, specific, and super barcodes. Plant Mol. Biol. 2021, 105, 215–228. [Google Scholar] [CrossRef]
| Samples | Ingredients | Thermal Processing or Additives |
|---|---|---|
| Product 1 | 10% chickpeas, 10% lathyrus seeds, 10% Beluga lentils, 10% red lentils, 10% red beans, 10% black-eyed beans, 10% white beans, 10% green beans, 10% crashed peas, 5% fagopyrum seeds, 5% black beans, 5% borlotti beans | no |
| Product 2 | 40% white beans, 30% borlotti beans, 10% lathyrus seeds, 10% black-eyed beans, 10% mung bean (Vigna radiata) | no |
| Product 3 | 38% white quinoa, 31% millet, 13% red lentils, 13% red quinoa, 4% dehydrated carots, 0.5% dehydrated leeks, Dehydrated parsley | no |
| Product 4 | Durum wheat semolina, 4% dehydrated tomatoes, 3% dehydrated spinach, traces of soya and mustard | yes |
| Product 5 | Red lentils flour | yes |
| Product 6 | 36.5% lentils, red pepper, corn, onion, carrots, zucchini, tomato, extra virgin oil, salt and black pepper | yes |
| Product 7 | 28–30% tomato juice solids, salt | yes |
| Product 8 | 60% peeled plum tomatoes, 40% slightly concentrated tomato juice, acidity regulator, citric acid | yes |
| Product 9 | beans, green beans, carrots, white turnip, salt | yes |
| Product 10 | sun dried tomatoes, sulfur dioxide, salt | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsolakidou, M.-D.; Nikoloudakis, N.; Tisseyre, C.; Knez, M.; Barilli, E.; Mattas, K.; Katsiotis, A. DNA Barcoding for Tracing Biodiversity in Mixed Crop Food Products: A Proof of Concept Within the BioValue Project. Foods 2025, 14, 3256. https://doi.org/10.3390/foods14183256
Tsolakidou M-D, Nikoloudakis N, Tisseyre C, Knez M, Barilli E, Mattas K, Katsiotis A. DNA Barcoding for Tracing Biodiversity in Mixed Crop Food Products: A Proof of Concept Within the BioValue Project. Foods. 2025; 14(18):3256. https://doi.org/10.3390/foods14183256
Chicago/Turabian StyleTsolakidou, Maria-Dimitra, Nikolaos Nikoloudakis, Cyril Tisseyre, Marija Knez, Eleonora Barilli, Konstadinos Mattas, and Andreas Katsiotis. 2025. "DNA Barcoding for Tracing Biodiversity in Mixed Crop Food Products: A Proof of Concept Within the BioValue Project" Foods 14, no. 18: 3256. https://doi.org/10.3390/foods14183256
APA StyleTsolakidou, M.-D., Nikoloudakis, N., Tisseyre, C., Knez, M., Barilli, E., Mattas, K., & Katsiotis, A. (2025). DNA Barcoding for Tracing Biodiversity in Mixed Crop Food Products: A Proof of Concept Within the BioValue Project. Foods, 14(18), 3256. https://doi.org/10.3390/foods14183256










