Stable Isotope Ratio Analysis for the Discrimination of the Geographic Origin of Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Sample Treatment
2.3. EA-IRMS Analysis
2.4. Data Handling and Statistical Analysis
3. Results
Stable Isotope Results for Rice Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| δ15NAIR (‰) | δ13CV-PDB (‰) | δ34SV-CDT (‰) | ||
|---|---|---|---|---|
| δ15NAIR (‰) | Pearson’s r | — | ||
| df | — | |||
| p-value | — | |||
| δ13CV-PDB (‰) | Pearson’s r | −0.114 | — | |
| df | 368 | — | ||
| p-value | 0.029 | — | ||
| δ34SV-CDT (‰) | Pearson’s r | 0.073 | −0.354 | — |
| df | 368 | 368 | — | |
| p-value | 0.163 | <0.001 | — |
| Model | Mean Accuracy | Mean Kappa |
|---|---|---|
| Classification Tree (ctree) | 0.931 | 0.892 |
| Recursive Partitioning (rpart) | 0.895 | 0.837 |
| Random Forest (rf) | 0.962 | 0.941 |
| k-Nearest Neighbors (knn) | 0.961 | 0.940 |
| Partial Least Squares (pls) | 0.869 | 0.798 |
References
- Camin, F.; Boner, M.; Bontempo, L.; Fauhl-Hassek, C.; Kelly, S.D.; Riedl, J.; Rossmann, A. Stable isotope techniques for verifying the declared geographical origin of food in legal cases. Trends Food Sci. Technol. 2017, 61, 176. [Google Scholar] [CrossRef]
- Thomatou, A.A.; Mazarakioti, E.C.; Zotos, A.; Kontogeorgos, A.; Patakas, A.; Ladavos, A. Application of Stable Isotope Analysis for Detecting the Geographical Origin of the Greek Currants “Vostizza”: A Preliminary Study. Foods 2023, 12, 1672. [Google Scholar] [CrossRef]
- Lo Feudo, G.; Naccarato, A.; Sindona, G.; Tagarelli, A. Investigating the origin of tomatoes and triple concentrated tomato pastes through multielement determination by inductively coupled plasma mass spectrometry and statistical analysis. J. Agric. Food Chem. 2010, 58, 3801. [Google Scholar] [CrossRef]
- Longobardi, F.; Casiello, G.; Cortese, M.; Perini, M.; Camin, F.; Catucci, L.; Agostiano, A. Discrimination of geographical origin of lentils (Lens culinaris Medik.) using isotope ratio mass spectrometry combined with chemometrics. Food Chem. 2015, 188, 343. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, J.K.; Prabakaran, M.; Yang, J.H.; Kim, S.H. Authenticity of rice (Oryza sativa L.) geographical origin based on analysis of C, N, O and S stable isotope ratios: A preliminary case report in Korea, China and Philippine. J. Sci. Food Agric. 2016, 96, 2433. [Google Scholar] [CrossRef] [PubMed]
- Kukusamude, C.; Puripunyavanich, V.; Kongsri, S. Combination of light stable isotopic and elemental signatures in Thai Hom Mali rice with chemometric analysis. Food Chem. X 2023, 17, 100613. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.; Marchesan, E. Phenolic compounds and antioxidant activity of rice. Braz. Arch. Biol. Technol. 2011, 54, 371. [Google Scholar] [CrossRef]
- Cheajesadagul, P.; Arnaudguilhem, C.; Shiowatana, J.; Siripinyanond, A.; Szpunar, J. Discrimination of geographical origin of rice based on multi-element fingerprinting by high resolution inductively coupled plasma mass spectrometry. Food Chem. 2013, 141, 3504. [Google Scholar] [CrossRef] [PubMed]
- Sripakdee, T.; Sricharoen, P.; Nateewattana, J.; Chansuvarn, W.; Amnuaywattanakul, T.; Othata, P.; Chanthai, S.; Limchoowong, N. Enhancing ultrasound-assisted extraction for evaluating antioxidant activity and total phenolic contents in Thai Hom Mali rice: A K-means clustering approach. J. Food Meas. Charact. 2025, 19, 5625–5639. [Google Scholar] [CrossRef]
- Kongsri, S.; Sricharoen, P.; Limchoowong, N.; Kukusamude, C. Tracing the Geographical Origin of Thai Hom Mali Rice in Three Contiguous Provinces of Thailand Using Stable Isotopic and Elemental Markers Combined with Multivariate Analysis. Foods 2021, 10, 2349. [Google Scholar] [CrossRef]
- Okonogi, S.; Kaewpinta, A.; Junmahasathien, T.; Yotsawimonwat, S. Effect of rice variety and modification on antioxidant and anti-inflammatory activities. Drug Discov. Ther. 2018, 12, 206. [Google Scholar] [CrossRef]
- Vlachos, A.; Arvanitoyannis, I. A Review of Rice Authenticity/Adulteration Methods and Results. Crit. Rev. Food Sci. Nutr. 2008, 48, 553. [Google Scholar] [CrossRef]
- Suzuki, Y.; Chikaraishi, Y.; Ogawa, N.; Ohkouchi, N.; Korenaga, T. Geographical origin of polished rice based on multiple element and stable isotope analyses. Food Chem. 2008, 109, 470. [Google Scholar] [CrossRef]
- Le Nguyen Doan, D.; Nguyen, Q.C.; Marini, F.; Biancolillo, A. Authentication of Rice (Oryza sativa L.) using Near Infrared Spectroscopy Combined with Different Chemometric Classification Strategies. Appl. Sci. 2021, 11, 362. [Google Scholar] [CrossRef]
- Panero, F.S.; Panero, P.S.; Panero, J.S.; Faria, F.S.E.D.V.; Rodriguez, A.F.R. Rapid and Green Method Forensic Authentication of Rice Using Near-Infrared Spectroscopy (NIRS). J. Agric. Sci. 2020, 12, 105. [Google Scholar] [CrossRef]
- Song, C.; Liu, J.; Wang, C.; Li, Z.; Zhang, D.; Li, P. Rapid identification of adulterated rice based on data fusion of near-infrared spectroscopy and machine vision. J. Food Meas. Charact. 2024, 18, 3881. [Google Scholar] [CrossRef]
- Sha, M.; Gui, D.; Zhang, Z.; Ji, X.; Shi, X.; Liu, J.; Zhang, D. Evaluation of sample pretreatment method for geographic authentication of rice using Raman spectroscopy. Food Meas. Charact. 2019, 13, 1705. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, J.; Wu, G.; Wang, Y.; Zhang, H.; Wang, L.; Qian, H.; Qi, X. Identification of rice varieties and determination of their geographical origin in China using Raman spectroscopy. J. Cereal Sci. 2018, 82, 175. [Google Scholar] [CrossRef]
- Chae, Y.K.; Kim, S.H. Discrimination of Rice Products by Geographical Origins and Cultivars by Two-Dimensional NMR Spectroscopy. Bull. Korean Chem. Soc. 2016, 37, 1612. [Google Scholar] [CrossRef]
- Huo, Y.; Kamal, G.M.; Wang, J.; Liu, H.; Zhang, G.; Hu, Z.; Anwar, F.; Du, H. 1H NMR-based metabolomics for discrimination of rice from different geographical origins of China. J. Cereal Sci. 2017, 76, 243. [Google Scholar] [CrossRef]
- Saeed, M.; Kim, J.S.; Kim, S.Y.; Ryu, J.E.; Ko, J.; Zaidi, S.F.A.; Seo, J.A.; Kim, Y.S.; Lee, D.Y.; Choi, H.K. Differentiation of Geographical Origin of White and Brown Rice Samples Using NMR Spectroscopy Coupled with Machine Learning Techniques. Metabolites 2022, 12, 1012. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, J.K.; Lee, K.J.; Park, S.K.; Lee, J.H.; Son, N.Y.; Jin, Y.I.; Kim, S.H. Geographic authentication of Asian rice (Oryza sativa L.) using multielemental and stable isotopic data combined with multivariate analysis. Food Chem. 2018, 240, 840. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, W.; Zhang, Y.; Chen, T.; Shao, S.; Zhou, L.; Yuan, Y.; Xie, T.; Rogers, K.M. Assuring food safety and traceability of polished rice from different production regions in China and Southeast Asia using chemometric models. Food Control 2019, 99, 1–10. [Google Scholar] [CrossRef]
- Maione, C.; Batista, B.L.; Campiglia, A.D.; Barbos, F., Jr.; Barbosa, R.M. Classification of geographic origin of rice by data mining and inductively coupled plasma mass spectrometry. Comput. Electron. Agric. 2016, 121, 101. [Google Scholar] [CrossRef]
- Wang, J.; Chen, T.; Zhang, W.; Zhao, Y.; Yang, S.; Chen, A. Tracing the geographical origin of rice by stable isotopic analyses combined with chemometrics. Food Chem. 2020, 313, 126093. [Google Scholar] [CrossRef]
- Katerinopoulou, K.; Kontogeorgos, A.; Salmas, C.E.; Patakas, A.; Ladavos, A. Geographical Origin Authentication of Agri-Food Products: A Review. Foods 2020, 9, 489. [Google Scholar] [CrossRef]
- Kelly, S.; Heaton, K.; Hoogewerff, J. Tracing the geographical origin of food: The application of multi-element and multi-isotope analysis. Trends Food Sci. Technol. 2005, 16, 555. [Google Scholar] [CrossRef]
- Gonzalvez, A.; Armenta, S.; De La Guardia, M. Trace-element composition and stable-isotope ratio for discrimination of foods with Protected Designation of Origin. Trends Anal. Chem. 2009, 28, 1295. [Google Scholar] [CrossRef]
- Mahne Opatic, A.; Necemer, M.; Lojen, S.; Masten, J.; Zlatic, E.; Šircelj, H.; Stopar, D.; Vidrih, R. Determination of geographical origin of commercial tomato through analysis of stable isotopes, elemental composition and chemical markers. Food Control 2018, 89, 133. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, S.-H.; Bong, Y.-S. Geographical origin authentication of onions using stable isotope ratio and compositions of C, H, O, N, and S. Food Control 2019, 101, 121. [Google Scholar] [CrossRef]
- Wu, Y.; Luo, D.; Dong, H.; Wan, J.; Luo, H.; Xian, Y.; Guo, X.; Qin, F.; Han, W.; Wang, L.; et al. Geographical origin of cereal grains based on element analyser-stable isotope ratio mass spectrometry (EA-SIRMS). Food Chem. 2015, 174, 553. [Google Scholar] [CrossRef]
- Kukusamude, C.; Kongsri, S. Investigating differences in light stable isotopes between Thai jasmine rice and Sungyod rice. J. Phys. Conf. Ser. 2017, 901, 012046. [Google Scholar] [CrossRef]
- Thomatou, A.-A.; Mazarakioti, E.C.; Zotos, A.; Kokkotos, E.; Kontogeorgos, A.; Patakas, A.; Ladavos, A. Stable Isotope Ratio Analysis for the Geographic Origin Discrimination of Greek Beans “Gigantes-Elefantes” (Phaseolus coccineus L.). Foods 2024, 13, 2107. [Google Scholar] [CrossRef] [PubMed]
- Field, A. Discovering Statistics Using SPSS; SAGE Publication: London, UK, 2005. [Google Scholar] [CrossRef]
- Mazarakioti, E.C.; Zotos, A.; Verykios, V.S.; Kokkotos, E.; Thomatou, A.A.; Kontogeorgos, A.; Patakas, A.; Ladavos, A. Multi-Elemental Analysis and Geographical Discrimination of Greek “Gigantes Elefantes” Beans Utilizing Inductively Coupled Plasma Mass Spectrometry and Machine Learning Models. Foods 2024, 13, 3015. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81. [Google Scholar] [CrossRef]
- Rokach, L.; Maimon, O. Decision trees. In Data Mining and Knowledge Discovery Handbook; Maimon, O., Rokach, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 165–192. [Google Scholar] [CrossRef]
- Kelly, S.; Baxter, M.; Chapman, S.; Rhodes, C.; Dennis, J.; Brereton, P. The application of isotopic and elemental analysis to determine the geographical origin of premium long grain rice. Eur. Food Res. Technol. 2002, 214, 72. [Google Scholar] [CrossRef]
- Kongsri, S.; Sola, P.; Kukusamude, C. A pilot study of stable isotope ratio and elemental compositions in Leb Nok rice in the south of Thailand. J. Phys. Conf. Ser. 2018, 1144, 012167. [Google Scholar] [CrossRef]
- Korenaga, T.; Musashi, M.; Nakashita, R.; Suzuki, Y. Statistical Analysis of Rice Samples for Compositions of Multiple Light Elements (H, C, N, and O) and Their Stable Isotopes. Anal. Sci. 2010, 26, 873. [Google Scholar] [CrossRef] [PubMed]
- Kukusamude, C.; Kongsri, S. Elemental and isotopic profiling of Thai jasmine rice (Khao Dawk Mali 105) for discrimination of geographical origins in Thung Kula Rong Hai area, Thailand. Food Control 2018, 91, 357. [Google Scholar] [CrossRef]
- Li, A.; Keely, B.; Chan, S.H.; Baxter, M.; Rees, G.; Kelly, S. Verifying the provenance of rice using stable isotope ratio and multi-element analyses: A feasibility study. Qual. Assur. Saf. Crop. Foods. 2015, 7, 343. [Google Scholar] [CrossRef]
- Zhi, L.; Yuan, W.; Yudi, H.; Wei, L.; Bin, L.; Guiyuan, M. Multi-stable isotope and multi-element origin traceability of rice from the main producing regions in Asia: A long-term investigation during 2017–2020. Food Chem. 2023, 412, 135417. [Google Scholar] [CrossRef] [PubMed]
- Wadood, S.A.; Chunlin, L.; Nie, J.; Rogers, K.M.; Mei, H.; Zhang, Y.; Shah, I.; Qamar, A.; Yuan, Y. Stable isotopic fingerprinting of authentic basmati rice from Pakistan. Food Control 2024, 157, 110166. [Google Scholar] [CrossRef]
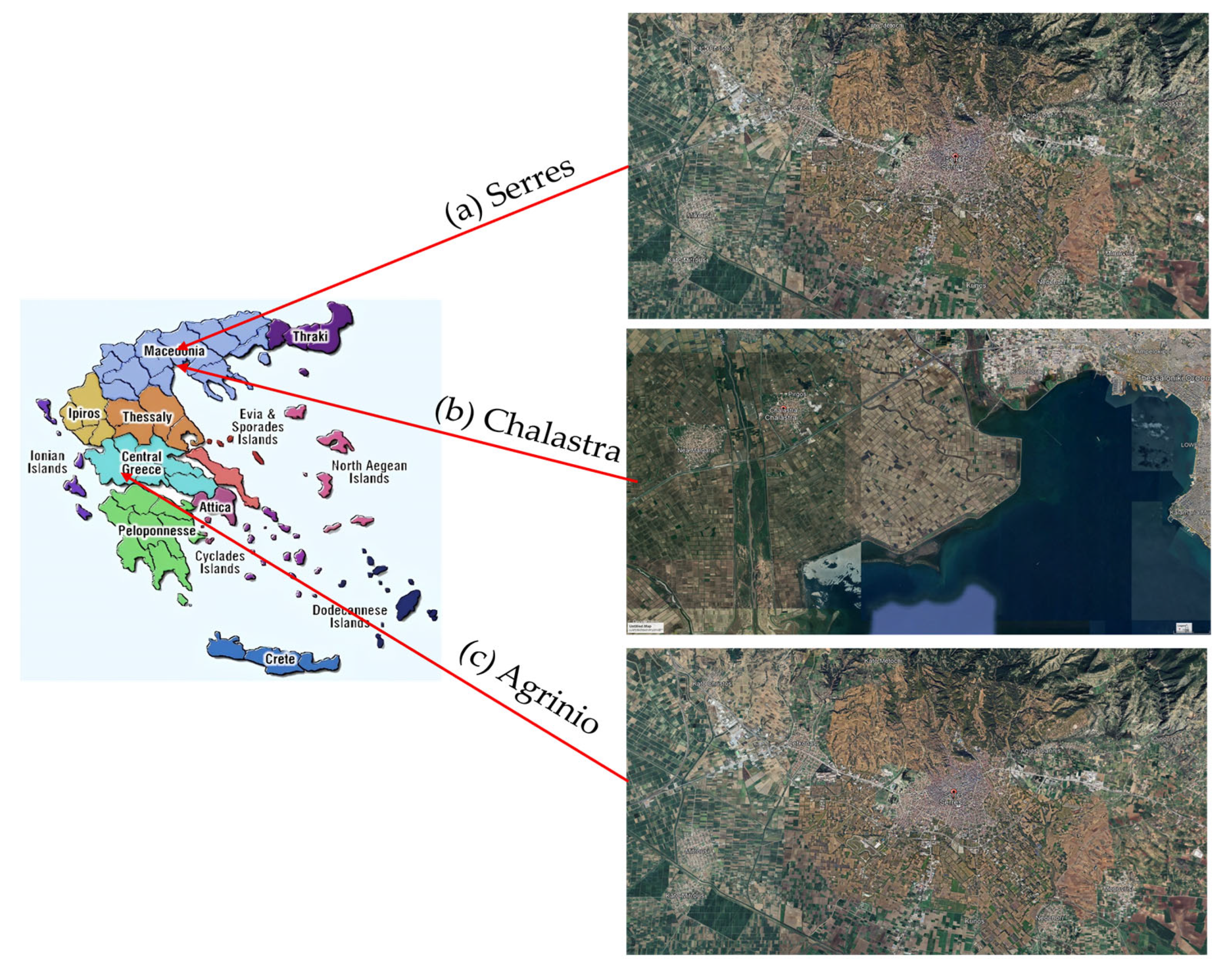

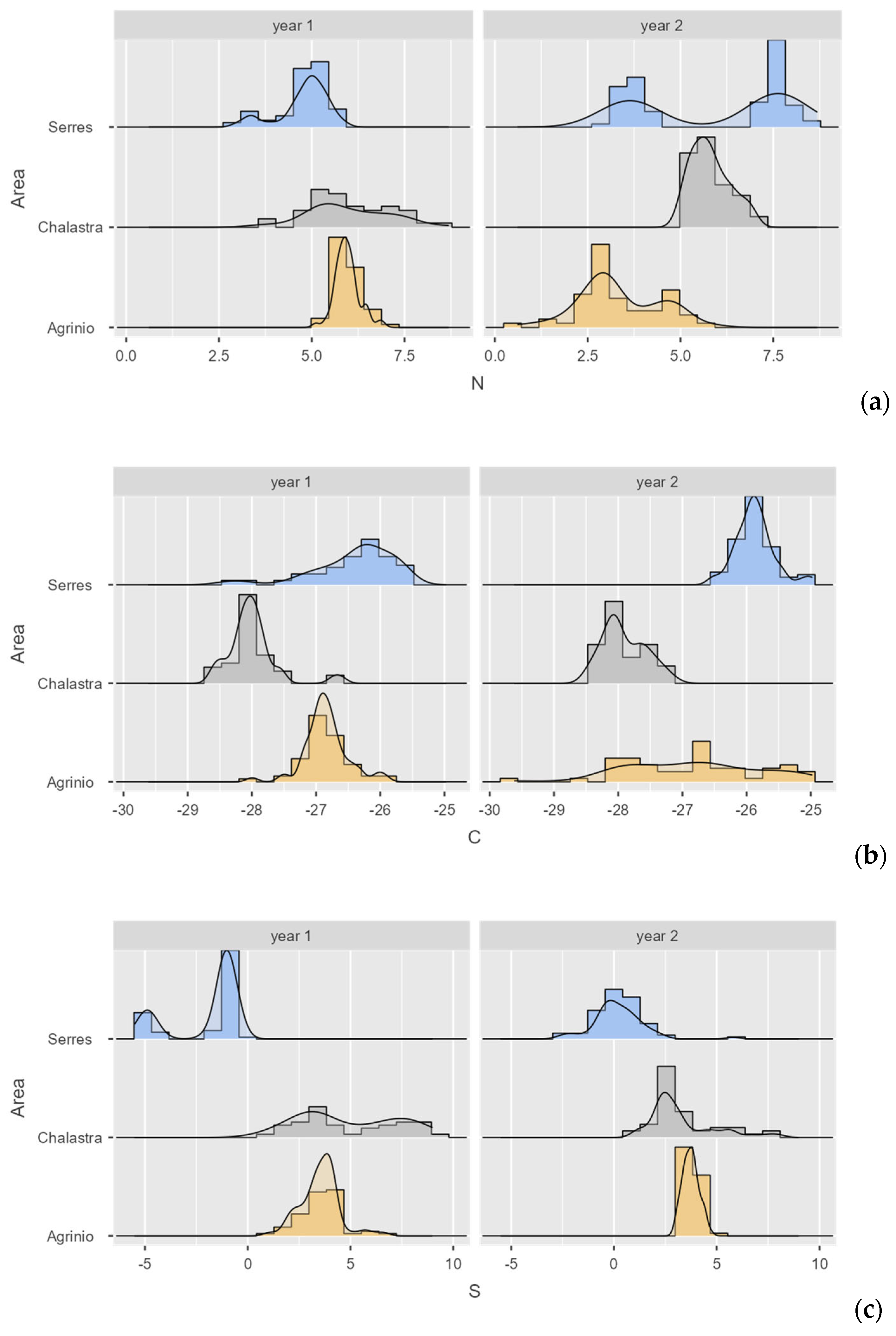
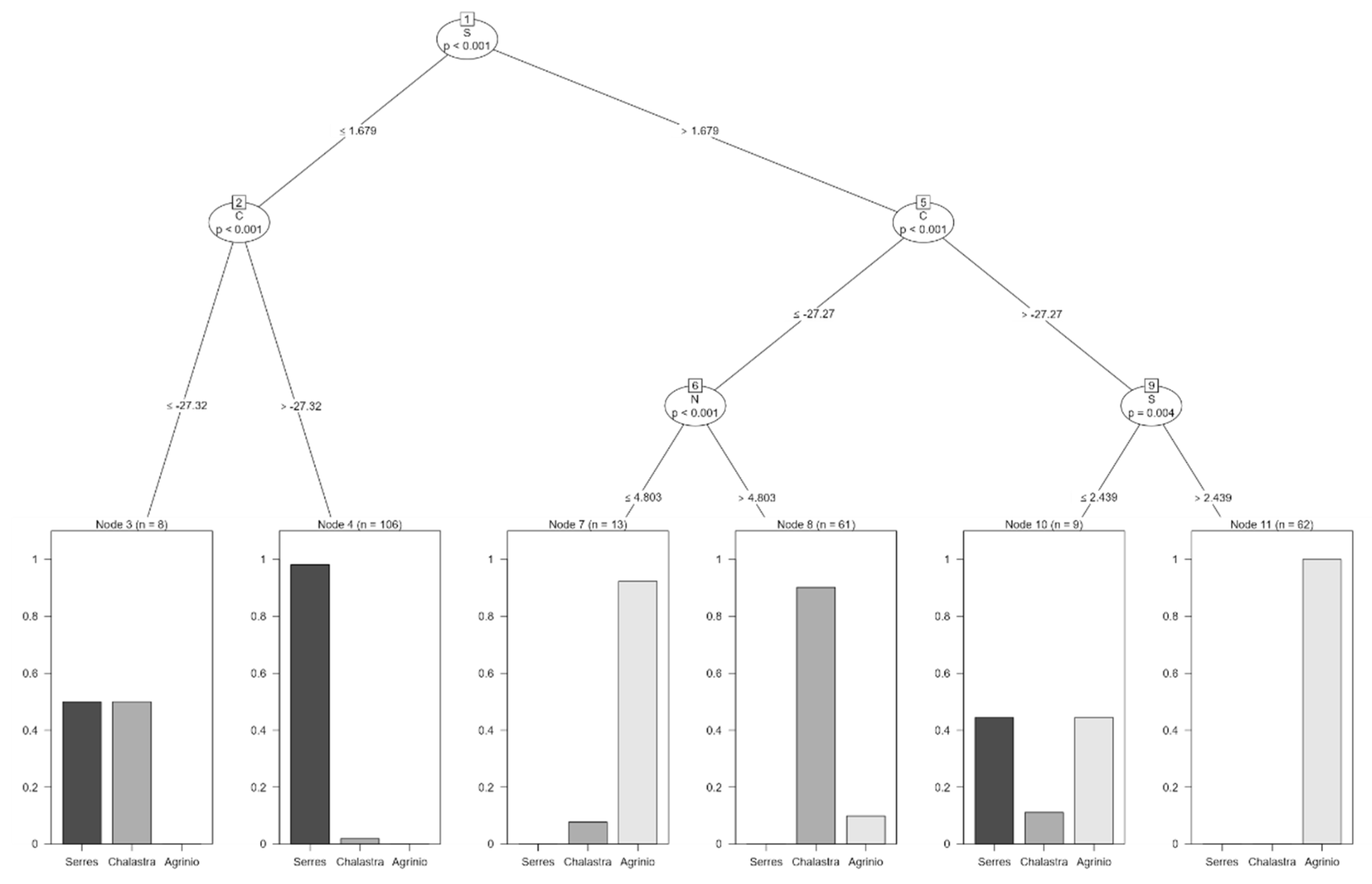
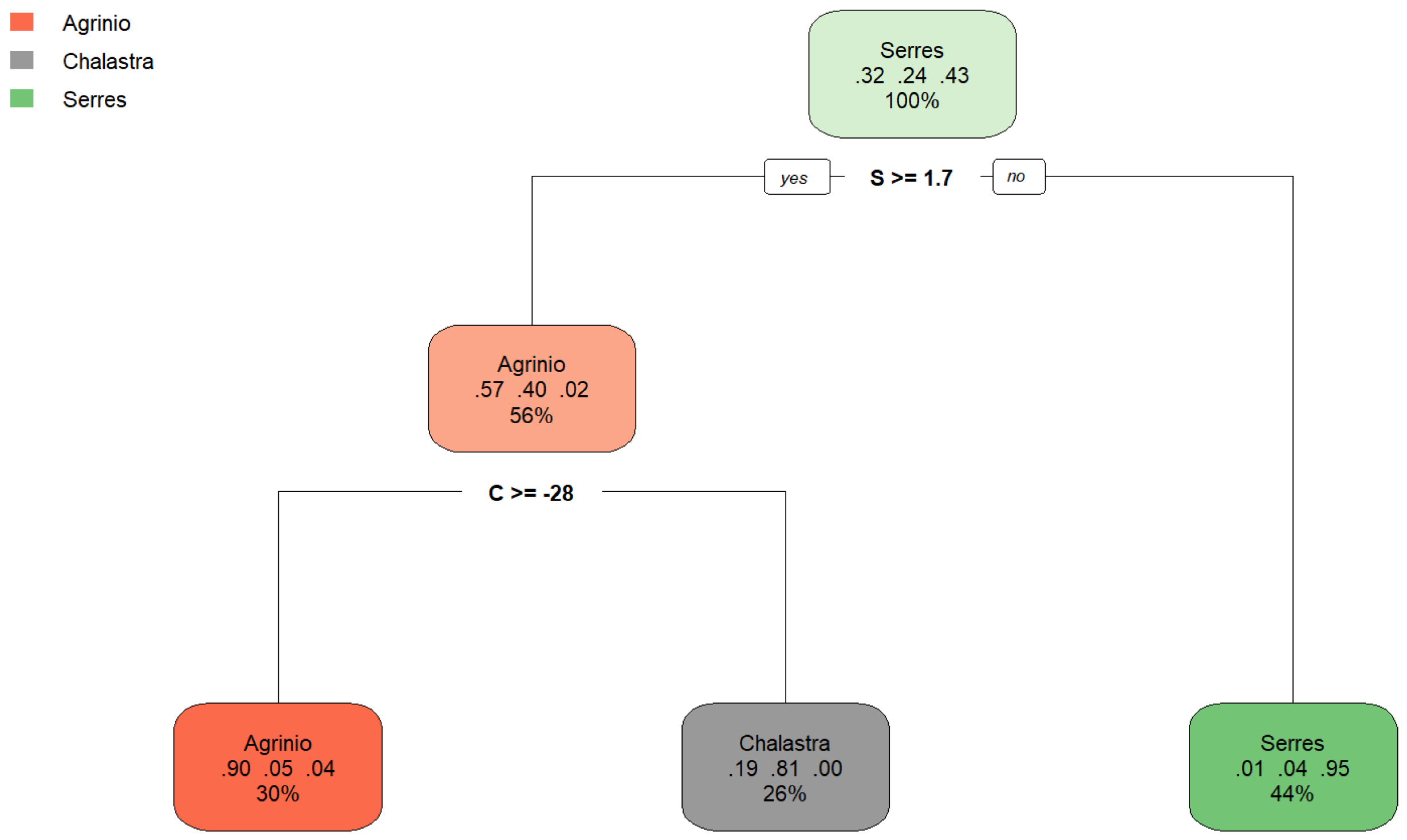
| δ (‰) | Area | N | Mean δ (‰) | Std. Error | Year |
|---|---|---|---|---|---|
| δ15NAIR (‰) | Agrinio | 60 | 5.95 | 0.323 | 2021 |
| Serres | 80 | 4.79 | 0.672 | 2021 | |
| Chalastra | 43 | 6.00 | 1.09 | 2023 | |
| δ13CV-PDB (‰) | Agrinio | 60 | −26.8 | 0.344 | 2021 |
| Serres | 80 | −26.4 | 0.628 | 2021 | |
| Chalastra | 43 | −28.0 | 0.387 | 2023 | |
| δ34SV-CDT (‰) | Agrinio | 60 | 3.51 | 0.947 | 2021 |
| Serres | 80 | −1.99 | 1.71 | 2021 | |
| Chalastra | 43 | 4.86 | 2.36 | 2023 | |
| δ15NAIR (‰) | Agrinio | 60 | 3.34 | 1.08 | 2022 |
| Serres | 80 | 5.88 | 2.02 | 2022 | |
| Chalastra | 47 | 5.81 | 0.523 | 2024 | |
| δ13CV-PDB (‰) | Agrinio | 60 | −26.8 | 1.01 | 2022 |
| Serres | 80 | −25.9 | 0.302 | 2022 | |
| Chalastra | 47 | −27.9 | 0.333 | 2024 | |
| δ34SV-CDT (‰) | Agrinio | 60 | 3.74 | 0.391 | 2022 |
| Serres | 80 | 0.183 | 1.21 | 2022 | |
| Chalastra | 47 | 3.23 | 1.57 | 2024 |
| Univariate Test | Dependent Variable δ (‰) | Sum of Squares | df | Mean Square | F Value | Sig. |
|---|---|---|---|---|---|---|
| Area | δ15NAIR (‰) | 82.82 | 2 | 41.412 | 30.33 | <0.001 |
| δ13CV-PDB (‰) | 189.72 | 2 | 94.861 | 295.71 | <0.001 | |
| δ34SV-CDT (‰) | 2005.74 | 2 | 1002.868 | 482.84 | <0.001 | |
| Year | δ15NAIR (‰) | 16.35 | 1 | 16.354 | 11.98 | <0.001 |
| δ13CV-PDB (‰) | 6.12 | 1 | 6.117 | 19.07 | <0.001 | |
| δ34SV-CDT (‰) | 35.22 | 1 | 35.223 | 16.96 | <0.001 | |
| Area x Year | δ15NAIR (‰) | 237.23 | 2 | 118.617 | 86.87 | <0.001 |
| δ13CV-PDB (‰) | 5.39 | 2 | 2.697 | 8.41 | <0.001 | |
| δ34SV-CDT (‰) | 214.45 | 2 | 107.227 | 51.63 | <0.001 | |
| Residuals | δ15NAIR (‰) | 497.03 | 364 | 1.365 | ||
| δ13CV-PDB (‰) | 116.77 | 364 | 0.321 | |||
| δ34SV-CDT (‰) | 756.04 | 364 | 2.077 |
| Value | F Value | DF1 | Df2 | Sig. | |
|---|---|---|---|---|---|
| Area | 1.122 | 154.7 | 6 | 726 | <0.001 |
| Year | 0.109 | 14.7 | 3 | 362 | <0.001 |
| Area x Year | 0.538 | 44.5 | 6 | 726 | <0.001 |
| Accuracy and kappa for the decision tree model | |||
| (A) With training set | Lower | Upper | |
| Accuracy 95% CI | 0.931 | 0.892 | 0.958 |
| Kappa | 0.892 | ||
| (B) With test set | Lower | Upper | |
| Accuracy 95% CI | 0.919 | 0.852 | 0.962 |
| Kappa | 0.874 | ||
| Prediction evaluation of the decision tree model | |||
| (A) With training set | |||
| Predicted | |||
| Serres | Chalastra | Agrinio | |
| Serres | 112 | 7 | 4 |
| Chalastra | 0 | 55 | 6 |
| Agrinio | 0 | 1 | 74 |
| (B) With test set | |||
| Predicted | |||
| Serres | Chalastra | Agrinio | |
| Serres | 47 | 1 | 4 |
| Chalastra | 0 | 24 | 1 |
| Agrinio | 1 | 2 | 31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomatou, A.-A.; Mazarakioti, E.C.; Zotos, A.; Kontogeorgos, A.; Patakas, A.; Ladavos, A. Stable Isotope Ratio Analysis for the Discrimination of the Geographic Origin of Rice (Oryza sativa L.). Foods 2025, 14, 3163. https://doi.org/10.3390/foods14183163
Thomatou A-A, Mazarakioti EC, Zotos A, Kontogeorgos A, Patakas A, Ladavos A. Stable Isotope Ratio Analysis for the Discrimination of the Geographic Origin of Rice (Oryza sativa L.). Foods. 2025; 14(18):3163. https://doi.org/10.3390/foods14183163
Chicago/Turabian StyleThomatou, Anna-Akrivi, Eleni C. Mazarakioti, Anastasios Zotos, Achilleas Kontogeorgos, Angelos Patakas, and Athanasios Ladavos. 2025. "Stable Isotope Ratio Analysis for the Discrimination of the Geographic Origin of Rice (Oryza sativa L.)" Foods 14, no. 18: 3163. https://doi.org/10.3390/foods14183163
APA StyleThomatou, A.-A., Mazarakioti, E. C., Zotos, A., Kontogeorgos, A., Patakas, A., & Ladavos, A. (2025). Stable Isotope Ratio Analysis for the Discrimination of the Geographic Origin of Rice (Oryza sativa L.). Foods, 14(18), 3163. https://doi.org/10.3390/foods14183163






