Comparing a Whole Grain Blend with Polished White Rice for Starch Digestibility and Gut Microbiota Fermentation in Diabetic Patients: An In Vitro Study
Abstract
1. Introduction
2. Experimental
2.1. Materials
2.2. In Vitro Simulated Digestion
2.2.1. Sample Preparation
2.2.2. Static Digestion Procedure
2.2.3. Starch Hydrolysis and Glycemic Index Estimation
2.2.4. Dialysis of Digested Products
2.3. In Vitro Fecal Fermentation
2.3.1. Participant Recruitment and Sample Collection
2.3.2. Fermentation Procedure
2.4. Short-Chain Fatty Acid (SCFA) Analysis
2.5. Non-Starch Polysaccharides (NSPs) Degradation Products Quantification
2.6. Metagenomic Analysis of Gut Microbiota
2.7. Statistical Analysis
3. Results and Discussion
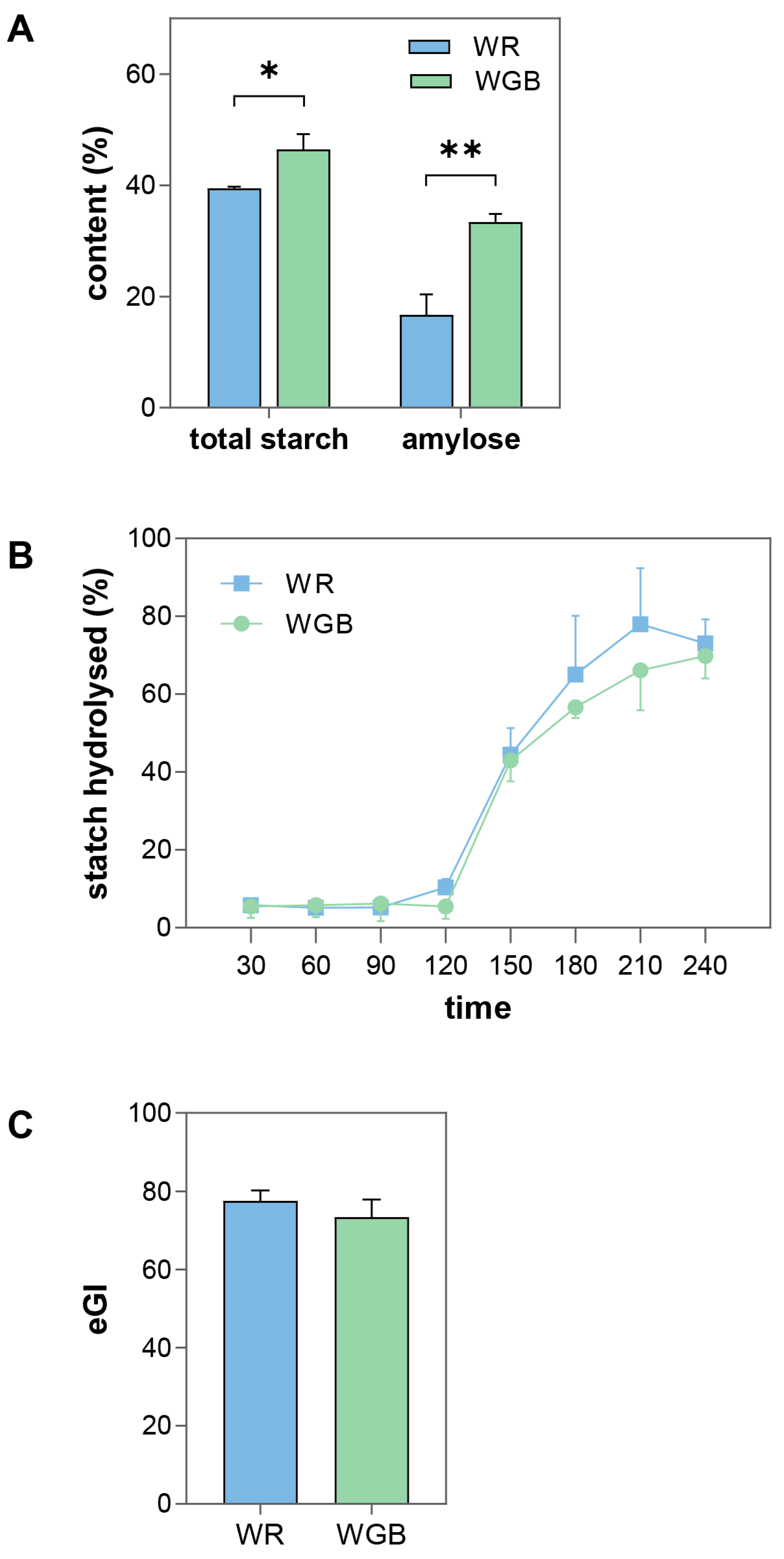
3.1. WGB Had Higher Amylose Content and Reduced Starch Digestibility and Glycemic Response Compared to WR
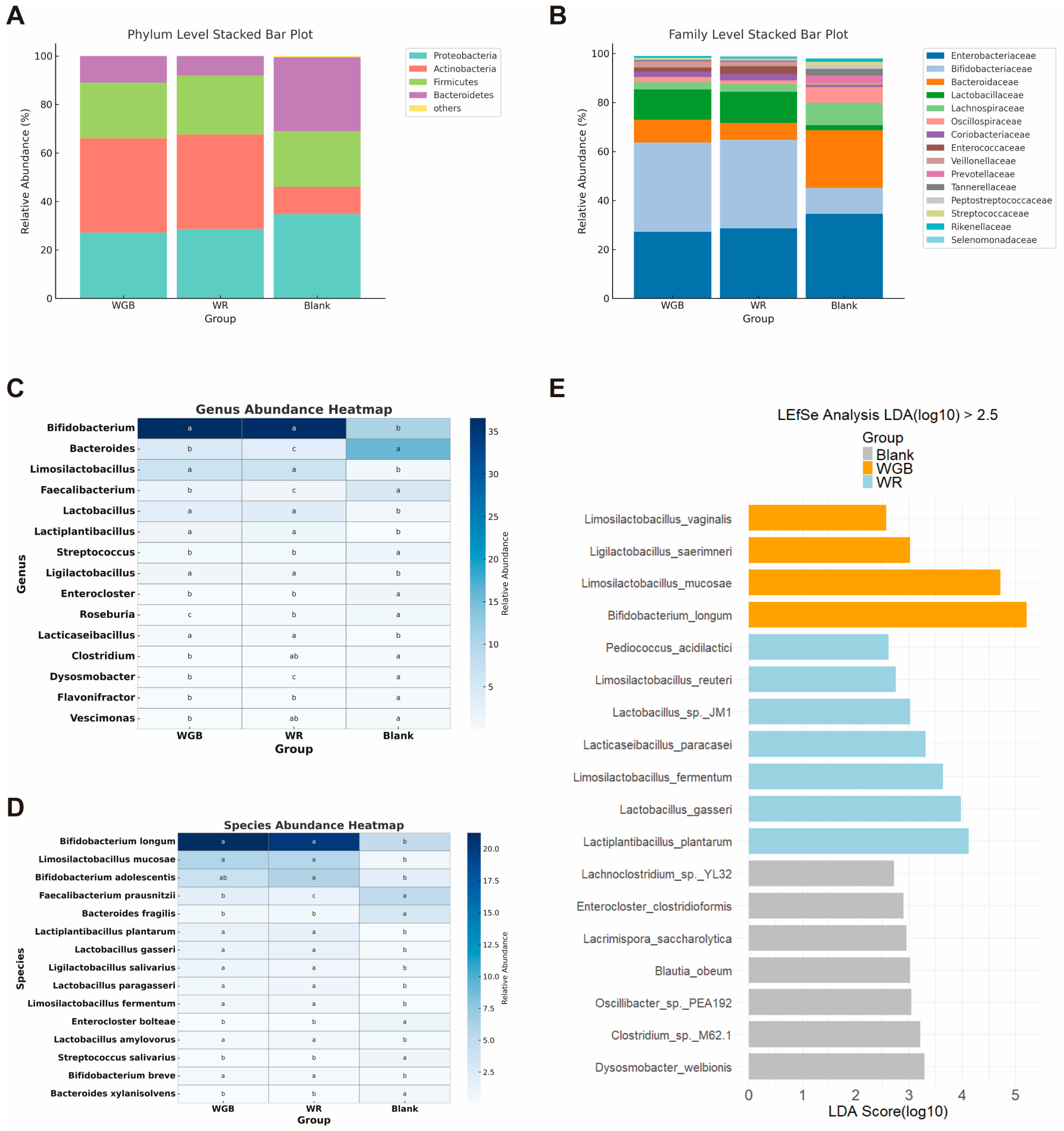
3.2. WGB Restored Microbial Diversity and Enriched Beneficial Taxa
3.2.1. α-Diversity and β-Diversity Analysis
3.2.2. Taxonomic Shifts in Gut Microbiota
3.3. WGB Modulated Microbial Metabolism: SCFA Production and NSP Degradation
3.3.1. SCFA Production and Functional Implications
3.3.2. NSP Degradation and Fermentable Sugar Composition
3.4. WGB Altered Gut Microbial Function: KEGG and CAZyme Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, E.; Lim, S.; Lamptey, R.; Webb, D.R.; Davies, M.J. Type 2 Diabetes. Lancet 2022, 400, 1803–1820. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Su, L.; Liang, B.; Tan, J.; Chen, Q.; Long, J.; Xie, J.; Wu, G.; Yan, Y.; Guo, X.; et al. Trends in Prevalence, Awareness, Treatment, and Control of Diabetes Mellitus in Mainland China from 1979 to 2012. Int. J. Endocrinol. 2013, 2013, 753150. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, J.; He, Y.; Jiang, B.; Wu, L.; Wang, Y.; Di, Z.; Zeng, J. Awareness, Treatment and Control of Type 2 Diabetes among Chinese Elderly and Its Changing Trend for Past Decade. BMC Public Health 2016, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wu, T.; Zhang, M.; Li, C.; Liu, Q.; Li, F. Prevalence, Awareness and Control of Type 2 Diabetes Mellitus and Risk Factors in Chinese Elderly Population. BMC Public Health 2022, 22, 1382. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, F.S.; Brand-Miller, J.C.; Foster-Powell, K.; Buyken, A.E.; Goletzke, J. International Tables of Glycemic Index and Glycemic Load Values 2021: A Systematic Review. Am. J. Clin. Nutr. 2021, 114, 1625–1632. [Google Scholar] [CrossRef] [PubMed]
- Cousin, E.; Schmidt, M.I.; Ong, K.L.; Lozano, R.; Afshin, A.; Abushouk, A.I.; Agarwal, G.; Agudelo-Botero, M.; Al-Aly, Z.; Alcalde-Rabanal, J.E.; et al. Burden of Diabetes and Hyperglycaemia in Adults in the Americas, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet Diabetes Endocrinol. 2022, 10, 655–667. [Google Scholar] [CrossRef] [PubMed]
- O’Hearn, M.; Lara-Castor, L.; Cudhea, F.; Miller, V.; Reedy, J.; Shi, P.; Zhang, J.; Wong, J.B.; Economos, C.D.; Micha, R.; et al. Incident Type 2 Diabetes Attributable to Suboptimal Diet in 184 Countries. Nat. Med. 2023, 29, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Venn, B.J.; Green, T.J. Glycemic Index and Glycemic Load: Measurement Issues and Their Effect on Diet-Disease Relationships. Eur. J. Clin. Nutr. 2007, 61 (Suppl. S1), S122–S131. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Qi, J.; Zou, Y. Association between Intake of White Rice and Incident Type 2 Diabetes—An Updated Meta-Analysis. Diabetes Res. Clin. Pract. 2021, 172, 108651. [Google Scholar] [CrossRef] [PubMed]
- P., N.P.V.; Joye, I.J. Dietary Fibre from Whole Grains and Their Benefits on Metabolic Health. Nutrients 2020, 12, 3045. [Google Scholar] [CrossRef]
- Hashimoto, M.; Hossain, S.; Matsuzaki, K.; Shido, O.; Yoshino, K. The Journey from White Rice to Ultra-High Hydrostatic Pressurized Brown Rice: An Excellent Endeavor for Ideal Nutrition from Staple Food. Crit. Rev. Food Sci. Nutr. 2022, 62, 1502–1520. [Google Scholar] [CrossRef] [PubMed]
- Rebeira, S.P.; Prasantha, B.D.R.; Jayatilake, D.V.; Dunuwila, G.R.; Piyasiri, C.H.; Herath, H.M.K.W.P. A Comparative Study of Dietary Fiber Content, In Vitro Starch Digestibility and Cooking Quality Characteristics of Pigmented and Non–Pigmented Traditional and Improved Rice (Oryza sativa L.). Food Res. Int. 2022, 157, 111389. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ding, M.; Sampson, L.; Willett, W.C.; Manson, J.E.; Wang, M.; Rosner, B.; Hu, F.B.; Sun, Q. Intake of Whole Grain Foods and Risk of Type 2 Diabetes: Results from Three Prospective Cohort Studies. BMJ 2020, 370, m2206. [Google Scholar] [CrossRef] [PubMed]
- Åberg, S.; Mann, J.; Neumann, S.; Ross, A.B.; Reynolds, A.N. Whole-Grain Processing and Glycemic Control in Type 2 Diabetes: A Randomized Crossover Trial. Diabetes Care 2020, 43, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Baars, D.P.; Fondevila, M.F.; Meijnikman, A.S.; Nieuwdorp, M. The Central Role of the Gut Microbiota in the Pathophysiology and Management of Type 2 Diabetes. Cell Host Microbe 2024, 32, 1280–1300. [Google Scholar] [CrossRef]
- Mei, Z.; Wang, F.; Bhosle, A.; Dong, D.; Mehta, R.; Ghazi, A.; Zhang, Y.; Liu, Y.; Rinott, E.; Ma, S.; et al. Strain-Specific Gut Microbial Signatures in Type 2 Diabetes Revealed by a Cross-Cohort Analysis of 8,117 Metagenomes. Nat. Med. 2024, 30, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; O’Mahony, A.K.; Dos Santos, R.S.; Marroquí, L.; Cotter, P.D. Gut Microbial Metabolic Signatures in Diabetes Mellitus and Potential Preventive and Therapeutic Applications. Gut Microbes 2024, 16, 2401654. [Google Scholar] [CrossRef] [PubMed]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short Chain Fatty Acids in Human Gut and Metabolic Health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T. How Important Are Fatty Acids in Human Health and Can They Be Used in Treating Diseases? Gut Microbes 2024, 16, 2420765. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’Toole, P.W.; O’Connor, E.M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Mok, B.; Chung, H.-J.; Park, H.Y.; Kim, H.-S. Heat-Treated Brown Rice Starch Structure and Effect on Short-Chain Fatty Acids and Mouse Intestinal Microbiota. Int. J. Biol. Macromol. 2024, 283, 137597. [Google Scholar] [CrossRef] [PubMed]
- Aalim, H.; Wang, D.; Luo, Z. Black Rice (Oryza sativa L.) Processing: Evaluation of Physicochemical Properties, in Vitro Starch Digestibility, and Phenolic Functions Linked to Type 2 Diabetes. Food Res. Int. 2021, 141, 109898. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Ren, C.; Li, L.; Huang, C.; Zhao, Q.; Zhong, Y.; Hu, Q.; Liao, W.; Xia, H.; Yang, L.; et al. Effects of Germinated Brown Rice and Germinated Black Rice on People with Type 2 Diabetes Mellitus Combined with Dyslipidaemia. Food Funct. 2024, 15, 6642–6656. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST Static in Vitro Simulation of Gastrointestinal Food Digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, W.; Chen, H.; Fang, X.; Han, Y.; Xie, M.; Gao, H. In Vitro Fecal Fermentation Characteristics of Bamboo Shoot (Phyllostachys Edulis) Polysaccharide. Food Chem. X 2021, 11, 100129. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Van Haute, M.J.; Rose, D.J. Processing Has Differential Effects on Microbiota-Accessible Carbohydrates in Whole Grains during In Vitro Fermentation. Appl. Environ. Microbiol. 2020, 86, e01705-20. [Google Scholar] [CrossRef] [PubMed]
- Qadir, N.; Wani, I.A. In-Vitro Digestibility of Rice Starch and Factors Regulating Its Digestion Process: A Review. Carbohydr. Polym. 2022, 291, 119600. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A Standardised Static in Vitro Digestion Method Suitable for Food—An International Consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tan, Y.; McClements, D.J. Applications of the INFOGEST In Vitro Digestion Model to Foods: A Review. Annu. Rev. Food Sci. Technol. 2023, 14, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Nadia, J.; Bronlund, J.; Singh, R.P.; Singh, H.; Bornhorst, G.M. Structural Breakdown of Starch-Based Foods during Gastric Digestion and Its Link to Glycemic Response: In Vivo and in Vitro Considerations. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2660–2698. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.-M.; Madalena, D.A.; Pinheiro, A.C.; Vicente, A.A. Rice in Vitro Digestion: Application of INFOGEST Harmonized Protocol for Glycemic Index Determination and Starch Morphological Study. J. Food Sci. Technol. 2020, 57, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, J.; Sun, Y.; Ji, H.; Liu, F.; Peng, X.; Zhong, Y.; Geng, F.; Nie, S. In Vitro Digestion of Eight Types of Wholegrains and Their Dietary Recommendations for Different Populations. Food Chem. 2022, 370, 131069. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Dou, B.; Pugh, J.E.; Lett, A.M.; Frost, G.S. The Impact of Starchy Food Structure on Postprandial Glycemic Response and Appetite: A Systematic Review with Meta-Analysis of Randomized Crossover Trials. Am. J. Clin. Nutr. 2021, 114, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chen, Y.; Feng, P.; Shen, J.; Fan, X.; Chen, Y.; Yu, W. Fine Structure of Starch Biomacromolecules and Digestibility: The Regulative Role of Amylose and Amylopectin in the Digestive Hydrolysis of Starch in Rice. Carbohydr. Polym. 2025, 350, 123040. [Google Scholar] [CrossRef] [PubMed]
- Tiozon, R.N., Jr.; Sartagoda, K.J.D.; Fernie, A.R.; Sreenivasulu, N. The Nutritional Profile and Human Health Benefit of Pigmented Rice and the Impact of Post-Harvest Processes and Product Development on the Nutritional Components: A Review. Crit. Rev. Food Sci. Nutr. 2023, 63, 3867–3894. [Google Scholar] [CrossRef] [PubMed]
- Ratseewo, J.; Warren, F.J.; Siriamornpun, S. The Influence of Starch Structure and Anthocyanin Content on the Digestibility of Thai Pigmented Rice. Food Chem. 2019, 298, 124949. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Xu, Y.; Jia, C.; Zhang, B.; Niu, M.; Zhao, S. Structural Changes of Rice Starch and Activity Inhibition of Starch Digestive Enzymes by Anthocyanins Retarded Starch Digestibility. Carbohydr. Polym. 2021, 261, 117841. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Rani, R.; Awana, M.; Pitale, D.; Kulshreshta, A.; Sharma, S.; Bollinedi, H.; Singh, A.; Singh, B.; Singh, A.K.; et al. Role of Nutraceutical Starch and Proanthocyanidins of Pigmented Rice in Regulating Hyperglycemia: Enzyme Inhibition, Enhanced Glucose Uptake and Hepatic Glucose Homeostasis Using In Vitro Model. Food Chem. 2021, 335, 127505. [Google Scholar] [CrossRef] [PubMed]
- Kraithong, S.; Theppawong, A.; Bunyameen, N.; Zhang, X.; Huang, R. Advancements in Understanding Low Starch Hydrolysis in Pigmented Rice: A Comprehensive Overview of Mechanisms. Food Chem. 2024, 439, 138079. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Devkota, L.; Flanagan, B.M.; Gu, Z.; Zhang, B.; Dhital, S. Plant Cell Wall Composition Modulates the Gut Microbiota and Metabolites in in-Vitro Fermentation. Carbohydr. Polym. 2023, 316, 121074. [Google Scholar] [CrossRef] [PubMed]
- Petakh, P.; Oksenych, V.; Kamyshnyi, A. The F/B Ratio as a Biomarker for Inflammation in COVID-19 and T2D: Impact of Metformin. Biomed. Pharmacother. 2023, 163, 114892. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Pan, T.; Li, L.; Wang, H.; Zhu, J.; Zhang, H.; Zhao, J.; Chen, W.; Lu, W. Bifidobacterium Longum Mediated Tryptophan Metabolism to Improve Atopic Dermatitis via the Gut-Skin Axis. Gut Microbes 2022, 14, 2044723. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Mao, B.; Tang, X.; Zhang, Q.; Zhao, J.; Zhang, H.; Chen, W.; Cui, S. Improvement of Inflammatory Bowel Disease by Lactic Acid Bacteria-Derived Metabolites: A Review. Crit. Rev. Food Sci. Nutr. 2025, 65, 1261–1278. [Google Scholar] [CrossRef] [PubMed]
- Gavzy, S.J.; Kensiski, A.; Lee, Z.L.; Mongodin, E.F.; Ma, B.; Bromberg, J.S. Bifidobacterium Mechanisms of Immune Modulation and Tolerance. Gut Microbes 2023, 15, 2291164. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Lin, Y.; Wu, Y.; Zhou, J.; Cao, L.; Chen, J.; Li, Y.; Tan, N.; Zhong, S. Bacteroides Fragilis Supplementation Deteriorated Metabolic Dysfunction, Inflammation, and Aorta Atherosclerosis by Inducing Gut Microbiota Dysbiosis in Animal Model. Nutrients 2022, 14, 2199. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Zhang, T.; Kang, S. Fecal Microbiota Composition, Their Interactions, and Metagenome Function in US Adults with Type 2 Diabetes According to Enterotypes. Int. J. Mol. Sci. 2023, 24, 9533. [Google Scholar] [CrossRef] [PubMed]
- Magdy Wasfy, R.; Mbaye, B.; Borentain, P.; Tidjani Alou, M.; Murillo Ruiz, M.L.; Caputo, A.; Andrieu, C.; Armstrong, N.; Million, M.; Gerolami, R. Ethanol-Producing Enterocloster Bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case–Control Culturomics Study. Microorganisms 2023, 11, 2437. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial Transformations of Human Bile Acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Álvarez-Quintero, R.; Velásquez-Mejía, E.P.; Sierra, J.A.; Corrales-Agudelo, V.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Higher Fecal Short-Chain Fatty Acid Levels Are Associated with Gut Microbiome Dysbiosis, Obesity, Hypertension and Cardiometabolic Disease Risk Factors. Nutrients 2018, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient Metabolism by the Human Gut Microbiome: Major Fermentation by-Products and Their Impact on Host Health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.K.; Rossi, M.; Bajka, B.; Whelan, K. Dietary Fibre in Gastrointestinal Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Leschonski, K.P.; Mortensen, M.S.; Hansen, L.B.S.; Krogh, K.B.R.M.; Kabel, M.A.; Laursen, M.F. Structure-Dependent Stimulation of Gut Bacteria by Arabinoxylo-Oligosaccharides (AXOS): A Review. Gut Microbes 2024, 16, 2430419. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q.; Hu, J.; Chen, H.; Geng, F.; Nie, S. Arabinoxylan Ameliorates Type 2 Diabetes by Regulating the Gut Microbiota and Metabolites. Food Chem. 2022, 371, 131106. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of Energy Metabolism by Long-Chain Fatty Acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Q.; Fu, R.; Zhao, M.; Xu, M. Comparing a Whole Grain Blend with Polished White Rice for Starch Digestibility and Gut Microbiota Fermentation in Diabetic Patients: An In Vitro Study. Foods 2025, 14, 2557. https://doi.org/10.3390/foods14152557
Du Q, Fu R, Zhao M, Xu M. Comparing a Whole Grain Blend with Polished White Rice for Starch Digestibility and Gut Microbiota Fermentation in Diabetic Patients: An In Vitro Study. Foods. 2025; 14(15):2557. https://doi.org/10.3390/foods14152557
Chicago/Turabian StyleDu, Qian, Ruisheng Fu, Ming Zhao, and Meihong Xu. 2025. "Comparing a Whole Grain Blend with Polished White Rice for Starch Digestibility and Gut Microbiota Fermentation in Diabetic Patients: An In Vitro Study" Foods 14, no. 15: 2557. https://doi.org/10.3390/foods14152557
APA StyleDu, Q., Fu, R., Zhao, M., & Xu, M. (2025). Comparing a Whole Grain Blend with Polished White Rice for Starch Digestibility and Gut Microbiota Fermentation in Diabetic Patients: An In Vitro Study. Foods, 14(15), 2557. https://doi.org/10.3390/foods14152557






