Dissecting the High Esterase/Lipase Activity and Probiotic Traits in Lactiplantibacillus plantarum B22: A Genome-Guided Functional Characterization
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Strains
2.2. Strains and Growth Conditions
2.3. Screening Esterases/Lipases-Producing Strains
2.4. Genome Sequencing and Analysis
2.4.1. DNA Extraction of L. plantarum B22
2.4.2. Library Construction
2.4.3. Gene Annotation and Analysis
2.4.4. Prediction and Analysis of Esterase/Lipase Genes in L. plantarum B22 Genome
2.4.5. Unraveling the Structural Features and Substrate Binding Mechanisms of Esterases/Lipases
2.5. Methodology for Probiotic Properties Evaluation of L. plantarum B22
2.5.1. Acid and Bile Resistance of L. plantarum B22
2.5.2. Gastrointestinal Tract Tolerance
2.5.3. Assays for Antioxidant Activities
2.5.4. Evaluation of Antibacterial Activity
2.6. Safety Analysis of L. plantarum B22
2.6.1. Detection of Harmful Metabolites of L. plantarum B22
- (1)
- Hemolysis experiment
- (2)
- Amino Acid Decarboxylase Test
2.6.2. Antibiotic Sensitivity Analysis of L. plantarum B22
2.7. Statistical Analysis
3. Results and Discussion
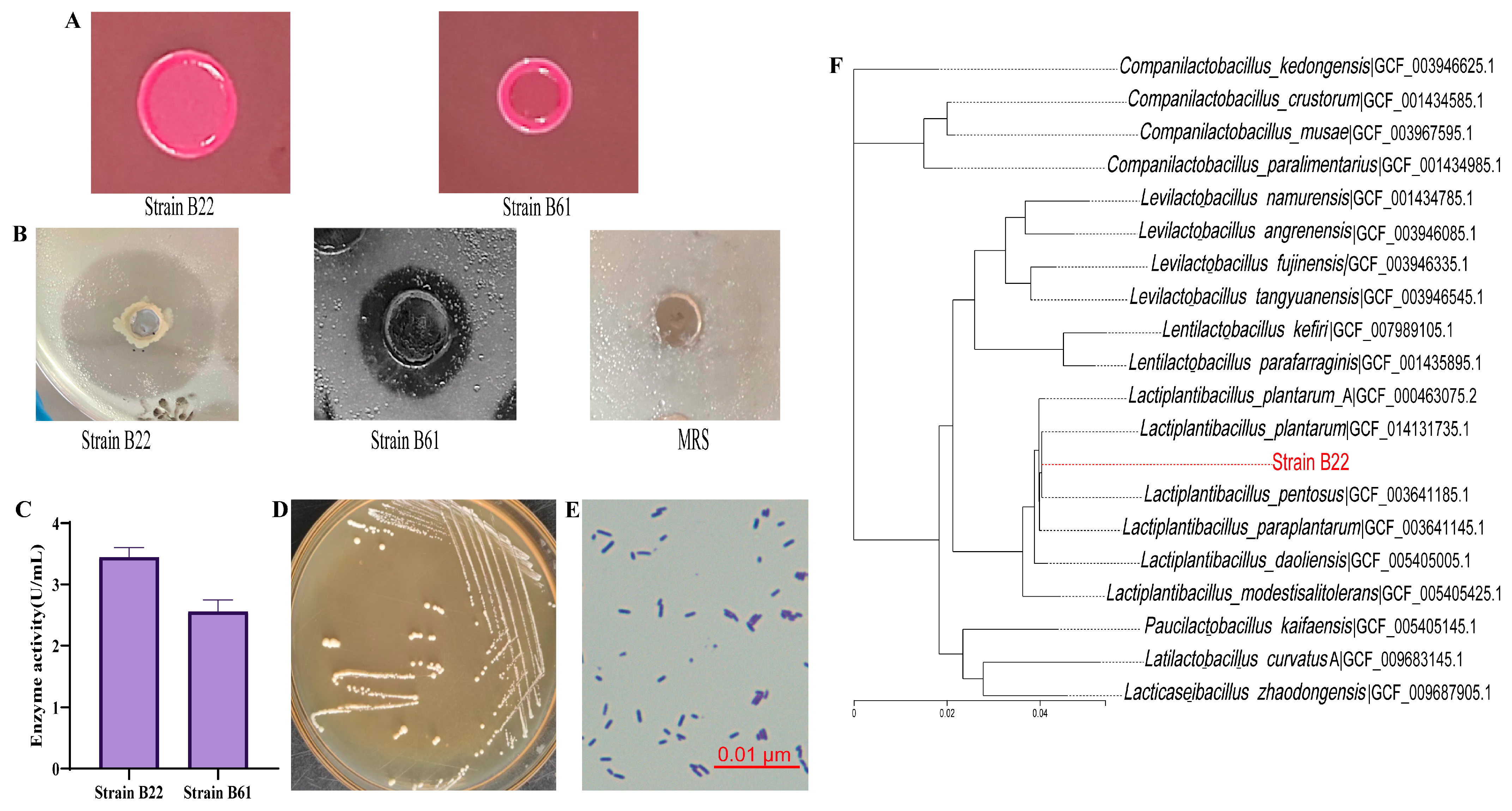
3.1. Screening and Identification of High-Esterase/Lipase-Producing LAB Strains
3.1.1. Screening of High-Esterase/Lipase-Producing LAB Strains
3.1.2. Morphological and Molecular Identification of High Esterase/Lipase-Producing LAB Strain
3.2. Elucidation of the Structure and Molecular Mechanism of Substrate Interactions of Esterase/Lipase by Genome-Wide Annotation and Molecular Docking
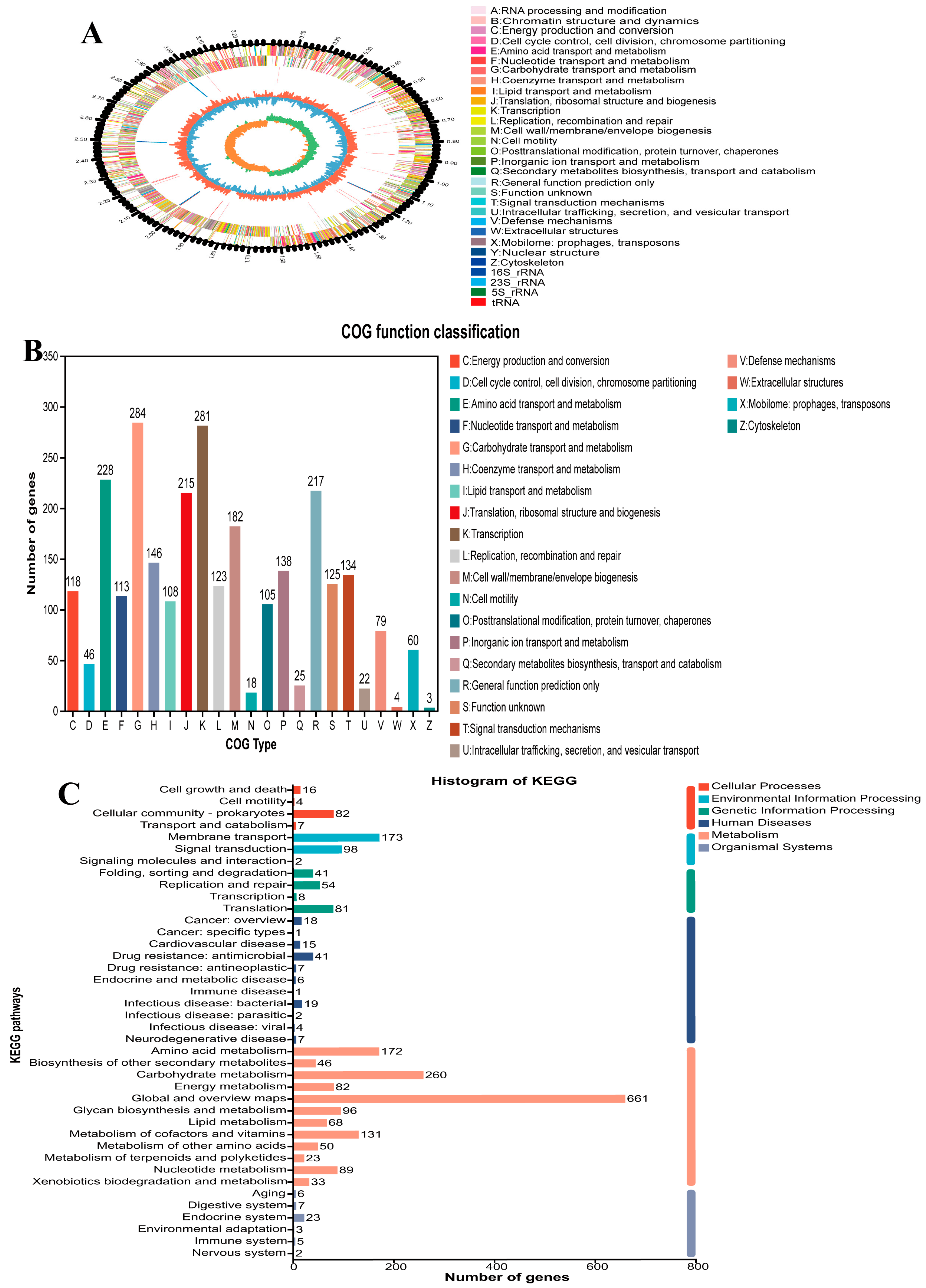
3.2.1. Genomic Characteristics and Functional Annotation of L. plantarum B22
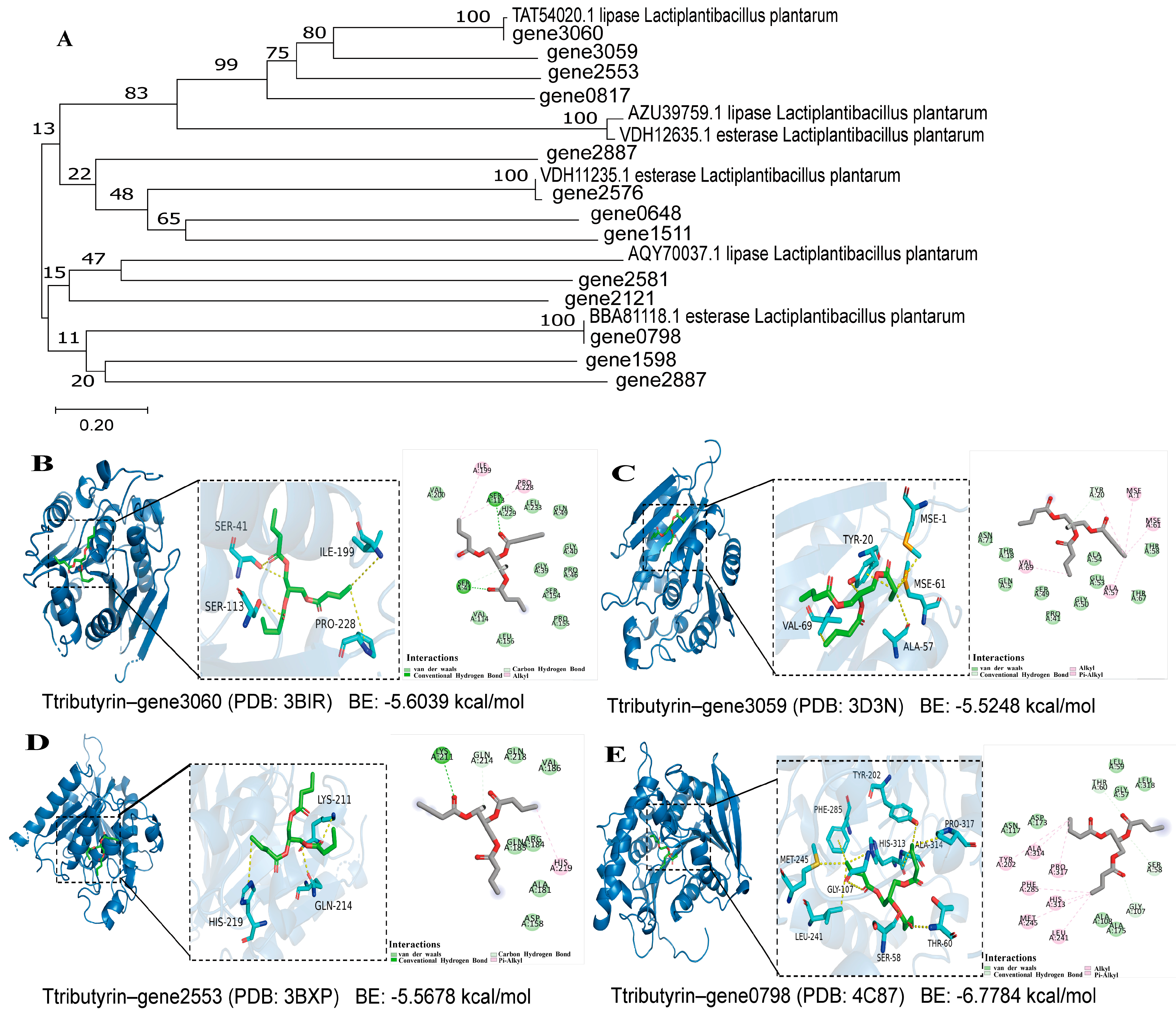
3.2.2. Identification of Esterase/Lipase Genes and Bioinformatics Analysis
3.2.3. Molecular Mechanism of Substrate Interactions of Esterase/Lipase
3.3. Evaluation of Probiotic Properties of L. plantarum B22
3.3.1. Acid and Bile Salt Tolerance
3.3.2. Gastrointestinal Survival Rate
3.3.3. Antioxidant Activity Analysis
3.3.4. Antimicrobial Properties
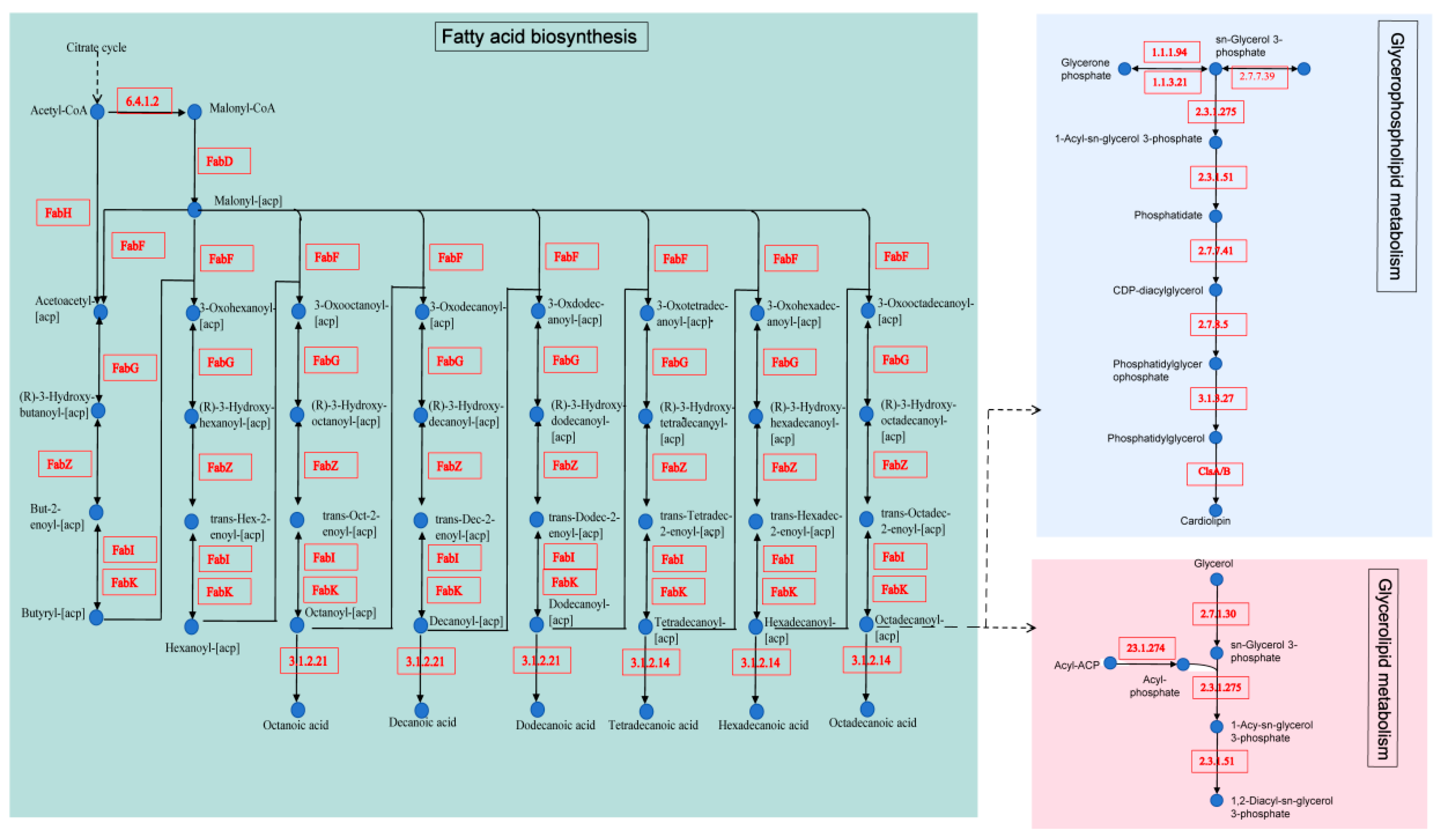
3.3.5. Reveals Key Metabolism Pathways in L. plantarum B22
3.4. Safety Evaluation of L. plantarum B22
3.4.1. Hemolytic Activity Analysis
3.4.2. Analysis of Harmful Metabolites
3.4.3. Antibiotic Resistance Testing
3.4.4. Virulence Factor Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, X.; Liu, P.; Tang, X.; Wang, T.; Xu, Z.; Hou, D.; Wu, D.; Han, N. Construction and analysis of a food-grade Lactiplantibacillus plantarum esterase/lipase overexpression system. LWT 2022, 163, 113539. [Google Scholar] [CrossRef]
- Zan, X.; Cui, F.; Sun, J.; Zhou, S.; Song, Y. Novel dual-functional enzyme Lip10 catalyzes lipase and acyltransferase activities in the oleaginous fungus Mucor circinelloides. J. Agric. Food Chem. 2019, 67, 13176–13184. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Ryu, B.H.; Kim, J.; Yoo, W.; An, D.R.; Kim, B.-Y.; Kwon, S.; Lee, S.; Wang, Y.; Kim, K.K.; et al. Characterization of a novel SGNH-type esterase from Lactobacillus plantarum. Int. J. Biol. Macromol. 2017, 96, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Chong, W.; Qi, Y.; Ji, L.; Zhang, Z.; Lu, Z.; Nian, B.; Hu, Y. Computer-aided tunnel engineering: A promising strategy for improving lipase applications in esterification reactions. ACS Catal. 2023, 14, 67–83. [Google Scholar] [CrossRef]
- Filho, D.G.; Silva, A.G.; Guidini, C.Z. Lipases: Sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 2019, 103, 7399–7423. [Google Scholar] [CrossRef]
- Salgado, C.A.; dos Santos, C.I.A.; Vanetti, M.C.D. Microbial lipases: Propitious biocatalysts for the food industry. Food Biosci. 2022, 45, 101509. [Google Scholar] [CrossRef]
- Zulaika, A.; Rahman, H.; Ningrum, S.S.; Maulida, A.F. Exploring microbial lipases: Screening and identification for biocatalyst potential in bioethanol synthesis from glycerol-based biodiesel waste. Results Eng. 2024, 23, 102427. [Google Scholar] [CrossRef]
- Gajendran, V.P.; Rajamani, S. Recent Advancements in Harnessing Lactic Acid Bacterial Metabolites for Fruits and Vegetables Preservation. Probiotics Antimicrob. Proteins 2024, 1–17. [Google Scholar] [CrossRef]
- Topçu, K.C.; Kaya, M.; Kaban, G. Probiotic properties of lactic acid bacteria strains isolated from pastırma. LWT 2020, 134, 110216. [Google Scholar] [CrossRef]
- Erem, E.; Kilic-Akyilmaz, M. The role of fermentation with lactic acid bacteria in quality and health effects of plant-based dairy analogues. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13402. [Google Scholar] [CrossRef]
- Wang, Y.; Han, J.; Wang, D.; Gao, F.; Zhang, K.; Tian, J.; Jin, Y. Research update on the impact of lactic acid bacteria on the substance metabolism, flavor, and quality characteristics of fermented meat products. Foods 2022, 11, 2090. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.-J.; Kim, J.; Lee, S.H.; Whon, T.W.; Sung, H.; Bae, J.-W.; Choi, Y.-E.; Roh, S.W. Role of combinated lactic acid bacteria in bacterial, viral, and metabolite dynamics during fermentation of vegetable food, kimchi. Food Res. Int. 2022, 157, 111261. [Google Scholar] [CrossRef]
- Zinjanab, M.S.; Golmakani, M.T.; Eskandari, M.H.; Toh, M.; Liu, S.Q. Natural flavor biosynthesis by lipase in fermented milk using in situ produced ethanol. J. Food Sci. Technol. 2021, 58, 1858–1868. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Paventi, G.; Di Martino, C. Biosynthesis of Gamma-Aminobutyric Acid (GABA) by Lactiplantibacillus plantarum in fermented food production. Curr. Issues Mol. Biol. 2023, 46, 200–220. [Google Scholar] [CrossRef]
- Esteban-Torres, M.; Reverón, I.; Santamaría, L.; Mancheño, J.M.; Rivas, B.d.L.; Muñoz, R. The Lp_3561 and Lp_3562 enzymes support a functional divergence process in the lipase/esterase toolkit from Lactobacillus plantarum. Front. Microbiol. 2016, 7, 1118. [Google Scholar] [CrossRef]
- Wang, K.; Froehlich, J.E.; Zienkiewicz, A.; Hersh, H.; Benning, C. A Plastid Phosphatidylglycerol Lipase Contributes to the Export of Acyl Groups from Plastids for Seed Oil Biosynthesis. Plant Cell 2017, 29, 1678–1696. [Google Scholar] [CrossRef]
- Majumder, D.; Dey, A.; Ray, S.; Bhattacharya, D.; Nag, M.; Lahiri, D. Use of genomics & proteomics in studying lipase producing microorganisms & its application. Food Chem. Mol. Sci. 2024, 9, 100218. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Y.; Wang, Y. Whole-genome sequencing of a potential ester-synthesizing bacterium isolated from fermented golden pomfret and identification of its lipase encoding genes. Foods 2022, 11, 1954. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Cho, Y.; Lee, Y.W.; Jung, W.H. Whole genome sequencing analysis of the cutaneous pathogenic yeast Malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff. Mycoses 2017, 60, 188–197. [Google Scholar] [CrossRef]
- Kuhn, H.W.; Lasseter, A.G.; Adams, P.P.; Avile, C.F.; Stone, B.L.; Akins, D.R.; Jewett, T.J.; Jewett, M.W.; Samuels, D.S. BB0562 is a nutritional virulence determinant with lipase activity important for Borrelia burgdorferi infection and survival in fatty acid deficient environments. PLOS Pathog. 2021, 17, e1009869. [Google Scholar] [CrossRef]
- Rampanti, G.; Cantarini, A.; Cardinali, F.; Milanović, V.; Garofalo, C.; Aquilanti, L.; Osimani, A. Technological and Enzymatic Characterization of Autochthonous Lactic Acid Bacteria Isolated from Viili Natural Starters. Foods 2024, 13, 1115. [Google Scholar] [CrossRef]
- Tsigkrimani, M.; Panagiotarea, K.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods 2022, 11, 459. [Google Scholar] [CrossRef]
- Yin, H.C.; Jiang, D.H.; Yu, T.F.; Jiang, X.J.; Liu, D. Characterization and functionality of Ligilactobacillus agilis 1003 isolated from chicken cecum against Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2024, 14, 1432422. [Google Scholar] [CrossRef] [PubMed]
- Apostolakos, I.; Paramithiotis, S.; Mataragas, M. Comparative genomic analysis reveals the functional traits and safety status of lactic acid bacteria retrieved from artisanal cheeses and raw sheep milk. Foods 2023, 12, 599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shang, Z.; Liu, Z.; Hu, X.; Yi, J. Flavor production in fermented chayote inoculated with lactic acid bacteria strains: Genomics and metabolomics based analysis. Food Res. Int. 2023, 163, 112224. [Google Scholar] [CrossRef]
- Yao, S.; Hao, L.; Zhou, R.; Jin, Y.; Huang, J.; Wu, C. Formation of Biofilm by Tetragenococcus halophilus Benefited Stress Tolerance and Anti-biofilm Activity Against S. aureus and S. Typhimurium. Front. Microbiol. 2022, 13, 819302. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.A.; Khattab, R.A.; Ragab, Y.M.; Hassan, M. Safety assessment of Enterococcus lactis strains complemented with comparative genomics analysis reveals probiotic and safety characteristics of the entire species. BMC Genom. 2023, 24, 667. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Yang, R.-S.; Lin, Y.-C.; Xin, W.-G.; Zhou, H.-Y.; Wang, F.; Zhang, Q.-L.; Lin, L.-B. Assessment of the safety and probiotic characteristics of Lactobacillus salivarius CGMCC20700 based on whole-genome sequencing and phenotypic analysis. Front. Microbiol. 2023, 14, 1120263. [Google Scholar] [CrossRef]
- Salwoom, L.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Shariff, F.M.; Convey, P.; Ali, M.S.M. New recombinant cold-adapted and organic solvent tolerant lipase from psychrophilic Pseudomonas sp. lsk25, isolated from signy island antarctica. Int. J. Mol. Sci. 2019, 20, 1264. [Google Scholar] [CrossRef]
- Chai, Y.; Ma, Q.; He, J.; Wei, G.; Huang, A. Rapid Revealing of Quorum Sensing (QS)-Regulated PLA, Biofilm and Lysine Targets of Lactiplantibacillus plantarum L3. Curr. Microbiol. 2024, 81, 303. [Google Scholar] [CrossRef]
- Tang, J.; Peng, X.; Liu, D.-M.; Xu, Y.-Q.; Liang, M.-H.; Xiong, J.; Wu, J.-J. Assessment of the safety and probiotic properties of Lactobacillus delbrueckii DMLD-H1 based on comprehensive genomic and phenotypic analysis. LWT 2023, 184, 115070. [Google Scholar] [CrossRef]
- Li, X.-H.; Huang, Y.-Y.; Lu, L.-M.; Zhao, L.-J.; Luo, X.-K.; Li, R.-J.; Dai, Y.-Y.; Qin, C.; Huang, Y.-Q.; Chen, H. Early genetic diagnosis of clarithromycin resistance in Helicobacter pylori. World J. Gastroenterol. 2021, 27, 3595–3608. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Xu, J.; Tang, N.; Chen, W.; Gu, Q.; Li, H. Screening, cloning, immobilization and application prospects of a novel β-glucosidase from the soil metagenome. Environ. Res. 2024, 244, 117676. [Google Scholar] [CrossRef]
- Liu, W.; Luo, X.; Tao, Y.; Huang, Y.; Zhao, M.; Yu, J.; Feng, F.; Wei, W. Ultrasound enhanced butyric acid-lauric acid designer lipid synthesis: Based on artificial neural network and changes in enzymatic structure. Ultrason. Sonochemistry 2022, 88, 106100. [Google Scholar] [CrossRef]
- Nanjaiah, M.; Rastogi, N.K.; Devappa, S. Study of the probiotic properties of Lacticaseibacillus casei subsp. casei NCIM 5752 and the optimization of whey-based media for the production of its biomass using response surface methodology. 3 Biotech 2024, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Wang, D.; Wang, T.; Wang, G.; Chai, Y.; Li, Y.; Mei, M.; Wang, H.; Huang, A. Probiotic potential and safety properties of Limosilactobacillus fermentum A51 with high exopolysaccharide production. Front. Microbiol. 2025, 16, 1498352. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Li, L.; Zheng, Y.; Liu, D.; Zhou, Q.; Fu, L. Characterization of Lactobacillus amylolyticus L6 as potential probiotics based on genome sequence and corresponding phenotypes. LWT 2018, 90, 460–468. [Google Scholar] [CrossRef]
- Wang, X.; Han, M.; Zhang, M.; Wang, Y.; Ren, Y.; Yue, T.; Gao, Z. In vitro evaluation of the hypoglycemic properties of lactic acid bacteria and its fermentation adaptability in apple juice. LWT 2021, 136, 110363. [Google Scholar] [CrossRef]
- Yang, S.-J.; Lee, J.-E.; Lim, S.-M.; Kim, Y.-J.; Lee, N.-K.; Paik, H.-D. Antioxidant and immune-enhancing effects of probiotic Lactobacillus plantarum 200655 isolated from kimchi. Food Sci. Biotechnol. 2019, 28, 491–499. [Google Scholar] [CrossRef]
- Lu, J.; Mao, Y.; Ma, T.; Liu, X.; Cheng, X.; Bai, Y.; Li, S. Screening and genome analysis of lactic acid bacteria with high exopolysaccharide production and good probiotic properties. Food Biosci. 2023, 56, 103211. [Google Scholar] [CrossRef]
- Li, Q.-Q.; Zeng, S.-P.; Liang, M.-H.; Yousaf, M.; Wu, Y.-P.; Tang, J.; Xiong, J.; Liu, D.-M. Safety and metabolism characteristics of Lacticaseibacillus rhamnosus LR-ZB1107-01 based on complete genome and corresponding phenotype. LWT 2024, 204, 116443. [Google Scholar] [CrossRef]
- Zareie, Z.; Moayedi, A.; Garavand, F.; Tabar-Heydar, K.; Khomeiri, M.; Maghsoudlou, Y. Probiotic properties, safety assessment, and aroma-generating attributes of some lactic acid bacteria isolated from Iranian traditional cheese. Fermentation 2023, 9, 338. [Google Scholar] [CrossRef]
- Shi, X.; Wang, X.; Hou, X.; Tian, Q.; Hui, M. Gene Mining and Flavour Metabolism Analyses of Wickerhamomyces anomalus Y-1 Isolated From a Chinese Liquor Fermentation Starter. Front. Microbiol. 2022, 13, 891387. [Google Scholar] [CrossRef]
- Griebeler, N.; Polloni, A.E.; Remonatto, D.; Arbter, F.; Vardanega, R.; Cechet, J.L.; Di Luccio, M.; de Oliveira, D.; Treichel, H.; Cansian, R.L.; et al. Isolation and screening of lipase-producing fungi with hydrolytic activity. Food Bioprocess Technol. 2011, 4, 578–586. [Google Scholar] [CrossRef]
- Kumar, A.; Joishy, T.; Das, S.; Kalita, M.C.; Mukherjee, A.K.; Khan, M.R. A Potential Probiotic Lactobacillus plantarum JBC5 Improves Longevity and Healthy Aging by Modulating Antioxidative, Innate Immunity and Serotonin-Signaling Pathways in Caenorhabditis elegans. Antioxidants 2022, 11, 268. [Google Scholar] [CrossRef]
- Xia, Y.; Wei, Z.-Y.; He, R.; Li, J.-H.; Wang, Z.-X.; Huo, J.-D.; Chen, J.-H. Hybrid de novo Genome Assembly of Erwinia sp. E602 and Bioinformatic Analysis Characterized a New Plasmid-Borne lac Operon Under Positive Selection. Front. Microbiol. 2021, 12, 783195. [Google Scholar] [CrossRef]
- Lei, X.; Fang, Z. GBDTCDA: Predicting circRNA-disease Associations Based on Gradient Boosting Decision Tree with Multiple Biological Data Fusion. Int. J. Biol. Sci. 2019, 15, 2911–2924. [Google Scholar] [CrossRef]
- Jiang, C.; Yan, H.; Shen, X.; Zhang, Y.; Wang, Y.; Sun, S.; Jiang, H.; Zang, H.; Zhao, X.; Hou, N.; et al. Genome Functional Analysis of the Psychrotrophic Lignin-Degrading Bacterium Arthrobacter sp. C2 and the Role of DyP in Catalyzing Lignin Degradation. Front. Microbiol. 2022, 13, 921549. [Google Scholar] [CrossRef]
- Ranasinghe, R.T.; Challand, M.R.; Ganzinger, K.A.; Lewis, B.W.; Softley, C.; Schmied, W.H.; Horrocks, M.H.; Shivji, N.; Chin, J.W.; Spencer, J.; et al. Detecting RNA base methylations in single cells by in situ hybridization. Nat. Commun. 2018, 9, 655. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, S.; Li, C.; Xiang, H.; Zhao, Y.; Chen, S.; Li, L.; Wu, Y. Application of Untargeted Metabolomics to Reveal the Taste-Related Metabolite Profiles during Mandarin Fish (Siniperca chuatsi) Fermentation. Foods 2022, 11, 944. [Google Scholar] [CrossRef]
- Rocha, B.M.d.O.; Sabino, Y.N.V.; de Almeida, T.C.; Palacio, F.B.; Rotta, I.S.; Dias, V.C.; da Silva, V.L.; Diniz, C.G.; Azevedo, V.A.d.C.; Brenig, B.; et al. Unlocking Probiotic Potential: Genomic Insights into Weissella paramesenteroides UFTM 2.6.1. Probiotics Antimicrob. Proteins 2024, 1–14. [Google Scholar] [CrossRef]
- Shinohara, S.; Gu, Y.; Yang, Y.; Furuta, Y.; Tanaka, M.; Yue, X.; Wang, W.; Kitano, M.; Kimura, H. Ethanol extracts of chickpeas alter the total lipid content and expression levels of genes related to fatty acid metabolism in mouse 3T3-L1 adipocytes. Int. J. Mol. Med. 2016, 38, 574–584. [Google Scholar] [CrossRef]
- Ramírez-Sánchez, O.; Pérez-Rodríguez, P.; Delaye, L.; Tiessen, A. Plant Proteins Are Smaller Because They Are Encoded by Fewer Exons than Animal Proteins. Genom. Proteom. Bioinform. 2016, 14, 357–370. [Google Scholar] [CrossRef]
- Califano, V.; Bloisi, F.; Aronne, A.; Federici, S.; Nasti, L.; Depero, L.E.; Vicari, L.R.M. Biosensor Applications of MAPLE Deposited Lipase. Biosensors 2014, 4, 329–339. [Google Scholar] [CrossRef]
- Liao, G.; Li, Y.; Wang, H.; Liu, Q.; Zhong, M.; Jia, D.; Huang, C.; Xu, X. Genome-wide identification and expression profiling analysis of sucrose synthase (SUS) and sucrose phosphate synthase (SPS) genes family in Actinidia chinensis and A. eriantha. BMC Plant Biol. 2022, 22, 215. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, H.; Liu, C.; Li, Q.; Niu, C. Prediction, biochemical characterization and application of key proteolytic enzymes from aspergillus oryzae BL18 in soy sauce fermentation. Food Res. Int. 2025, 211, 116382. [Google Scholar] [CrossRef]
- Yi, Z.; Xie, J. Genomic analysis of two representative strains of Shewanella putrefaciens isolated from bigeye tuna: Biofilm and spoilage-associated behavior. Foods 2022, 11, 1261. [Google Scholar] [CrossRef]
- Kusada, H.; Arita, M.; Tohno, M.; Tamaki, H. Bile Salt Hydrolase Degrades β-Lactam Antibiotics and Confers Antibiotic Resistance on Lactobacillus paragasseri. Front. Microbiol. 2022, 13, 858263. [Google Scholar] [CrossRef]
- Xu, T.; Chen, H.; Li, J.; Hong, S.; Shao, L.; Zheng, X.; Zou, Q.; Wang, Y.; Guo, S.; Jiang, J. Implications for Cation Selectivity and Evolution by a Novel Cation Diffusion Facilitator Family Member From the Moderate Halophile Planococcus dechangensis. Front. Microbiol. 2019, 10, 607. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, P.; Cheng, Y.; Liu, J.; Chen, X.; Zhang, T.; Gao, T.; Zhou, R.; Li, L. The Feed Additive Potassium Diformate Prevents Salmonella enterica Serovar Pullorum Infection and Affects Intestinal Flora in Chickens. Antibiotics 2022, 11, 1265. [Google Scholar] [CrossRef]
- Choi, T.-R.; Park, Y.-L.; Song, H.-S.; Lee, S.M.; Park, S.L.; Lee, H.S.; Kim, H.-J.; Bhatia, S.K.; Gurav, R.; Lee, Y.K.; et al. Effects of a Δ-9-fatty acid desaturase and a cyclopropane-fatty acid synthase from the novel psychrophile Pseudomonas sp. B14-6 on bacterial membrane properties. J. Ind. Microbiol. Biotechnol. 2020, 47, 1045–1057. [Google Scholar] [CrossRef]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Behbahani, B.A.; Jooyandeh, H.; Taki, M.; Falah, F. Evaluation of the probiotic, anti-bacterial, anti-biofilm, and safety properties of Lacticaseibacillus paracasei B31-2. LWT 2024, 207, 116676. [Google Scholar] [CrossRef]
- Xu, X.; Luo, D.; Bao, Y.; Liao, X.; Wu, J. Characterization of Diversity and Probiotic Efficiency of the Autochthonous Lactic Acid Bacteria in the Fermentation of Selected Raw Fruit and Vegetable Juices. Front. Microbiol. 2018, 9, 2539. [Google Scholar] [CrossRef]
- Treven, P.; Paveljšek, D.; Matijašić, B.B.; Lorbeg, P.M. The Effect of Food Matrix Taken with Probiotics on the Survival of Commercial Probiotics in Simulation of Gastrointestinal Digestion. Foods 2024, 13, 3135. [Google Scholar] [CrossRef]
- Liu, D.-M.; Huang, Y.-Y.; Liang, M.-H. Analysis of the probiotic characteristics and adaptability of Lactiplantibacillus plantarum DMDL 9010 to gastrointestinal environment by complete genome sequencing and corresponding phenotypes. LWT 2022, 158, 113129. [Google Scholar] [CrossRef]
- Jabłońska-Trypuć, A.; Wydro, U.; Wołejko, E.; Rodziewicz, J.; Butarewicz, A. Possible Protective Effects of TA on the Cancerous Effect of Mesotrione. Nutrients 2020, 12, 1343. [Google Scholar] [CrossRef]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free. Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef]
- Murgas, C.J.; Green, S.P.; Forney, A.K.; Korba, R.M.; An, S.-S.; Kitten, T.; Lucas, H.R. Intracellular Metal Speciation in Streptococcus sanguinis Establishes SsaACB as Critical for Redox Maintenance. ACS Infect. Dis. 2020, 6, 1906–1921. [Google Scholar] [CrossRef]
- Yang, S.; Liu, L.; Wang, J.; Guo, S.; Liu, G.; Chen, X.; Deng, X.; Tu, M.; Tao, Y.; Rao, Y. Antimicrobial activity against Staphylococcus aureus and genome features of Lactiplantibacillus plantarum LR-14 from Sichuan pickles. Arch. Microbiol. 2022, 204, 637. [Google Scholar] [CrossRef]
- Martín, I.; Barbosa, J.; Pereira, S.I.; Rodríguez, A.; Córdoba, J.J.; Teixeira, P. Study of lactic acid bacteria isolated from traditional ripened foods and partial characterization of their bacteriocins. LWT 2023, 173, 114300. [Google Scholar] [CrossRef]
- Hong, N.T.X.; Linh, N.T.H.; Baruah, K.; Thuy, D.T.B.; Phuoc, N.N. The Combined Use of Pediococcus pentosaceus and Fructooligosaccharide Improves Growth Performance, Immune Response, and Resistance of Whiteleg Shrimp Litopenaeus vannamei Against Vibrio parahaemolyticus. Front. Microbiol. 2022, 13, 826151. [Google Scholar] [CrossRef]
- Lee, Y.; Jang, A.; Jeong, M.-C.; Park, N.; Park, J.; Lee, W.C.; Cheong, C.; Kim, Y. Structural Characterization of an ACP from Thermotoga maritima: Insights into Hyperthermal Adaptation. Int. J. Mol. Sci. 2020, 21, 2600. [Google Scholar] [CrossRef]
- Guo, C.; Han, L.; Li, M.; Yu, L. Seabuckthorn (Hippophaë rhamnoides) Freeze-Dried Powder Protects against High-Fat Diet-Induced Obesity, Lipid Metabolism Disorders by Modulating the Gut Microbiota of Mice. Nutrients 2020, 12, 265. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Song, W.; Huang, Y.; Mu, Y.; Schmitz, W.; Zhang, S.; Lin, H.; Chen, H.; Ye, F.; et al. The Molecular Basis of Catalysis by SDR Family Members Ketoacyl-ACP Reductase FabG and Enoyl-ACP Reductase FabI in Type-II Fatty Acid Biosynthesis. Angew. Chem. 2023, 135, e202313109. [Google Scholar] [CrossRef]
- Martel, A.; Pasmans, F.; Hellebuyck, T.; Haesebrouck, F.; Vandamme, P. Devriesea agamarum gen. nov., sp. nov., a novel actinobacterium associated with dermatitis and septicaemia in agamid lizards. Int. J. Syst. Evol. Microbiol. 2008, 58, 2206–2209. [Google Scholar] [CrossRef]
- Sohanang, F.S.N.; Coton, M.; Debaets, S.; Coton, E.; Tatsadjieu, L.N.; Mohammadou, B.A. Bacterial diversity of traditional fermented milks from Cameroon and safety and antifungal activity assessment for selected lactic acid bacteria. LWT 2021, 138, 110635. [Google Scholar] [CrossRef]
- Chen, T.; Shao, Y.; Zhang, Y.; Zhao, Y.; Han, M.; Gai, Z. In vitro and in vivo genome-based safety evaluation of Lacticaseibacillus rhamnosus LRa05. Food Chem. Toxicol. 2024, 186, 114600. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q.; Lv, J.; Sun, X.; Cao, Y.; Yu, K.; Miao, C.; Zhang, Z.-S.; Yao, Z.; Wang, Q. Alpha-hemolysin of uropathogenic Escherichia coli induces GM-CSF-mediated acute kidney injury. Mucosal Immunol. 2020, 13, 22–33. [Google Scholar] [CrossRef]
- Ou, D.; Ling, N.; Wang, X.; Zou, Y.; Dong, J.; Zhang, D.; Shen, Y.; Ye, Y. Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui. Foods 2022, 11, 2177. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.-S.; Yu, H.-Y.; Kwon, M.; Kim, K.-K.; In, G.; Hong, S.-K.; Kim, S.-K. Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics. Foods 2022, 11, 1707. [Google Scholar] [CrossRef]
- Peres, C.M.; Alves, M.; Hernandez-Mendoza, A.; Moreira, L.; Silva, S.; Bronze, M.R.; Vilas-Boas, L.; Peres, C.; Malcata, F.X. Novel isolates of lactobacilli from fermented Portuguese olive as potential probiotics. LWT 2014, 59, 234–246. [Google Scholar] [CrossRef]
- Ashiq, J.; Hussain, A.; Gilani, M.A.; Riaz, S.; Nawaz, M.H. Ultrasensitive detection of histamine in spoiled meat employing silver nanoparticles decorated Perylene: An experimental-computational conjugation. Food Chem. 2025, 464, 141673. [Google Scholar] [CrossRef]
- Natrella, G.; Vacca, M.; Minervini, F.; Faccia, M.; De Angelis, M. A Comprehensive Review on the Biogenic Amines in Cheeses: Their Origin, Chemical Characteristics, Hazard and Reduction Strategies. Foods 2024, 13, 2583. [Google Scholar] [CrossRef]
- Abarquero, D.; Bodelón, R.; Flórez, A.B.; Fresno, J.M.; Renes, E.; Mayo, B.; Tornadijo, M.E. Technological and safety assessment of selected lactic acid bacteria for cheese starter cultures design: Enzymatic and antimicrobial activity, antibiotic resistance and biogenic amine production. LWT 2023, 180, 114709. [Google Scholar] [CrossRef]
- Guan, Y.; Cui, Y.; Qu, X.; Jing, K. Safety and robustness aspects analysis of Lactobacillus delbrueckii ssp. bulgaricus LDB-C1 based on the genome analysis and biological tests. Arch. Microbiol. 2021, 203, 3955–3964. [Google Scholar] [CrossRef]
- Luz, C.; Calpe, J.; Quiles, J.M.; Torrijos, R.; Vento, M.; Gormaz, M.; Mañes, J.; Meca, G. Probiotic characterization of Lactobacillus strains isolated from breast milk and employment for the elaboration of a fermented milk product. J. Funct. Foods 2021, 84, 104599. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, W.; Kwok, L.-Y.; Menghe, B. In vitro evalution of antibiotic resistance of Lactobacillus bulgaricus strains isolated from traditional dairy products. Czech J. Food Sci. 2019, 37, 36–43. [Google Scholar] [CrossRef]
- Ryser, L.T.; Arias-Roth, E.; Perreten, V.; Irmler, S.; Bruggmann, R. Genetic and Phenotypic Diversity of Morganella morganii Isolated From Cheese. Front. Microbiol. 2021, 12, 738492. [Google Scholar] [CrossRef] [PubMed]
- Leitão, J.H. Microbial Virulence Factors. Int. J. Mol. Sci. 2020, 21, 5320. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, L.; Nan, W.; Lu, J.; Zhang, S.; Ujiroghene, O.J.; Pang, X.; Lv, J. Whole-genome sequencing and genomic-based acid tolerance mechanisms of Lactobacillus delbrueckii subsp. bulgaricus LJJ. Appl. Microbiol. Biotechnol. 2020, 104, 7631–7642. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Pan, Y.; Liu, H. Comparison on the Growth Variability of Vibrio parahaemolyticus Coupled With Strain Sources and Genotypes Analyses in Simulated Gastric Digestion Fluids. Front. Microbiol. 2020, 11, 212. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, J.; Nie, X.; Chitrakar, B.; Gao, J.; Sang, Y. Mutual adhesion of Lactobacillus spp. to intestinal cells: A review of perspectives on surface layer proteins and cell surface receptors. Int. J. Biol. Macromol. 2024, 282, 137031. [Google Scholar] [CrossRef] [PubMed]
- Araújo, L.; Papa-Ezdra, R.; Ávila, P.; Iribarnegaray, V.; Bado, I.; Telechea, H.; Garcia-Fulgueiras, V.; Vignoli, R. Great Plasticity in a Great Pathogen: Capsular Types, Virulence Factors and Biofilm Formation in ESBL-Producing Klebsiella pneumoniae from Pediatric Infections in Uruguay. Antibiotics 2024, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, Z.; Salari, A.; Khanzadi, S.; Rhim, J.; Shamloo, E. Preparation of milk-based probiotic lactic acid bacteria biofilms: A new generation of probiotics. Food Sci. Nutr. 2023, 11, 2915–2924. [Google Scholar] [CrossRef]
- Nguyen, M.-T.; Luqman, A.; Bitschar, K.; Hertlein, T.; Dick, J.; Ohlsen, K.; Bröker, B.; Schittek, B.; Götz, F. Staphylococcal (phospho)lipases promote biofilm formation and host cell invasion. Int. J. Med Microbiol. 2018, 308, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Qian, M.; Cheng, F.; Wang, Y.; Han, J.; Xu, Y.; Zhang, K.; Tian, J.; Jin, Y. The effect of lactic acid bacteria on lipid metabolism and flavor of fermented sausages. Food Biosci. 2023, 56, 103172. [Google Scholar] [CrossRef]
- Nieuwboer, M.v.D.; van Hemert, S.; Claassen, E.; de Vos, W.M. Lactobacillus plantarum WCFS1 and its host interaction: A dozen years after the genome. Microb. Biotechnol. 2016, 9, 452–465. [Google Scholar] [CrossRef]

| Attributes | Values |
|---|---|
| Genome Size (bp) | 3,290,520 |
| G + C Content (%) | 44.57 |
| Coding Gene Number | 3148 |
| Coding Gene Average Length (bp) | 893.53 |
| rRNA | 16 |
| 5S rRNA | 6 |
| 16S rRNA | 5 |
| 23S rRNA | 5 |
| tRNA | 69 |
| sRNA | 42 |
| Plasmids | 1 |
| Gene ID | Annotation | Number of Amino Acids | Molecular Weight (Da) | Theoretical pI | Instability Index | Extinction Coefficients | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Alpha Helix (Hh, %) | Extended Strand (Ee, %) | Random Coil (Cc, %) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| gene0648 | Esterase/lipase | 248 | 27,852.40 | 5.67 | 36.95 | 26,930 | 85.40 | −0.215 | 45.56 | 16.53 | 37.90 |
| gene0798 | Acetyl esterase/lipase | 346 | 37,837.99 | 4.85 | 38.44 | 41,370 | 97.34 | 0.047 | 43.79 | 12.13 | 44.08 |
| gene0817 | Acetyl esterase/lipase | 229 | 24,892.17 | 5.39 | 26.37 | 51,910 | 95.15 | −0.022 | 29.55 | 21.21 | 49.24 |
| gene1511 | lipase activity | 250 | 28,778.94 | 8.00 | 35.86 | 41,370 | 93.24 | −0.255 | 42.80 | 15.20 | 42.00 |
| gene1598 | Lipase_GDSL_2;Lipase_GDSL | 314 | 34,504.56 | 9.88 | 21.70 | 32,890 | 84.78 | −0.326 | 46.50 | 12.42 | 41.08 |
| gene2121 | Lipase_GDSL_2;Lipase_GDSL | 712 | 79,929.89 | 4.80 | 21.78 | 83,200 | 91.01 | −0.195 | 28.99 | 19.14 | 51.87 |
| gene2581 | Lipase_GDSL_2;Lipase_GDSL | 233 | 25,483.04 | 8.98 | 11.94 | 36,440 | 97.51 | −0.182 | 43.78 | 13.73 | 42.49 |
| gene2576 | Monoacylglycerol lipase ABHD6 | 246 | 26,816.43 | 5.78 | 41.34 | 21,890 | 94.84 | −0.028 | 38.62 | 18.29 | 43.9 |
| gene2553 | Acetyl esterase/lipase | 276 | 30,490.71 | 6.33 | 31.11 | 57,995 | 90.94 | −0.049 | 27.17 | 18.84 | 53.99 |
| gene2578 | Acetyl esterase/lipase | 469 | 50,662.08 | 6.05 | 21.78 | 66,935 | 87.29 | −0.113 | 33.26 | 14.50 | 52.24 |
| gene3059 | Acetyl esterase/lipase | 278 | 31,534.87 | 5.45 | 36.32 | 55,475 | 80.04 | −0.274 | 28.06 | 17.63 | 54.32 |
| gene3060 | Acetyl esterase/lipase | 278 | 30,806.92 | 5.17 | 42.24 | 50,880 | 86.69 | 0.014 | 25.90 | 19.78 | 54.32 |
| gene2887 | Lipase_GDSL_2;Lipase_GDSL | 213 | 23,106.85 | 5.12 | 28.43 | 32,890 | 81.64 | −0.054 | 29.58 | 15.02 | 55.40 |
| Gene ID | Gene Name | Annotation | Gene ID | Gene Name | Annotation |
|---|---|---|---|---|---|
| Universal Stress Family Protein | Bile Salt Resistance | ||||
| gene0955 | uspA | MULTISPECIES: universal stress protein | gene1457 | cfa | cyclopropane-fatty-acyl-phospholipid synthase family protein |
| gene1140 | - | MULTISPECIES: universal stress protein | gene2753 | cfa | cyclopropane-fatty-acyl-phospholipid synthase family protein |
| gene1462 | - | MULTISPECIES: universal stress protein | gene1067 | perM | AI-2E family transporter |
| gene1498 | - | universal stress protein | gene1941 | perM | MULTISPECIES: AI-2E family transporter |
| gene2395 | uspA | MULTISPECIES: universal stress protein | Oxidative stress | ||
| gene2317 | uspA | universal stress protein | gene3072 | katE | catalase |
| gene2070 | uspA | MULTISPECIES: universal stress protein | gene0205 | btuE | MULTISPECIES: glutathione peroxidase |
| gene2609 | - | universal stress protein | gene2956 | yfeX | Dyp-type peroxidase |
| gene2523 | uspA | MULTISPECIES: universal stress protein | gene0068 | arsC | MULTISPECIES: arsenate reductase (thioredoxin) |
| gene3149 | - | MULTISPECIES: universal stress protein | gene0217 | trxA | MULTISPECIES: thioredoxin |
| Acid stress | gene0617 | trxB | MULTISPECIES: thioredoxin-disulfide reductase | ||
| gene2091 | atpA | MULTISPECIES: F0F1 ATP synthase subunit alpha | gene1009 | trxA | MULTISPECIES: thioredoxin family protein |
| gene2088 | atpC | MULTISPECIES: F0F1 ATP synthase subunit epsilon | gene2016 | trxA | MULTISPECIES: thioredoxin |
| gene2090 | atpG | MULTISPECIES: F0F1 ATP synthase subunit gamma | gene2308 | - | MULTISPECIES: thioredoxin family protein |
| gene2095 | atpB | F0F1 ATP synthase subunit A | gene2960 | trxA | MULTISPECIES: thioredoxin family protein |
| gene2089 | atpD | MULTISPECIES: F0F1 ATP synthase subunit beta | gene2056 | tpx | thiol peroxidase |
| gene2094 | atpE | MULTISPECIES: F0F1 ATP synthase subunit C | gene0252 | mntH | manganese transport protein |
| gene2093 | atpF | MULTISPECIES: F0F1 ATP synthase subunit B | gene0895 | mntB | manganese ABC transporter, permease protein |
| gene0181 | nhaC | Na(+)/H(+) antiporter NhaC | gene1575 | ppaC | manganese-dependent inorganic pyrophosphatase |
| gene2896 | nhaC | Na+/H+ antiporter NhaC | |||
| Bacteriocin | Organic acid | ||||
| gene0376 | - | MULTISPECIES: two-peptide bacteriocin plantaricin JK subunit PlnK | gene0469 | ldh | MULTISPECIES: L-lactate dehydrogenase |
| gene0388 | - | MULTISPECIES: two-peptide bacteriocin plantaricin EF subunit PlnF | gene0716 | ldhA | MULTISPECIES: D-2-hydroxyacid dehydrogenase |
| gene0389 | - | MULTISPECIES: two-peptide bacteriocin plantaricin EF subunit PlnE | |||
| gene0377 | - | MULTISPECIES: two-peptide bacteriocin plantaricin JK subunit PlnJ | |||
| Antibiotic | Specifications (μg/Piece) | Inhibition Zones Diameter (mm) | Antibiotic Susceptibility | Antibiotic | Specifications (μg/Piece) | Inhibition Zones Diameter (mm) | Antibiotic Susceptibility |
|---|---|---|---|---|---|---|---|
| Penicillin | 1 | 27.60 ± 2.84 | S | Vancomycin | 30 | 0.00 ± 0.00 | R |
| Piperacillin | 100 | 35.27 ± 0.33 | S | Chloramphenicol | 30 | 0.00 ± 0.00 | R |
| Ampicillin | 10 | 31.89 ± 3.16 | S | Cefalexin | 30 | 0.00 ± 0.00 | R |
| Kanamycin | 30 | 17.28 ± 0.46 | I | Cephazolin | 30 | 0.00 ± 0.00 | R |
| Streptomycin | 10 | 0.00 ± 0.00 | R | Cefuroxim | 30 | 41.55 ± 0.72 | S |
| Gentamicin | 10 | 18.71 ± 3.12 | I | Ceftazidime | 30 | 25.51 ± 2.80 | S |
| Amikacin | 10 | 0.00 ± 0.00 | R | Ceftiaxone | 30 | 39.43 ± 1.31 | S |
| Tetracycline | 30 | 17.59 ± 1.10 | I | Cefoperazone | 30 | 24.61 ± 0.99 | S |
| Doxycycline | 30 | 0.00 ± 0.00 | R | Erythromycin | 15 | 27.42 ± 1.46 | S |
| Minocycline | 30 | 40.09 ± 3.46 | S | Lincomycin | 2 | 17.38 ± 1.71 | I |
| Gene ID | Location | VFcategory | Related Genes | Gene ID | Location | VFcategory | Related Genes |
|---|---|---|---|---|---|---|---|
| gene1376 | Chromosome | Stress survival | recN | gene1391 | Chromosome | Immune modulation | rpe |
| gene1574 | Chromosome | Stress survival | msrA | gene0980 | Chromosome | Immune modulation | rfbD |
| gene3072 | Chromosome | Stress survival | katA | gene0700 | Chromosome | Immune modulation | oatA |
| gene0637 | Chromosome | Stress survival | clpP | gene3134 | Chromosome | Immune modulation | msbA |
| gene1086 | Chromosome | Stress survival | clpE | gene0620 | Chromosome | Immune modulation | manB |
| gene0833 | Chromosome | Stress survival | clpC | gene0428 | Chromosome | Immune modulation | lpxA |
| gene3034 | Chromosome | Stress survival | bsh | gene0990 | Chromosome | Immune modulation | kfiC |
| gene1707 | Chromosome | Regulation | relA | gene0614 | Chromosome | Immune modulation | hasC |
| gene0051 | Chromosome | Regulation | phoP | gene1761 | Chromosome | Immune modulation | gtrB |
| gene1321 | Chromosome | Regulation | mprA | gene1319 | Chromosome | Immune modulation | gndA |
| gene2213 | Chromosome | Regulation | bvrR | gene2993 | Chromosome | Immune modulation | galE |
| gene2701 | Chromosome | Nutritional/Metabolic factor | shuU | gene1633 | Chromosome | Immune modulation | ddrA |
| gene1477 | Chromosome | Nutritional/Metabolic factor | pvdH | gene0321 | Chromosome | Immune modulation | cpsK |
| gene2371 | Chromosome | Nutritional/Metabolic factor | purCD | gene0966 | Chromosome | Immune modulation | cpsI |
| gene1281 | Chromosome | Nutritional/Metabolic factor | narH | gene0664 | Chromosome | Immune modulation | cpsG |
| gene2930 | Chromosome | Nutritional/Metabolic factor | mgtB | gene1743 | Chromosome | Immune modulation | cpsB |
| gene2648 | Chromosome | Nutritional/Metabolic factor | iroC | gene0996 | Chromosome | Immune modulation | cps4I |
| gene2783 | Chromosome | Nutritional/Metabolic factor | fbpC | gene1561 | Chromosome | Immune modulation | bcs1′ |
| gene1718 | Chromosome | Nutritional/Metabolic factor | dhbF | gene2003 | Chromosome | Biofilm | bopD |
| gene2358 | Chromosome | Nutritional/Metabolic factor | carB | gene1864 | Chromosome | Adherence | tufA |
| gene2359 | Chromosome | Nutritional/Metabolic factor | carA | gene1542 | Chromosome | Adherence | pavA |
| gene1434 | Chromosome | Motility | flmH | gene3148 | Chromosome | Adherence | lap |
| gene2249 | Chromosome | Immune modulation | wbuZ | gene2078 | Chromosome | Adherence | IlpA |
| gene0976 | Chromosome | Immune modulation | wbtL | gene0590 | Chromosome | Adherence | groEL |
| gene0802 | Chromosome | Immune modulation | wbtH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, Y.; Li, Z.; Zheng, W.; Yang, X.; He, J.; Hu, S.; Shi, J.; Li, Y.; Wei, G.; Huang, A. Dissecting the High Esterase/Lipase Activity and Probiotic Traits in Lactiplantibacillus plantarum B22: A Genome-Guided Functional Characterization. Foods 2025, 14, 2354. https://doi.org/10.3390/foods14132354
Chai Y, Li Z, Zheng W, Yang X, He J, Hu S, Shi J, Li Y, Wei G, Huang A. Dissecting the High Esterase/Lipase Activity and Probiotic Traits in Lactiplantibacillus plantarum B22: A Genome-Guided Functional Characterization. Foods. 2025; 14(13):2354. https://doi.org/10.3390/foods14132354
Chicago/Turabian StyleChai, Yunmei, Zhenzhu Li, Wentao Zheng, Xue Yang, Jinze He, Shaomei Hu, Jindou Shi, Yufang Li, Guangqiang Wei, and Aixiang Huang. 2025. "Dissecting the High Esterase/Lipase Activity and Probiotic Traits in Lactiplantibacillus plantarum B22: A Genome-Guided Functional Characterization" Foods 14, no. 13: 2354. https://doi.org/10.3390/foods14132354
APA StyleChai, Y., Li, Z., Zheng, W., Yang, X., He, J., Hu, S., Shi, J., Li, Y., Wei, G., & Huang, A. (2025). Dissecting the High Esterase/Lipase Activity and Probiotic Traits in Lactiplantibacillus plantarum B22: A Genome-Guided Functional Characterization. Foods, 14(13), 2354. https://doi.org/10.3390/foods14132354







