Evaluation of the Functional Properties and Edible Safety of Concocted Xanthii Fructus Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of XFP
2.2.1. Alkaline Extraction and Acid Precipitation (AARP/AACP)
2.2.2. Salting Method (SRP/SCP)
2.3. Characterization of XFP
2.3.1. XFP Preparation and Electrophoretic Analysis
2.3.2. Chemical Composition
2.3.3. Amino Acid Composition
2.4. Spectral Structure
2.4.1. Circular Dichroism (CD)
2.4.2. Fluorescence Spectroscopy
2.4.3. Fourier Transform Infrared Spectrum (FTIR)
2.4.4. Ultraviolet Spectrum (UV)
2.5. Functional Properties
2.5.1. Solubility
2.5.2. Water/Oil Absorption Capacity (WAC/OAC)
2.5.3. Thermal Stability
2.5.4. Emulsion Activity Index (EAI) and Emulsion Stability Index (ESI)
2.5.5. Foaming Capacity (FC) and Foaming Stability (FS)
2.6. Safety Assessment Using Animal Models
2.6.1. Animals and Drug Administration
2.6.2. Sample Collection
2.7. UHPLC-Q-Exactive Orbitrap MS Analysis
2.7.1. UHPLC and HRMS Parameters
2.7.2. Sample Preparation
2.8. Gut Microbiota Sample Processing
2.9. Data Processing and Statistical Analyses
3. Results and Discussion
3.1. Characterization of XFP
3.1.1. XFP Profile
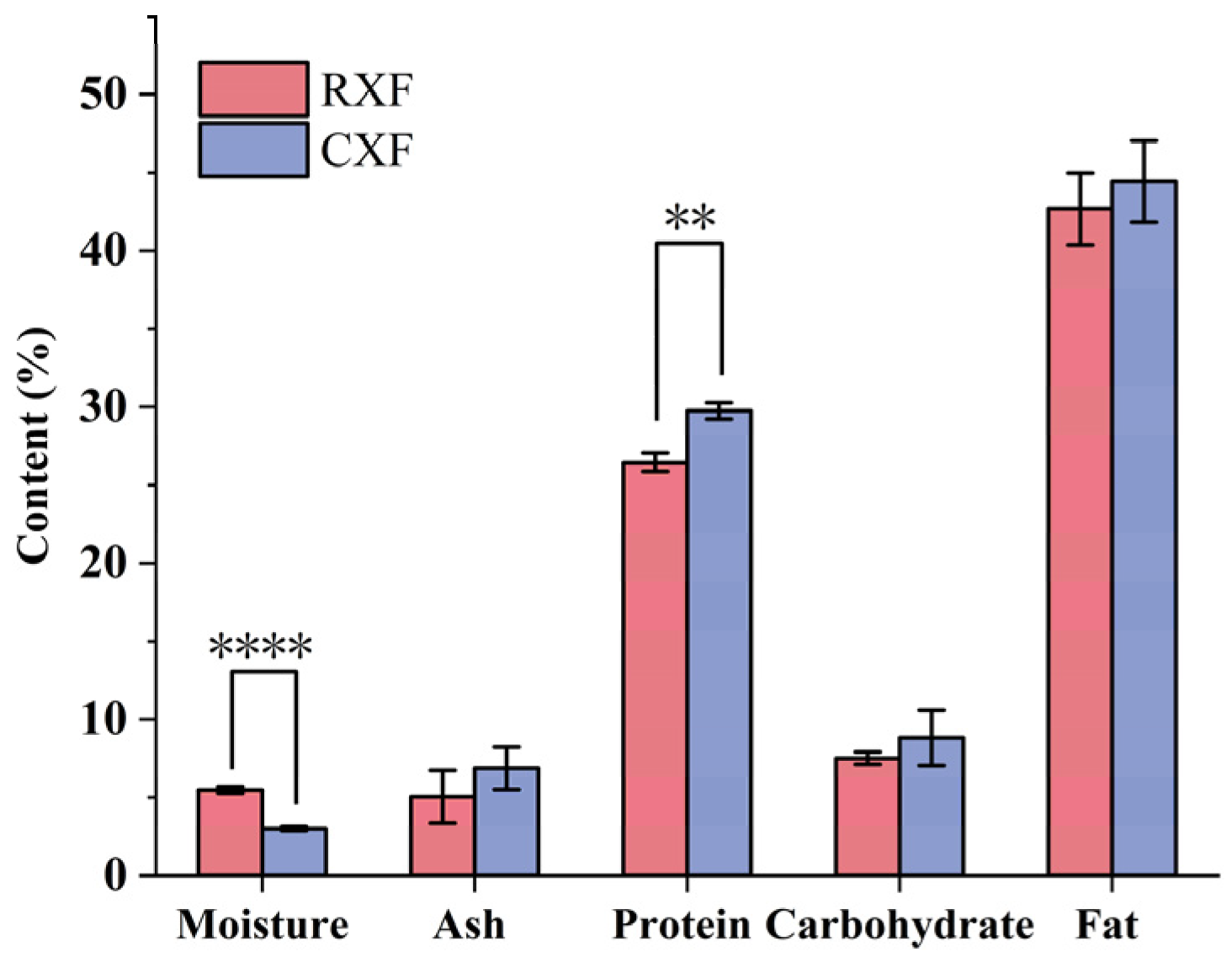
3.1.2. Proximate Analysis of XF Seeds
3.1.3. Amino Acid Profile of XFP
3.2. Structural Characteristics of XFP Extracted at Different pHs
3.2.1. CD Spectroscopy
3.2.2. Fluorescence Spectrum
3.2.3. FTIR Analysis
3.2.4. UV Analysis
3.3. Functional Properties of XFP
3.3.1. Solubility of XFP
3.3.2. WAC/OAC
3.3.3. Thermal Stability of XFP
3.3.4. EAI and ESI
3.3.5. FC and FS
3.4. Animal Experiments
3.4.1. Body Weight
3.4.2. Organ Index
3.4.3. Biochemical Indices
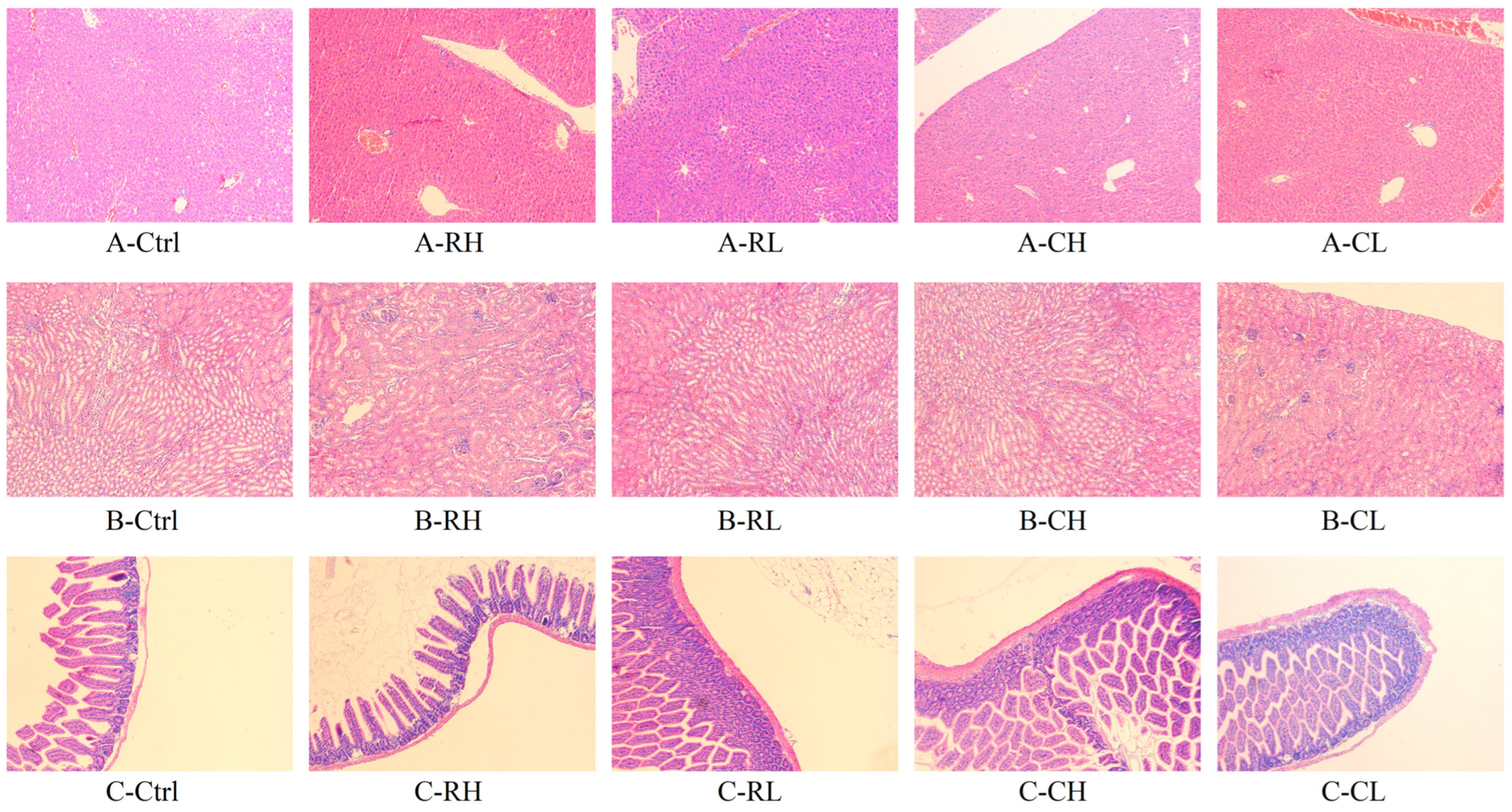
3.4.4. HE Staining Sections
3.4.5. TEM Observations
3.5. Gut Microbiota Analysis
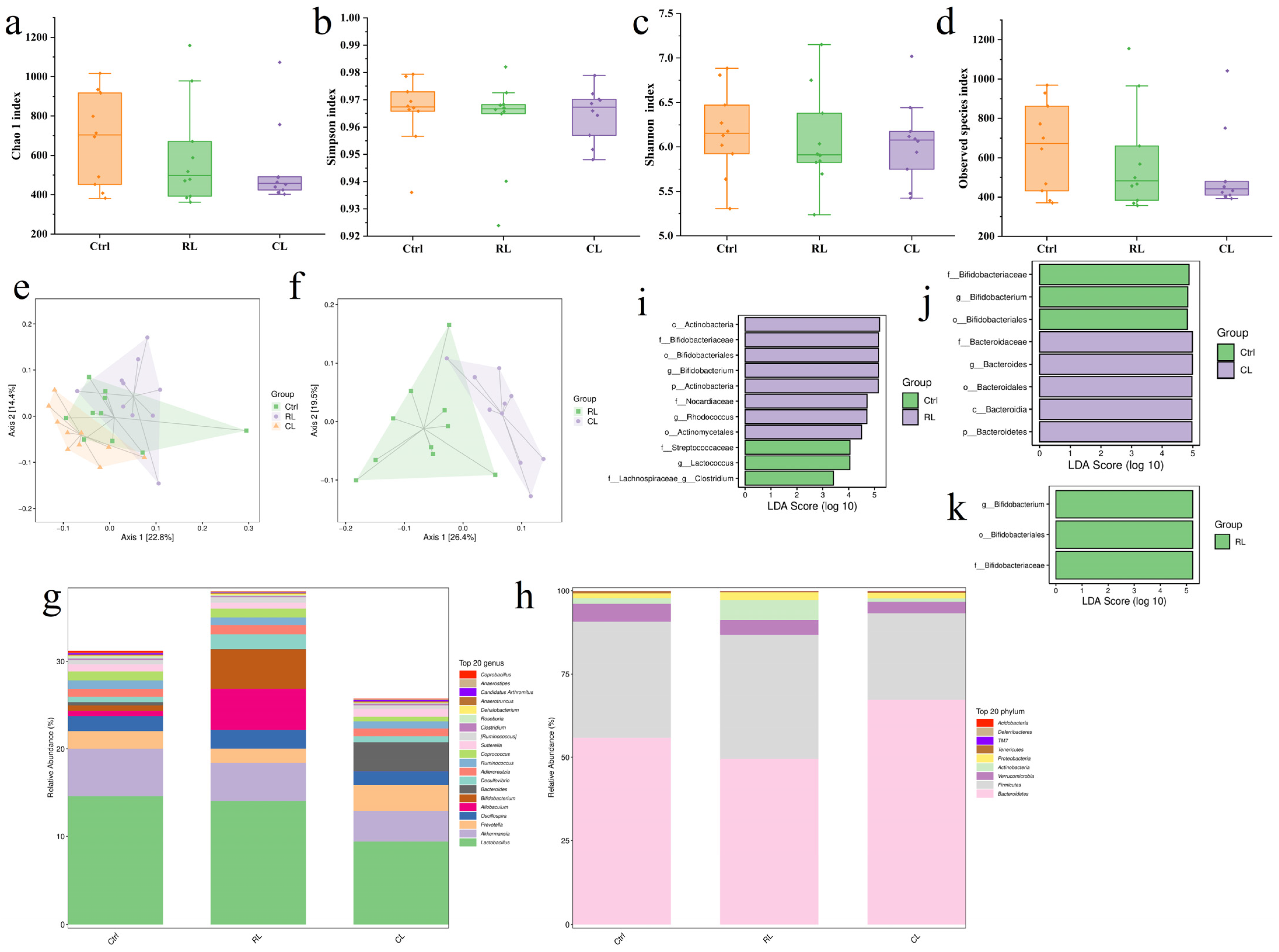
3.5.1. Alpha Diversity Analysis
3.5.2. Beta Diversity Analysis
3.5.3. Microbial Community Composition Analysis
3.5.4. Differential Microbial Community Analysis
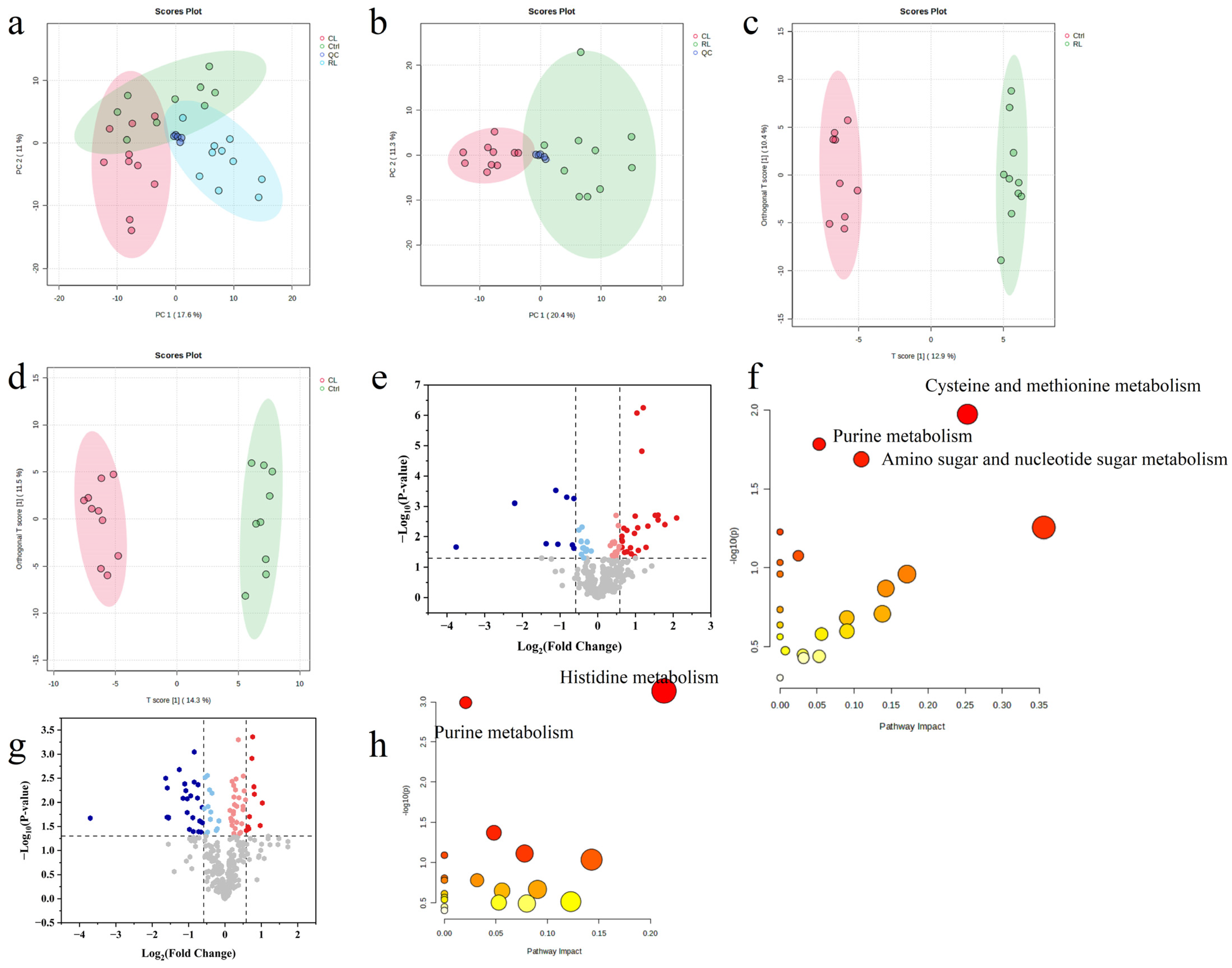
3.6. Untargeted Metabolomics and Chemometrics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kocyigit, E.; Kocaadam-Bozkurt, B.; Bozkurt, O.; Ağagündüz, D.; Capasso, R. Plant Toxic Proteins: Their Biological Activities, Mechanism of Action and Removal Strategies. Toxins 2023, 15, 356. [Google Scholar] [CrossRef] [PubMed]
- Hertzler, S.R.; Lieblein-Boff, J.C.; Weiler, M.; Allgeier, C. Plant Proteins: Assessing Their Nutritional Quality and Effects on Health and Physical Function. Nutrients 2020, 12, 3704. [Google Scholar] [CrossRef] [PubMed]
- Berrazaga, I.; Micard, V.; Gueugneau, M.; Walrand, S. The Role of the Anabolic Properties of Plant- versus Animal-Based Protein Sources in Supporting Muscle Mass Maintenance: A Critical Review. Nutrients 2019, 11, 1825. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zou, P.-R.; Hu, F.; Zhu, W.; Wei, Z.-J. Updates on Plant-Based Protein Products as an Alternative to Animal Protein: Technology, Properties, and Their Health Benefits. Molecules 2023, 28, 4016. [Google Scholar] [CrossRef]
- Yan, S.; Bhawal, R.; Yin, Z.; Lee, Y.C. Recent advances in proteomics and metabolomics in plants. Mol. Hortic. 2022, 2, 17. [Google Scholar] [CrossRef]
- Karabulut, G.; Goksen, G.; Mousavi Khaneghah, A. Plant-based protein modification strategies towards challenges. J. Agric. Food Res. 2024, 15, 101017. [Google Scholar] [CrossRef]
- Ma, K.K.; Greis, M.; Lu, J.; Nolden, A.A.; McClements, D.J.; Kinchla, A.J. Functional Performance of Plant Proteins. Foods 2022, 11, 594. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, L.; Yang, C.; Sun, Z.; Xu, X.; Li, L.; Zang, H. Research on the Structure of Peanut Allergen Protein Ara H1 Based on Aquaphotomics. Front. Nutr. 2021, 8, 696355. [Google Scholar] [CrossRef]
- Sultana, M.S.; Mazarei, M.; Jurat-Fuentes, J.L.; Hewezi, T.; Millwood, R.J.; Stewart, C.N. Overexpression of Soybean Trypsin Inhibitor Genes Decreases Defoliation by Corn Earworm (Helicoverpa zea) in Soybean (Glycine max) and Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1129454. [Google Scholar] [CrossRef]
- Liu, L.; Yu, H.; Wu, H.; Yang, X.; Pan, Y.; Chen, Y.; Wang, K.; Wang, W.; Zhang, W.; Jin, Y.; et al. Toxic proteins from Croton tiglium L. exert a proinflammatory effect by inducing release of proinflammatory cytokines and activating the p38-MAPK signaling pathway. Mol. Med. Rep. 2017, 16, 631–638. [Google Scholar] [CrossRef]
- Meneguelli de Souza, L.C.; de Carvalho, L.P.; Araújo, J.S.; de Melo, E.J.T.; Machado, O.L.T. Cell Toxicity by Ricin and Elucidation of Mechanism of Ricin Inactivation. Int. J. Biol. Macromol. 2018, 113, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Jetzt, A.E.; Li, X.-P.; Tumer, N.E.; Cohick, W.S. Toxicity of Ricin a Chain Is Reduced in Mammalian Cells by Inhibiting Its Interaction with the Ribosome. Toxicol. Appl. Pharmacol. 2016, 310, 120–128. [Google Scholar] [CrossRef]
- Moshiri, M.; Hamid, F.; Etemad, L. Ricin Toxicity: Clinical and Molecular Aspects. Rep. Biochem. Mol. Biol. 2016, 4, 60–65. [Google Scholar]
- Yuan, S.-M. Nutriology, Pharmacology and Cardiovascular Effects of Xanthium Sibiricum. Prog. Nutr. 2020, 22, 370–377. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, X.-J.; Yang, L.; Zhang, J.-X.; Hou, A.-J.; Man, W.-J.; Wang, S.; Yang, B.-Y.; Chan, K.; Wang, Q.-H.; et al. The fruits of Xanthium sibiricum Patr: A review on phytochemistry, pharmacological activities, and toxicity. World J. Tradit. Chin. Med. 2020, 6, 408–422. [Google Scholar] [CrossRef]
- Zhou, S.; Wen, Y.; Duan, Y.; Li, Q.; Gao, Y.; Yu, X. Functional properties and composition of new “nut” oil obtained from Xanthium sibiricum seeds. Eur. J. Lipid Sci. Technol. 2022, 124, 2100135. [Google Scholar] [CrossRef]
- Pawar, S.; Hole, J.; Bankar, M.; Channapattana, S.; Srinidhi, C. Studies on Xanthium strumarium L. Seed Oil: Biodiesel Synthesis and Process Optimization. Mater. Today Proc. 2022, 66, 2169–2177. [Google Scholar] [CrossRef]
- Pi, X.; Zhu, L.; Liu, J.; Zhang, B. Effect of Thermal Processing on Food Allergenicity: Mechanisms, Application, Influence Factor, and Future Perspective. J. Agric. Food Chem. 2024, 72, 20225–20240. [Google Scholar] [CrossRef]
- Tian, Y.; Rao, H.; Zhang, K.; Tao, S.; Xue, W. Effects of Different Thermal Processing Methods on the Structure and Allergenicity of Peanut Allergen Ara h 1. Food Sci. Nutr. 2018, 6, 1706–1714. [Google Scholar] [CrossRef]
- Zhang, L.; Qu, H.; Xie, M.; Shi, T.; Shi, P.; Yu, M. Effects of Different Cooking Methods on Phenol Content and Antioxidant Activity in Sprouted Peanut. Molecules 2023, 28, 4684. [Google Scholar] [CrossRef]
- Beyer, K.; Morrow, E.; Li, X.M.; Bardina, L.; Bannon, G.A.; Burks, A.W.; Sampson, H.A. Effects of Cooking Methods on Peanut Allergenicity. J. Allergy Clin. Immunol. 2001, 107, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cheng, J.; Xue, F. Effects of Restrictive Enzymatic Modification on the Structure and Functional Characteristics of Prunus japonica Thunb. Separation Protein. Nanjing Univ. Tradit. Chin. Med. 2023, 39, 888–894. [Google Scholar] [CrossRef]
- Liu, S.; Xie, Y.; Li, B.; Li, S.; Yu, W.; Ye, A.; Guo, Q. Structural Properties of Quinoa Protein Isolate: Impact of Neutral to High Alkaline Extraction pH. Foods 2023, 12, 2589. [Google Scholar] [CrossRef]
- Yi-Shen, Z.; Shuai, S.; FitzGerald, R. Mung Bean Proteins and Peptides: Nutritional, Functional and Bioactive Properties. Food Nutr. Res. 2018, 62, 10-29219. [Google Scholar] [CrossRef]
- Perler, B.K.; Friedman, E.S.; Wu, G.D. The role of the gut microbiota in the relationship between diet and human health. Annu. Rev. Physiol. 2023, 85, 449–468. [Google Scholar] [CrossRef]
- Wan, Z.; Zheng, J.; Zhu, Z.; Sang, L.; Zhu, J.; Luo, S.; Zhao, Y.; Wang, R.; Zhang, Y.; Zuo, K.; et al. Intermediate role of gut microbiota in vitamin B nutrition and its influences on human health. Front. Nutr. 2022, 9, 1031502. [Google Scholar] [CrossRef] [PubMed]
- Afzaal, M.; Saeed, F.; Shah, Y.A. Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 2022, 13, 999001. [Google Scholar] [CrossRef]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Gut Microbiota Influence in Type 2 Diabetes Mellitus (T2DM). Gut Pathog. 2021, 13, 50. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Wang, X.; Peng, J.; Cai, P.; Xia, Y.; Yi, C.; Shang, A.; Akanyibah, F.A.; Mao, F. The Emerging Role of the Gut Microbiota and Its Application in Inflammatory Bowel Disease. Biomed. Pharmacother. 2024, 179, 117302. [Google Scholar] [CrossRef]
- Fontaine, F.; Turjeman, S.; Callens, K. The intersection of undernutrition, microbiome, and child development in the first years of life. Nat. Commun. 2023, 14, 3554. [Google Scholar] [CrossRef] [PubMed]
- Zoghi, S.; Sadeghpour Heravi, F.; Nikniaz, Z.; Shirmohamadi, M.; Moaddab, S.Y.; Ebrahimzadeh Leylabadlo, H. Gut microbiota and childhood malnutrition: Understanding the link and exploring therapeutic interventions. Eng. Life Sci. 2023, 24, 2300070. [Google Scholar] [CrossRef]
- Iddrisu, I.; Monteagudo-Mera, A.; Poveda, C.; Pyle, S.; Shahzad, M.; Andrews, S.; Walton, G.E. Malnutrition and gut microbiota in children. Nutrients 2021, 13, 2727. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Y.; Mostafa, I.; Hibberd, M.C.; Das, S.; Mahfuz, M.; Naila, N.N.; Islam, M.M.; Huq, S.; Alam, M.A.; Zaman, M.U.; et al. A Microbiota-Directed Food Intervention for Undernourished Children. N. Engl. J. Med. 2021, 384, 1517–1528. [Google Scholar] [CrossRef]
- Gehrig, J.L.; Venkatesh, S.; Chang, H.-W.; Hibberd, M.C.; Kung, V.L.; Cheng, J.; Chen, R.Y.; Subramanian, S.; Cowardin, C.A.; Meier, M.F.; et al. Effects of Microbiota-Directed Foods in Gnotobiotic Animals and Undernourished Children. Science 2019, 365, eaau4732. [Google Scholar] [CrossRef]
- Bielik, V.; Kolisek, M. Bioaccessibility and bioavailability of minerals in relation to a healthy gut microbiome. Int. J. Mol. Sci. 2021, 22, 6803. [Google Scholar] [CrossRef]
- Pajarillo, E.A.B.; Lee, E.; Kang, D.-K. Trace metals and animal health: Interplay of the gut microbiota with iron, manganese, zinc, and copper. Anim. Nutr. 2021, 7, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J.-N.; Sun, B.-H.; Liu, Q.; Ma, J.; Zhang, Q.; Liu, Y.-X.; Chen, N.; Chen, F. The role of genotype and diet in shaping gut microbiome in a genetic vitamin A deficient mouse model. J. Genet. Genom. 2022, 49, 155–164. [Google Scholar] [CrossRef]
- Otten, A.T.; Bourgonje, A.R.; Peters, V.; Alizadeh, B.Z.; Dijkstra, G.; Harmsen, H.J.M. Vitamin C Supplementation in Healthy Individuals Leads to Shifts of Bacterial Populations in the Gut—A Pilot Study. Antioxidants 2021, 10, 1278. [Google Scholar] [CrossRef]
- Hibberd, M.C.; Wu, M.; Rodionov, D.A.; Li, X.; Cheng, J.; Griffin, N.W.; Barratt, M.J.; Giannone, R.J.; Hettich, R.L.; Osterman, A.L.; et al. The Effects of Micronutrient Deficiencies on Bacterial Species from the Human Gut Microbiota. Sci. Transl. Med. 2017, 9, eaal4069. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Ding, J.; Yuan, S.; Yu, H.; Guo, Y.; Cheng, Y.; Qian, H.; Yao, W. A self-adhesive glutenin-based coating cross-linked by genipin for suppressing microplastic shedding in harsh environments. Green Chem. 2025. [Google Scholar] [CrossRef]
- Cerk, K.; Aguilera-Gómez, M. Microbiota analysis for risk assessment: Evaluation of hazardous dietary substances and its potential role on the gut microbiome variability and dysbiosis. EFSA J. 2022, 20, e200404. [Google Scholar] [CrossRef] [PubMed]
- Kortesniemi, M.; Noerman, S.; Kårlund, A.; Raita, J.; Meuronen, T.; Koistinen, V.; Landberg, R.; Hanhineva, K. Nutritional metabolomics: Recent developments and future needs. Curr. Opin. Chem. Biol. 2023, 77, 102400. [Google Scholar] [CrossRef]
- Souza, A.L.; Patti, G.J. A Protocol for Untargeted Metabolomic Analysis: From Sample Preparation to Data Processing. Methods Mol. Biol. 2021, 2276, 357–382. [Google Scholar] [CrossRef]
- Pérez-Míguez, R.; Castro-Puyana, M.; Sánchez-López, E.; Plaza, M.; Marina, M.L. Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform. Molecules 2020, 25, 887. [Google Scholar] [CrossRef]
- Li, R.; Sun, Z.; Zhao, Y.; Li, L.; Yang, X.; Cen, J.; Chen, S.; Li, C.; Wang, Y. Application of UHPLC-Q-TOF-MS/MS Metabolomics Approach to Investigate the Taste and Nutrition Changes in Tilapia Fillets Treated with Different Thermal Processing Methods. Food Chem. 2021, 356, 129737. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, L.-L.; Li, W.; Zhang, Y.; Zhang, Y.; Liu, F.; Zou, L. Application of Metabolomics for Revealing the Interventional Effects of Functional Foods on Metabolic Diseases. Food Chem. 2022, 367, 130697. [Google Scholar] [CrossRef]
- Yuan, L.; Jiang, F.; Cao, X.; Liu, Y.; Xu, Y.-J. Metabolomics Reveals the Toxicological Effects of Polar Compounds from Frying Palm Oil. Food Funct. 2020, 11, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Quan, W.; Jiao, Y.; Li, Y.; Xue, C.; Liu, G.; Wang, Z.; Qin, F.; He, Z.; Zeng, M.; Chen, J. Metabolic Changes from Exposure to Harmful Maillard Reaction Products and High-Fat Diet on Sprague-Dawley Rats. Food Res. Int. 2021, 141, 110129. [Google Scholar] [CrossRef]
- National Pharmacopoeia Committee. General Rule 0213. In Pharmacopoeia of the People’s Republic of China, 2020th ed.; Part IV; National Pharmacopoeia Committee, Ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- National Pharmacopoeia Committee. General Rule 0731. In Pharmacopoeia of the People’s Republic of China, 2020th ed.; Part IV; National Pharmacopoeia Committee, Ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Bala, E.; Dey, S.; Patra, S.; Singha, S. Assessment of Microwave Drying for Rapid and Convenient Analysis of Medicinal Plants for Quality Assurance. Phytochem. Anal. 2024, 35, 903–922. [Google Scholar] [CrossRef]
- GB 5009.4-2023; National Health Commission & State Administration for Market Regulation. Determination of Ash in Foods. National Food Safety Standard. Standards Press of China: Beijing, China, 2023.
- National Pharmacopoeia Committee. General Rule 0713. In Pharmacopoeia of the People’s Republic of China, 2020th ed.; Part IV; National Pharmacopoeia Committee, Ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- National Pharmacopoeia Committee. General Rule 3113. In Pharmacopoeia of the People’s Republic of China, 2020th ed.; Part IV; National Pharmacopoeia Committee, Ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Rezvankhah, A.; Yarmand, M.S.; Ghanbarzadeh, B.; Mirzaee, H. Generation of Bioactive Peptides from Lentil Protein: Degree of Hydrolysis, Antioxidant Activity, Phenol Content, ACE-Inhibitory Activity, Molecular Weight, Sensory, and Functional Properties. Food Meas. 2021, 15, 5021–5035. [Google Scholar] [CrossRef]
- Mekonnen, T.H.; Mussone, P.G.; Stashko, N.; Choi, P.Y.; Bressler, D.C. Recovery and Characterization of Proteinacious Material Recovered from Thermal and Alkaline Hydrolyzed Specified Risk Materials. Process Biochem. 2013, 48, 885–892. [Google Scholar] [CrossRef]
- Craine, E.B.; Murphy, K.M. Seed composition and amino acid profiles for quinoa grown in Washington State. Front. Nutr. 2020, 7, 126. [Google Scholar] [CrossRef]
- Du, M.; Xie, J.; Gong, B.; Xu, X.; Tang, W.; Li, X.; Li, C.; Xie, M. Extraction, Physicochemical Characteristics and Functional Properties of Mung Bean Protein. Food Hydrocoll. 2018, 76, 131–140. [Google Scholar] [CrossRef]
- Dakhili, S.; Abdolalizadeh, L.; Hosseini, S.M.; Shojaee-Aliabadi, S.; Mirmoghtadaie, L. Quinoa Protein: Composition, Structure and Functional Properties. Food Chem. 2019, 299, 125161. [Google Scholar] [CrossRef]
- Dimina, L.; Rémond, D.; Huneau, J.-F.; Mariotti, F. Combining plant proteins to achieve amino acid profiles adapted to various nutritional objectives—An exploratory analysis using linear programming. Front. Nutr. 2022, 8, 809685. [Google Scholar] [CrossRef]
- Gorissen, S.H.M.; Crombag, J.J.R.; Senden, J.M.G.; Waterval, W.A.H.; Bierau, J.; Verdijk, L.B.; van Loon, L.J.C. Protein content and amino acid composition of commercially available plant-based protein isolates. Amino Acids 2018, 50, 1685–1695. [Google Scholar] [CrossRef]
- Siddiqi, R.A.; Singh, T.P.; Rani, M.; Sogi, D.S.; Bhat, M.A. Diversity in grain, flour, amino acid composition, protein profiling, and proportion of total flour proteins of different wheat cultivars of North India. Front. Nutr. 2020, 7, 141. [Google Scholar] [CrossRef]
- Ashadullin, D.F.; Ashadullin, D.F.; Vasilova, N.Z.; Zuev, E.V.; Hajrullina, A.R. Amino Acid Content in the Spring Common Wheat Grains. Russ. Agric. Sci. 2023, 49, 265–270. [Google Scholar] [CrossRef]
- Olakanmi, S.J.; Jayas, D.S.; Paliwal, J. Implications of Blending Pulse and Wheat Flours on Rheology and Quality Characteristics of Baked Goods: A Review. Foods 2022, 11, 3287. [Google Scholar] [CrossRef]
- Kalman, D.S. Amino acid composition of an organic brown rice protein concentrate and isolate compared to soy and whey concentrates and isolates. Foods 2014, 3, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and methods for circular dichroism spectroscopy of proteins: A tutorial review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Vlasova, I.M.; Zhuravleva, V.V.; Saletsky, A.M. Denaturation of bovine serum albumin initiated by sodium dodecyl sulfate as monitored via the intrinsic fluorescence of the protein. Russ. J. Phys. Chem. B 2014, 8, 385–390. [Google Scholar] [CrossRef]
- Žoldák, G.; Jancura, D.; Sedlák, E. The fluorescence intensities ratio is not a reliable parameter for evaluation of protein unfolding transitions. Protein Sci. 2017, 26, 1236–1239. [Google Scholar] [CrossRef]
- Silverstein, R.M.; Bassler, G.C. Spectrometric Identification of Organic Compounds. J. Chem. Educ. 1962, 39, 546. [Google Scholar] [CrossRef]
- Li, T.; Cheng, Z.; Cao, L.; Jiang, X.; Fan, L. Data of fluorescence, UV-vis absorption and FTIR spectra for the study of interaction between two food colourants and BSA. Data Brief 2016, 8, 755–783. [Google Scholar] [CrossRef] [PubMed]
- Jahan, K.; Ashfaq, A.; Islam, R.U.; Younis, K.; Yousuf, O. Optimization of Ultrasound-Assisted Protein Extraction from Defatted Mustard Meal and Determination of Its Physical, Structural, and Functional Properties. J. Food Process. Preserv. 2022, 46, e16764. [Google Scholar] [CrossRef]
- Li, Y.; Tian, Y.; Deng, L.; Dai, T.; Liu, C.; Chen, J. High Energy Media Mill Modified Pea Dietary Fiber: Physicochemical Property and Its Mechanism in Stabilizing Pea Protein Beverage. Food Hydrocoll. 2024, 147, 109392. [Google Scholar] [CrossRef]
- Yu, X.; Huang, S.; Yang, F.; Qin, X.; Nie, C.; Deng, Q.; Huang, F.; Xiang, Q.; Zhu, Y.; Geng, F. Effect of Microwave Exposure to Flaxseed on the Composition, Structure and Techno-Functionality of Gum Polysaccharides. Food Hydrocoll. 2022, 125, 107447. [Google Scholar] [CrossRef]
- Shi, B.; Guo, X.; Liu, H. Dissecting Maillard reaction production in fried foods: Formation mechanisms, sensory characteristic attribution, control strategy, and gut homeostasis regulation. Food Chem. 2024, 438, 137994. [Google Scholar] [CrossRef]
- Zhang, S.; Holmes, M.; Ettelaie, R.; Sarkar, A. Pea protein microgel particles as Pickering stabilisers of oil-in-water emulsions: Responsiveness to pH and ionic strength. Food Hydrocoll. 2020, 102, 105583. [Google Scholar] [CrossRef]
- Li, M.; Huang, J.; Chen, Y.; Liu, C.; Wu, X. Protein from Red Adzuki Bean: Extraction Optimization, Glycosylation Modification and Physicochemical Properties of Glycation Products. J. Food Meas. Charact. 2024, 18, 4229–4245. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Ma, X.; Zheng, Y.; Hu, H.; Jiao, B.; McClements, D.J.; Wang, Q.; Shi, A. Interfacial and Foaming Properties of Plant and Microbial Proteins: Comparison of Structure-Function Behavior of Different Proteins. Food Chem. 2025, 463, 141431. [Google Scholar] [CrossRef]
- Han, Y.; Zhu, L.; Karrar, E.; Qi, X.; Zhang, H.; Wu, G. Pickering foams stabilized by protein-based particles: A review of characterization, stabilization, and application. Trends Food Sci. Technol. 2023, 133, 148–159. [Google Scholar] [CrossRef]
- Yanqing, L.; Yuyang, H.; Xiaoqi, D.; Zhimin, L.; Wentao, L.; Guang, Z.; Ying, Z.; Xiuqing, Z. Effect of Enzymatic Hydrolysis Followed after Extrusion Pretreatment on the Structure and Emulsibility of Soybean Protein. Process Biochem. 2022, 116, 173–184. [Google Scholar] [CrossRef]
- Li, N.; Girard, A.L. Impact of pH and temperature on whey protein-proanthocyanidin interactions and foaming properties. Food Hydrocoll. 2023, 134, 108100. [Google Scholar] [CrossRef]
- Janssen, F.; Monterde, V.; Wouters, A.G.B. Relevance of the Air–Water Interfacial and Foaming Properties of (Modified) Wheat Proteins for Food Systems. Compr. Rev. Food Sci. Food Saf. 2023, 22, 1517–1554. [Google Scholar] [CrossRef]
- Antalis, T.M.; Shea-Donohue, T.; Vogel, S.N.; Sears, C.; Fasano, A. Mechanisms of disease: Protease functions in intestinal mucosal pathobiology. Nat. Clin. Pract. Gastroenterol. Hepatol. 2007, 4, 393–402. [Google Scholar] [CrossRef]
- Lkhagva, E.; Chung, H.J.; Hong, J. The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol. 2021, 21, 44. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The power of microbiome studies: Some considerations on which alpha and beta metrics to use and how to report results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef]
- Maraki, S.; Mavromanolaki, V.E.; Kasimati, A.; Stafylaki, D.; Scoulica, E. Clinical and Microbiological Characteristics of Nocardiosis: A 5-Year Single-Center Study in Crete, Greece. Acta Microbiol. Immunol. Hung. 2023, 70, 239–245. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Wang, Q.-W.; Zheng, H.; Yang, Y.; Chang, X.; Du, Z.; Hang, Z.-N.; Li, Z.-S.; Liao, Z. Distinct Microbial Signatures and Their Predictive Value in Recurrent Acute Pancreatitis: Insights from 5-Region 16S rRNA Gene Sequencing. Front. Immunol. 2025, 16, 1558983. [Google Scholar] [CrossRef]
- Mao, H.; Fan, Y.; Tan, F.; Long, X. The effects of Bifidobacterium animalis QC08 on reducing uric acid level and providing renal protection in mice with hyperuricemia. Front. Microbiol. 2025, 16, 1529626. [Google Scholar] [CrossRef]
- Sarwar Gilani, G.; Wu Xiao, C.; Cockell, K.A. Impact of antinutritional factors in food proteins on the digestibility of protein and the bioavailability of amino acids and on protein quality. Br. J. Nutr. 2012, 108, S315–S332. [Google Scholar] [CrossRef]
- Allison, C.; Macfarlane, G.T. Influence of pH, nutrient availability, and growth rate on amine production by Bacteroides fragilis and Clostridium perfringens. Appl. Environ. Microbiol. 1989, 55, 2894–2898. [Google Scholar] [CrossRef]
- Li, Z.; Rasmussen, T.S.; Rasmussen, M.L.; Li, J.; Henríquez Olguín, C.; Kot, W.; Nielsen, D.S.; Jensen, T.E. The Gut Microbiome on a Periodized Low-Protein Diet Is Associated with Improved Metabolic Health. Front. Microbiol. 2019, 10, 709. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Y.; Li, Z.; Liu, X. Untargeted metabolomics profiles delineate metabolic alterations in mouse plasma during lung carcinoma development using UPLC-QTOF/MS in MSE mode. R. Soc. Open Sci. 2018, 5, 181143. [Google Scholar] [CrossRef]
- Hong, L.-L.; Cui, D.-X.; Wang, H.-D.; Jing, Q.; Li, X.; Hu, Y.; Yao, Y.-Q.; Gao, X.-M.; Guo, D.-A.; Yang, W.-Z. Recent advances in traditional Chinese medicine metabolism: Sample pre-treatment, MS-oriented analytical strategies and typical applications. TrAC Trends Anal. Chem. 2025, 189, 118269. [Google Scholar] [CrossRef]
- Ding, S.; Chen, M.; Liao, Y. Serum metabolic profiles of Chinese women with perimenopausal obesity explored by the untargeted metabolomics approach. Front. Endocrinol. 2021, 12, 637317. [Google Scholar] [CrossRef]
- Yang, H.-B.; Xu, Y.-Y.; Zhao, X.-N.; Zou, S.-W.; Zhang, Y.; Zhang, M.; Li, J.-T.; Ren, F.; Wang, L.-Y.; Lei, Q.-Y. Acetylation of MAT IIα Represses Tumour Cell Growth and Is Decreased in Human Hepatocellular Cancer. Nat. Commun. 2015, 6, 6973. [Google Scholar] [CrossRef]
- Kobayashi, S.; Sato, M.; Kasakoshi, T.; Tsutsui, T.; Sugimoto, M.; Osaki, M.; Okada, F.; Igarashi, K.; Hiratake, J.; Homma, T.; et al. Cystathionine is a novel substrate of cystine/glutamate transporter: Implications for immune function. J. Biol. Chem. 2015, 290, 8778–8788. [Google Scholar] [CrossRef]
- McBean, G.J. The Transsulfuration Pathway: A Source of Cysteine for Glutathione in Astrocytes. Amino Acids 2012, 42, 199–205. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Z.; Ma, S.; Zhang, H.; Kong, F.; He, Y.; Yang, X.; Wang, Y.; Xu, H.; Yang, A.; et al. Ratio of S-Adenosylmethionine to S-Adenosylhomocysteine as a Sensitive Indicator of Atherosclerosis. Mol. Med. Rep. 2016, 14, 289–300. [Google Scholar] [CrossRef]
- Xiao, J.; You, Y.; Chen, X.; Tang, Y.; Chen, Y.; Liu, Q.; Liu, Z.; Ling, W. Higher S-Adenosylhomocysteine and Lower Ratio of S-Adenosylmethionine to S-Adenosylhomocysteine Were More Closely Associated with Increased Risk of Subclinical Atherosclerosis than Homocysteine. Front. Nutr. 2022, 9, 918698. [Google Scholar] [CrossRef]
- Yamaoka, T.; Kondo, M.; Honda, S.; Iwahana, H.; Moritani, M.; Ii, S.; Yoshimoto, K.; Itakura, M. Amidophosphoribosyltransferase Limits the Rate of Cell Growth-Linked De Novo Purine Biosynthesis in the Presence of Constant Capacity of Salvage Purine Biosynthesis. J. Biol. Chem. 1997, 272, 17719–17725. [Google Scholar] [CrossRef]
- Plümper, E.; Freiburg, M.; Heckmann, K. Inhibition of Pair Formation by Concanavalin a and Concanavalin A-Binding Glycoconjugates in Euplotes Octocarinatus. J. Eukaryotic Microbiol. 1995, 42, 166–173. [Google Scholar] [CrossRef]
- Liu, Y.; Jarman, J.B.; Low, Y.S.; Augustijn, H.E.; Huang, S.; Chen, H.; DeFeo, M.E.; Sekiba, K.; Hou, B.-H.; Meng, X.; et al. A Widely Distributed Gene Cluster Compensates for Uricase Loss in Hominids. Cell 2023, 186, 3400–3413.e20. [Google Scholar] [CrossRef]




| SRP | SCP | AARP | AACP | |
|---|---|---|---|---|
| Asp | 7.14 ± 0.17 | 6.07 ± 0.51 * | 6.35 ± 0.07 | 6.85 ± 0.31 |
| Thr | 2.60 ± 0.06 | 2.08 ± 0.01 ** | 2.10 ± 0.02 | 2.09 ± 0.09 |
| Glu | 27.32 ± 0.41 | 28.16 ± 0.15 * | 15.06 ± 0.20 | 16.67 ± 0.82 # |
| Ser | 3.40 ± 0.41 | 3.35 ± 0.03 | 2.93 ± 0.03 | 3.11 ± 0.16 |
| Pro | 3.27 ± 0.04 | 3.03 ± 0.05 ** | 3.15 ± 0.03 | 3.42 ± 0.17 |
| Gly | 4.97 ± 0.03 | 4.74 ± 0.03 ** | 3.32 ± 0.05 | 3.57 ± 0.15 |
| Ala | 2.93 ± 0.04 | 2.21 ± 0.05 ** | 3.32 ± 0.02 | 3.57 ± 0.18 |
| Val | 4.24 ± 0.08 | 3.42 ± 0.02 ** | 3.31 ± 0.05 | 3.51 ± 0.12 |
| Met | 2.30 ± 0.05 | 2.13 ± 0.10 | 0.82 ± 0.02 | 0.98 ± 0.04 ## |
| Ile | 3.47 ± 0.08 | 2.84 ± 0.04 ** | 2.82 ± 0.08 | 3.09 ± 0.17 |
| Leu | 6.19 ± 0.12 | 5.37 ± 0.05 ** | 4.06 ± 0.06 | 4.27 ± 0.15 |
| Tyr | 1.93 ± 0.03 | 1.38 ± 0.00 ** | 1.87 ± 0.02 | 1.91 ± 0.07 |
| Phe | 3.18 ± 0.08 | 2.61 ± 0.02 ** | 3.47 ± 0.06 | 3.77 ± 0.19 |
| Lys | 3.91 ± 0.02 | 2.91 ± 0.04 ** | 1.71 ± 0.02 | 1.51 ± 0.08 # |
| His | 2.10 ± 0.04 | 1.92 ± 0.03 ** | 1.68 ± 0.02 | 1.78 ± 0.06 # |
| Arg | 8.90 ± 0.04 | 8.59 ± 0.02 ** | 6.21 ± 0.12 | 6.66 ± 0.28 |
| Cys | 3.57 ± 0.04 | 3.66 ± 0.04 * | 0.16 ± 0.00 | 0.20 ± 0.01 ## |
| Trp | 0.61 ± 0.01 | 0.34 ± 0.01 ** | 1.00 ± 0.03 | 1.08 ± 0.02 ## |
| All | 92.01 ± 0.90 | 84.80 ± 0.18 ** | 65.34 ± 1.49 | 70.05 ± 3.25 |
| Organ Index | Control | RH | RL | CH | CL |
|---|---|---|---|---|---|
| Liver (g per 100 g BW) | 4.57 ± 0.68 | 4.81 ± 0.31 | 4.72 ± 0.38 | 4.55 ± 0.22 | 4.35 ± 0.19 |
| Kidney (g per 100 g BW) | 1.86 ± 0.28 | 1.91 ± 0.20 | 1.81 ± 0.16 | 1.81 ± 0.16 | 1.80 ± 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Wan, Z.; Han, F.; Fan, X.; Hao, Y.; Wei, F.; Liu, Q. Evaluation of the Functional Properties and Edible Safety of Concocted Xanthii Fructus Protein. Foods 2025, 14, 1913. https://doi.org/10.3390/foods14111913
Dong Y, Wan Z, Han F, Fan X, Hao Y, Wei F, Liu Q. Evaluation of the Functional Properties and Edible Safety of Concocted Xanthii Fructus Protein. Foods. 2025; 14(11):1913. https://doi.org/10.3390/foods14111913
Chicago/Turabian StyleDong, Yuchen, Zihao Wan, Fuguo Han, Xuemei Fan, Yanli Hao, Fang Wei, and Qingfei Liu. 2025. "Evaluation of the Functional Properties and Edible Safety of Concocted Xanthii Fructus Protein" Foods 14, no. 11: 1913. https://doi.org/10.3390/foods14111913
APA StyleDong, Y., Wan, Z., Han, F., Fan, X., Hao, Y., Wei, F., & Liu, Q. (2025). Evaluation of the Functional Properties and Edible Safety of Concocted Xanthii Fructus Protein. Foods, 14(11), 1913. https://doi.org/10.3390/foods14111913







